Introduction
Pancreatic cancer (PC) is an aggressive malignant
tumor with a 5-year survival rate of <5% (1,2). Although
considerable efforts have been made in improving the effectiveness
of surgical resection and chemotherapy for PC, the remission and
survival rates remain poor (3,4). The most
significant challenge for PC therapy is that the underlying
molecular mechanism of its pathogenesis is largely unknown.
Therefore, it remains an urgent priority to determine the molecular
mechanisms underlying the development of PC.
MicroRNAs (miRNAs/miRs) are a class of non-coding
RNAs consisting of 20–22 nucleotides in length, which regulate gene
expression via directly binding to target mRNAs (5,6). Previous
studies have reported the role of numerous miRNAs in the
tumorigenesis of PC (7–10). For example, a previous study
demonstrated that miR-221 induced autophagy and promoted PC cell
apoptosis via downregulating the expression of histone deacetylase
1 (11). In another study, the
expression levels of miR-661 were revealed to be upregulated in PC
tissues, which predicted a poor clinical outcome in patients with
PC (12). By inhibiting
Beclin-1-induced autophagy, miR-216a also increased the
radiosensitivity of PC cells (13).
Moreover, miR-148a played a tumor-suppressive role in PC via
suppressing the Wnt/β-catenin signaling pathway and inhibiting
epithelial-mesenchymal transition (14). Insulin-like growth factor (IGF)-1 was
revealed to be abundantly expressed in PC tissues, and could
activate insulin/IGF-related signalling pathways to regulate the
proliferation, migration and invasion of PC cells (15). Numerous previous studies have
indicated that IGF-1 may function as an oncogene in PC and
represent an effective target for treatment (16–18).
The present study aimed to investigate the
expression levels of miR-7515 in PC tissues and cell lines, and to
determine whether its effects on the proliferation, migration and
invasion of PC cells were mediated via regulation of IGF-1. The
results of the present study may provide novel insights into
whether miR-7515 and its target gene, IGF-1, may represent novel
biomarkers for the diagnosis and clinical treatment of PC.
Materials and methods
Patients
PC and adjacent normal tissues were obtained from 82
patients with PC (age range, 35–73 years; 42 males and 40 females)
at the Department of Hepatobiliary Surgery, Guizhou Medical
University (Guiyang, China) between March 2017 and April 2020. None
of patients enrolled in the present study had received neoadjuvant
chemotherapy, radiotherapy or immunotherapy prior to surgery.
Written informed consent was obtained from all patients prior to
participation and the experimental protocol was approved (approval
no. 31) by the Human Research Ethics Review Committee of Guizhou
Medical University (Guiyang, China).
Cell lines and culture
The human PC cell lines, AsPC-1, BXPC-3, SW1990 and
PANC-1, and a normal pancreatic epithelial cell line, HPDE, were
obtained from the American Type Culture Collection. The cells were
cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.),
and maintained in an environment with 5% CO2 at 37°C.
PD98059, a MEK inhibitor, was purchased from PharMingen; BD
Biosciences. A solution of PD98059 (50 µM) in dimethyl sulfoxide
(DMSO; Sigma-Aldrich; Merck KGaA) was prepared and used after
diluting with medium for each assay. DMSO was also used as a blank
control.
Cell transfection
The miR-7515 inhibitor (GCCUGUUAGCAUGAAAAAA) and
miR-negative control (NC) inhibitor (CGGAACAGAUCAUACUUGCCUU) were
obtained from Shanghai GeneChem Co., Ltd. A 2nd-generation
lentiviral system was used, miR-7515 expression lentivirus was
generated by subcloning the PCR amplified full length human
miR-7515 cDNA into the pMSCV retrovirus plasmid by transient
transfection of 293T (GeneCopoeia, Inc.). Empty pMSCV retrovirus
plasmid was used as a negative control for miR-7515 expression
lentivirus. AsPC-1 and BXPC-3 cells were seeded into 6-well plates
at a density of 1×105 cells/well. After the cells had
adhered, a lentivirus was added according to manufacturer's
protocol (2 µg/ml; MOI=5 for BXPC-3; and MOI=10 for AsPC-1). To
obtain stably overexpressing miR-7515 cells or stable knockdown
cells, cells were selected for 14 days using 1 g/ml puromycin 48 h
after transfection. miR-7515 overexpression lentivirus and the
lentivirus miRNA NC (LV-miR-NC) were obtained from GeneCopoeia,
Inc. The IGF-1 overexpression and NC plasmids (5 µg/ml) were
obtained from Sangon Biotech Co., Ltd. Cells were transfected 48 h
after transfection at 37°C with the oligonucleotides and plasmids
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. After
48 h of transfection, the cells were collected for further use in
the following experiments.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was extracted from PC tissues and cell
lines using TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). mRNA was reverse-transcribed into cDNA using the SuperScript
IV Reverse Transcriptase kit (Thermo Fisher Scientific, Inc.),
while miRNA was reverse transcribed into cDNA using TaqMan™
MicroRNA Reverse Transcription kit (Thermo Fisher Scientific,
Inc.). qPCR was subsequently performed using a PowerUp™ SYBR™ Green
Master Mix (Thermo Fisher Scientific, Inc.). All the kits were used
according to the manufacturer's instructions. GAPDH was used as the
reference gene for IGF-1 expression, while U6 was used as the
reference gene for miR-7515 expression. The following primer
sequences were used for the qPCR: miR-7515,
5′-AGAAGGGAAGATGGTGAC-3′; IGF-1 forward,
5′-GCTCTTCAGTTCGTGTGTGGA-3′ and reverse,
5′-GCCTCCTTAGATCACAGCTCC-3′; GAPDH forward,
5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse,
5′-GGCTGTTGTCATACTTCTCATGG-3′; U6 forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′. The reaction conditions were as
follows: 95°C for 10 min, followed by 40 cycles at 95°C for 1 sec
and 60°C for 60 sec. Relative expression levels of the target mRNA
or miRNA were determined using the 2−ΔΔCq method
(19).
Cell Counting Kit-8 (CCK-8) assay
PC cells were seeded into 96-well plates at a
density of 2×103 cells/well and incubated at 37°C for 6,
24, 48, 72 or 96 h. Following incubation, the medium was removed
and 100 µl fresh DMEM containing 10 µl CCK-8 reagent (Wuhan Boster
Biological Technology, Ltd.) was added to each well. The cells were
subsequently incubated with 5% CO2 at 37°C for 2 h. The
absorbance of each well was measured at a wavelength of 450 nm
using a microplate spectrophotometer (Bio-Rad Laboratories,
Inc.).
Colony formation assay
Following transfection, PC cells were plated into
6-well plates at a density of 1×103 cells/well.
Following 14 days of incubation at 37°C, the medium was removed and
the cell colonies were fixed with 4% paraformaldehyde for 15 min at
room temperature (Wuhan Servicebio Technology Co., Ltd.) and
stained with 0.5% crystal violet for 30 min at room temperature
(Wuhan Servicebio Technology Co., Ltd.). The cells were washed with
PBS to remove the excess crystal violet and images of the colonies
were captured and the number of colonies (≥10 mm2) was
counted under the naked eye.
Cell cycle distribution analysis
Cell cycle distribution was determined using flow
cytometric analysis. Briefly, PC cells were harvested, washed with
cold PBS and fixed with 70% ethyl alcohol at 4°C for 24 h. The PC
cells were subsequently collected by centrifugation at 4°C, 300 × g
for 30 min and resuspended in staining solution containing
propidium iodide (Invitrogen; Thermo Fisher Scientific, Inc.). and
1X binding buffer at room temperature for 30 min. Following a
30-min incubation in the dark, the samples were analyzed using a
flow cytometer (BD Biosciences). The results of the cell cycle
distribution were analyzed using FlowJo software version 7.4.1
(FlowJo LLC).
Wound healing assay
PC cells were cultured in DMEM supplemented with 10%
FBS at 37°C in 6-well plates until they reached 95% confluence.
Then, artificial wounds were produced by scratching the cell
monolayer with a 200-µl pipette tip. After removing the nonadherent
cells with PBS, the cells were cultured in the serum-free DMEM and
cultured for 24 h. The wound area was visualized at 0 and 24 h
under a phase-contrast microscope at a magnification of ×100.
Transwell invasion assay
PC cells (1×105) were suspended in 200 µl
serum-free DMEM and plated into the upper chambers of a Transwell
plate (8 µm pore size; Corning, Inc.), which was precoated at 37°C
with 100 µl Matrigel (BD Biosciences). A total of 700 µl DMEM
supplemented with 10% FBS was plated into the lower chambers of the
Transwell plate to act as a chemoattractant. Following incubation
at 37°C for 24 h, the non-invasive cells were removed with a cotton
swab, and the invasive cells were fixed for 15 min at room
temperature with 4% paraformaldehyde and stained for 30 min at room
temperature with 0.5% crystal violet solution. Invasive cells were
visualized (magnification, ×40) in five randomly selected fields of
view and the number of invasive cells/field was calculated under a
phase-contrast microscope.
Animal studies
A total of 10 male BALB/c nude mice (weight, 16–18
g; 4–6 weeks old) were purchased from the Experimental Animal
Centre of Guizhou Medical University (Guiyang, China). All nude
mice were allowed free access to food and water and maintained on a
12-h light/dark cycle, with controlled temperature (22.5±2°C) and
humidity (45±5%). To determine tumor cell proliferation,
1×106 with 100 µl of PANC-1 cell suspension
overexpressing miR-7515 or transfected with LV-miR-NC were
subcutaneously injected into the right axilla of the BALB/c nude
mice. Tumor growth was assessed once a week, and after 5 weeks, the
mice were anesthetized via an intraperitoneal injection of 50 mg/kg
sodium pentobarbital and euthanized by cervical dislocation. Tumor
tissues were collected for subsequent analysis; the maximum tumor
volume observed in the present study was 400 mm3.
For live metastasis detection, 1×107
PANC-1 cells overexpressing miR-7515 or transfected with LV-miR-NC
were resuspended in 100 µl PBS and injected into the spleen of the
mice (n=5 mice/group). The health of nude mice was observed once a
day, and after 12 weeks, mice were anesthetized via an
intraperitoneal injection of 50 mg/kg sodium pentobarbital solution
and euthanized by cervical dislocation. The liver tissues were
separated and the metastatic foci were counted. Furthermore,
metastasis foci in the liver tissues were also detected using
hematoxylin and eosin (H&E) staining. Paraffin-embedded
specimens were cut into serial sections of 5-µm thickness with a
microtome. Sections were deparaffinized in xylene, rehydrated
through a graded ethanol series and stained for 30 min at room
temperature automatically with H&E. Then images were captured
using an upright light microscope (magnification, ×200; Olympus
Corporation). All animal experiments were performed in strict
accordance with the Guide for the Care and Use of Laboratory
Animals (20). The animal
experimental protocols were approved (approval no. 2000025) by the
Ethics Committee of Guizhou Medical University (Guiyang,
China).
Immunohistochemical analysis
Tissues were fixed in 4% paraformaldehyde for 30 min
at room temperature in formalin, embedded in paraffin and cut into
2-µm sections. The sections were incubated at 65°C for 30 min,
deparaffinized with xylene and rehydrated using a gradient series
of ethanol. Antigen retrieval was performed using sodium citrate
(Wuhan Servicebio Technology Co., Ltd.) then endogenous peroxidase
activity was blocked for 15 min at room temperature with 3%
H2O2 (Wuhan Servicebio Technology Co., Ltd.).
The sections were subsequently blocked for non-specific binding
with 5% BSA for 15 min at room temperature (Wuhan Servicebio
Technology Co., Ltd.), and incubated overnight at 4°C with
anti-Ki67 (1:200; cat. no. 27309-1-AP; ProteinTech Group, Inc.) or
anti-proliferating cell nuclear antigen (PCNA; 1:200; cat. no.
10205-2-AP; ProteinTech Group, Inc.) primary antibodies. Following
the incubation, the sections were washed with PBS three times for 5
min and incubated with a goat anti-mouse/rabbit poly-HRP secondary
antibody (1:500; cat. no. PR30009; ProteinTech Group, Inc.) for 2
h. A diaminobenzidine (DAB) kit (Wuhan Boster Biological
Technology, Ltd.) was used to visualize the antibody-antigen
binding. Images were captured using an upright metallurgical
microscope (×200; Olympus Corporation).
Bioinformatics analysis
The expression of miR-7515 in pancreatic cancer
tissues and adjacent tissues was first assessed by The Cancer
Genome Atlas (TCGA) (https://www.cancer.gov/types) and Genotype-Tissue
Expression (GTex) (http://commonfund.nih.gov/GTEx/) databases. The
parameters were as follows: LogFC >1 and a P-value <0.05. The
online database, TargetScan 7.2 (http://www.targetscan.org/vert_72/), was used to
predict the target genes of miR-7515. The Database for Annotation,
Visualization and Integrated Discovery (https://david.ncifcrf.gov/) was used to determine the
significantly enriched Kyoto Encyclopedia of Genes and Genomes
signaling pathway enrichment of miR-7515 target genes. Finally,
RStudio software (version: 8.1.2; IBM Corp.) was used to visualize
the pathway enrichment. Target genes of miR-7515 in the most
significantly enriched pathway were further analyzed.
Dual-luciferase reporter assay
SW1990 and PANC-1 cells were plated into 24-well
plates and cultured with 5% CO2 at 37°C until they
reached 70% confluence. Then, the psiCHECK-2/IGF-1 3′-untranslated
region (UTR) wild-type (WT) and psiCHECK-2/IGF-1 3′-UTR mutant type
(Mut) reporter plasmids (Shanghai GeneChem Co., Ltd.) were
transfected into 5×105 SW1990 and PANC-1 cells
overexpressing miR-7515 or transfected with the LV-miR-NC using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.).
Following 24 h of transfection, SW1990 and PANC-1 cells were
collected, and the fluorescence activity was detected using a Dual
Luciferase Reporter assay system (Promega Corporation).
Subsequently, the luciferase activity was normalized to the firefly
luciferase internal control.
Western blotting
Total protein was extracted from PC cells using RIPA
lysis buffer (Wuhan Servicebio Technology Co., Ltd.) supplemented
with 1% PMSF (Wuhan Servicebio Technology Co., Ltd.). Total protein
was quantified using a BCA method and separated via 10% SDS-PAGE
(Melone pharmaceutical Co., Ltd). The separated proteins (30 µg per
lane) were subsequently transferred onto PVDF membranes (BD
Biosciences) and blocked with 5% BSA for 2 h at room temperature.
The membranes were then incubated with the following primary
antibodies overnight at 4°C: Anti-IGF1 (1:1,000; cat. no. A12305;
ABclonal Biotech Co., Ltd.), p-c-Raf (1:1,000; product code
ab150365; Abcam), anti-c-Raf (1:1,000; product no. 53745; Cell
Signaling Technology, Inc.), p-ribosomal protein S6 kinase α-1
(p-p90Rsk; 1:1,000; product no. 12032; Cell Signaling Technology,
Inc.), anti-p90Rsk (1:1,000; product code ab32114; Abcam),
anti-p-MEK1/2 (1:1,000; product no. 2338; Cell Signaling
Technology, Inc.), anti-MEK1/2 (1:1,000; product no. 4694; Cell
Signaling Technology, Inc.), anti-p-ERK1/2 (1:1,000; product no.
8544; Cell Signaling Technology, Inc.), anti-ERK1/2 (1:1,000; cat.
no. 16443-1-AP; ProteinTech Group, Inc.), anti-cyclin D1 (1:1,000;
cat. no. 26939-1-AP; ProteinTech Group, Inc.), anti-CDK2 (1:1,000;
cat. no. 10122-1-AP; ProteinTech Group, Inc.), anti-MMP2 (1:1,000;
cat. no. 10373-2-AP; ProteinTech Group, Inc.), anti-MMP9 (1:1,000;
cat. no. 10375-2-AP; ProteinTech Group, Inc.) and anti-GAPDH
(1:1,000; cat. no. 60004-1-Ig; ProteinTech Group, Inc.). Following
the primary antibody incubation, the membranes were washed with
TBST (TBS with 0.1% Tween-20) thrice and incubated with secondary
antibodies at room temperature (1:5,000; cat. nos. BA1038 and
BA1039; Wuhan Boster Biological Technology, Ltd.) for 2 h. Protein
bands were visualized using ECL reagent (Wuhan Boster Biological
Technology, Ltd.) on a Gel imager (Bio-Rad Laboratories, Inc.). The
total c-Raf, p90Rsk, MEK and ERK1/2 antibodies were used on the
same blots as the respective phosphorylated proteins after
stripping (Bio-Rad Laboratories, Inc.). Image-Pro Plus 5.1 software
was used to analyze the expression of protein, while GAPDH was used
as a loading control.
Statistical analysis
All experiments were repeated 3 times and the data
was averaged. The data are presented as the mean ± standard
deviation (SD). Statistical analysis was performed using SPSS 20.0
software (IBM Corp.). The association between miR-7515 expression
and clinicopathological characteristics of patients with PC was
analyzed using a χ2 test. The correlation between
miR-7515 and IGF-1 expression levels was analyzed using Pearson's
correlation analysis. Comparisons were performed using a one-way
ANOVA combined with LSD-t-test or a paired Student's t-test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-7515 expression levels are
downregulated in PC
TCGA and GTex databases were first used to analyze
the expression levels of miR-7515 in human normal and PC tissues.
The results revealed that miR-7515 expression levels were
significantly downregulated in PC tissues (Fig. 1A). Next, the expression of miR-7515 in
82 PC and adjacent normal tissues was determined. RT-qPCR analysis
demonstrated that the expression levels of miR-7515 were also
downregulated in PC tissues compared with adjacent normal tissues
(Fig. 1B). To determine the
prognostic value of miR-7515, patients were divided into two groups
(high and low expression levels of miR-7515) using the median
expression value of 2.7; tissues with miR-7515 expression of
>2.7 were defined as high expression, while tissues with
miR-7515 expression of <2.7 were defined as low expression. The
results revealed that low expression levels of miR-7515 predicted a
lower survival rate (Fig. 1C). The
expression levels of miR-7515 were also revealed to be negatively
associated with tumor size, lymph node metastasis, TNM stage,
distant metastasis, perineural invasion and blood vessel invasion
(Table I). In addition, the
expression levels of miR-7515 were significantly downregulated in
PC cells compared with the normal pancreatic epithelial cell line,
HPDE (Fig. 1D).
 | Table I.Association between miR-7515
expression levels and the clinicopathological characteristics of
patients with PC. |
Table I.
Association between miR-7515
expression levels and the clinicopathological characteristics of
patients with PC.
|
|
| miR-7515
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Features | n | Low | High | χ2 | P-value |
|---|
| All cases | 82 | 41 | 41 |
|
|
| Age (years) |
|
|
| 0.44 | 0.507 |
|
<60 | 39 | 21 | 18 |
|
|
|
≥60 | 43 | 20 | 23 |
|
|
| Sex |
|
|
| 0.195 | 0.695 |
|
Males | 42 | 22 | 20 |
|
|
|
Females | 40 | 19 | 21 |
|
|
| Tumor size
(cm) |
|
|
| 7.13 | 0.008 |
|
<2 | 36 | 12 | 24 |
|
|
| ≥2 | 46 | 29 | 17 |
|
|
| Lymph node
metastasis |
|
|
| 10.63 | 0.001 |
|
Negative | 28 | 7 | 21 |
|
|
|
Positive | 54 | 34 | 20 |
|
|
| TNM stage |
|
|
| 12.424 | <0.001 |
| I and
II | 27 | 6 | 21 |
|
|
| III and
IV | 55 | 35 | 20 |
|
|
| Distant
metastasis |
|
|
| 4.481 | 0.027 |
|
Negative | 40 | 15 | 25 |
|
|
|
Positive | 42 | 26 | 16 |
|
|
| Perineural
invasion |
|
|
| 14.233 | <0.001 |
|
Negative | 45 | 14 | 31 |
|
|
|
Positive | 37 | 27 | 10 |
|
|
| Blood vessel
invasion |
|
|
| 8.264 | 0.004 |
|
Negative | 39 | 13 | 26 |
|
|
|
Positive | 43 | 28 | 15 |
|
|
miR-7515 regulates PC cell
proliferation in vitro
To determine the effects of miR-7515 on PC cell
proliferation, miR-7515 overexpression lentivirus and miR-7515
inhibitor were used to construct miR-7515-overexpressing and
-knockdown cells (Fig. 2A). CCK-8
assays were performed, and the results demonstrated that the
overexpression of miR-7515 significantly decreased the
proliferation rate of SW1990 and PANC-1 cells, while miR-7515
knockdown significantly increased the proliferation rate of AsPC-1
and BXPC-3 cells (Fig. 2B).
Similarly, the results of the colony formation assays revealed that
overexpression of miR-7515 decreased the colony formation of SW1990
and PANC-1 cells, while the knockdown of miR-7515 exerted the
opposite effects on AsPC-1 and BXPC-3 cells (Fig. 2C). Furthermore, the overexpression of
miR-7515 induced SW1990 and PANC-1 cell cycle arrest in the
G1 phase, while miR-7515 knockdown promoted the
progression from the G1 phase to the S phase in AsPC-1
and BXPC-3 cells (Fig. 2D). These
results indicated that miR-7515 may regulate the proliferation of
PC cells in vitro.
miR-7515 regulates PC cell migration
and invasion in vitro
The effects of miR-7515 on PC cell migration and
invasion were subsequently investigated. The results of the wound
healing assay revealed that the migration rate was significantly
decreased in the cells overexpressing miR-7515, while the knockdown
of miR-7515 increased the cell migration rate (Fig. 3A). Moreover, the results of the
Transwell assays demonstrated that the invasive abilities of SW1990
and PANC-1 cells overexpressing miR-7515 were decreased, while the
invasive abilities of AsPC-1 and BXPC-3 cells were increased
following knockdown of miR-7515 compared with the respective NC
cells (Fig. 3B). These results
indicated that miR-7515 may regulate PC cell migration and invasion
in vitro.
Overexpression of miR-7515 inhibits PC
cell proliferation and metastasis in vivo
The effects of miR-7515 on PC cell proliferation and
metastasis in vivo were also determined. Establishment of a
xenograft tumor model indicated that the tumors overexpressing
miR-7515 grew slower compared with the tumors derived from cells
transfected with LV-miR-NC (Fig. 4A and
B), and also expressed downregulated levels of Ki67 and PCNA
(Fig. 4C). The liver metastasis model
revealed that the overexpression of miR-7515 inhibited the
metastasis of PANC-1 cells in vivo compared with the tumors
derived from cells transfected with LV-miR-NC (Fig. 4D-F). Furthermore, the findings
indicated that mice with tumors overexpressing miR-7515 had a lower
death rate compared with the mice with tumors derived from cells
transfected with LV-miR-NC (Fig. 4G).
These results indicated that miR-7515 may play a tumor-suppressive
role in PC.
miR-7515 targets IGF-1 and regulates
Ras/Raf/MEK/ERK signaling pathway
To determine the molecular mechanism underlying the
role of miR-7515 in PC, bioinformatics analysis was performed. The
results demonstrated that the target genes of miR-7515 were most
significantly enriched in ‘Ras signaling pathway’ (Fig. 5A). Among the genes, IGF-1 was
identified as a target gene of miR-7515 and had a high binding
score for its involvement in the Ras signaling pathway (Fig. 5B). As previous studies revealed that
the Ras signaling pathway played a key role in the progression of
PC (21–23), it was hypothesized that miR-7515 may
regulate IGF-1 expression in PC cells. Results from the dual
luciferase reporter assay demonstrated that the relative luciferase
activity was significantly decreased in the cells co-transfected
with the psiCHECK-2/IGF-1 3′-UTR WT plasmid and miR-7515
overexpression lentivirus, while the relative luciferase activity
was not altered in the SW1990 and PANC-1 cells co-transfected with
the psiCHECK-2/IGF-1 3′-UTR Mut plasmid and miR-7515 overexpression
lentivirus (Fig. 5C). RT-qPCR and
western blot analysis demonstrated that miR-7515 overexpression
significantly downregulated the expression levels of IGF-1, while
the knockdown of miR-7515 upregulated the expression levels of
IGF-1 in SW1990 and PANC-1 (Fig. 5D and
E). Similarly, SW1990 and PANC-1cells overexpressing miR-7515
had decreased expression levels of p-c-Raf, p-p90Rsk, p-MEK and
p-ERK1/2 compared with the LV-miR-NC-transfected cells, while the
knockdown of miR-7515 in SW1990 and PANC-1 cells upregulated the
expression levels of p-c-Raf, p-p90Rsk, p-MEK and p-ERK1/2
(Fig. 5E). Moreover, the expression
levels of IGF-1 were revealed to be upregulated in PC tissues
compared with adjacent normal tissues (Fig. 6A), particularly in tumors from
patients with III–IV stage disease (Fig.
6B). miR-7515 expression was also revealed to be negatively
correlated with IGF-1 expression in PC tissues (Fig. 6C). Furthermore, the expression levels
of IGF-1 were significantly upregulated in PC cell lines compared
with the normal pancreatic epithelium cell line, HPDE (Fig. 6D and E). These results indicated that
miR-7515 may target IGF-1 and the downstream Ras/Raf/MEK/ERK
signaling pathway.
 | Figure 5.miR-7515 targets IGF-1 and regulates
the Ras/Raf/MEK/ERK signaling pathway. (A) Kyoto Encyclopedia of
Genes and Genomes signaling pathway enrichment analysis of the
target genes of miR-7515. (B) Binding site between miR-7515 and
IGF-1 is presented. (C) Dual luciferase reporter assays were
performed to validate the binding between miR-7515 and IGF-1. (D)
Reverse transcription-quantitative PCR was performed to analyze the
expression levels of IGF-1 in SW1990 and PANC-1 cells transfected
with the miR-7515 overexpression lentivirus and miR-7515 inhibitor.
(E) Western blotting was used to analyze the expression levels of
IGF-1, p-c-Raf, c-Raf, p-p90RSK, p90RSK, p-MEK, MEK, p-ERK1/2 and
ERK1/2 in SW1990 and PANC-1 cells transfected with the miR-7515
overexpression lentivirus and miR-7515 inhibitor. *P<0.05 and
**P<0.01. miR, microRNA; IGF-1, insulin like growth factor 1;
p-, phosphorylated; p90RSK, ribosomal protein S6 kinase α-1; NC,
negative control; LV, lentivirus. |
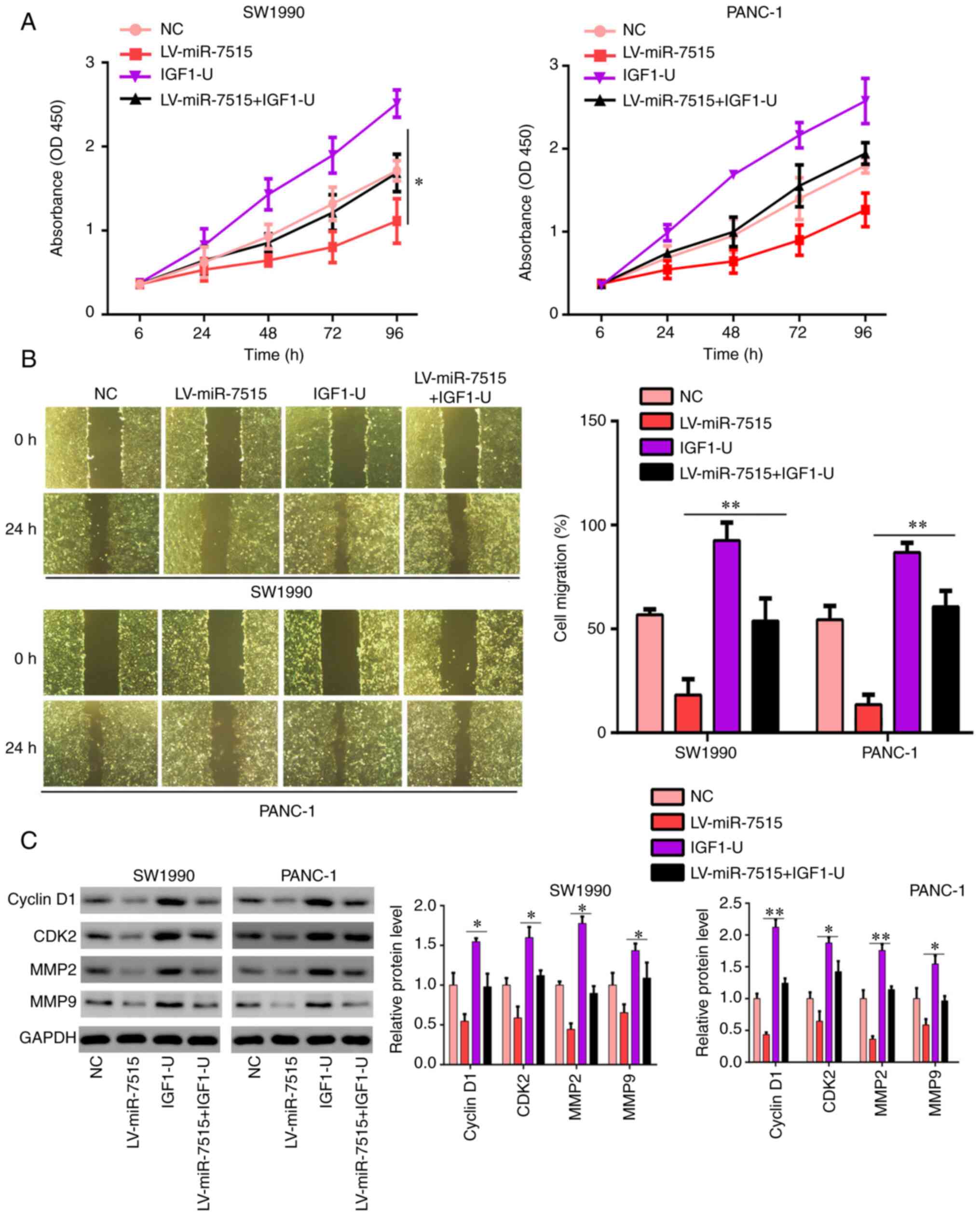
Overexpression of IGF-1 reverses the
inhibitory effects of miR-7515 overexpression
To determine whether miR-7515 regulated the
proliferation migration and invasion of PC cells via regulating
IGF-1, the miR-7515 overexpression lentivirus and IGF-1
overexpression plasmid were co-transfected in SW1990 and PANC-1
cells (Fig. 7A). The results
demonstrated that IGF-1 overexpression relieved the inhibitory
effects of miR-7515 overexpression on the Ras/Raf/MEK/ERK signaling
pathway (Fig. 7B). The results of the
CCK-8 assay revealed that overexpression of IGF-1 in SW1990 and
PANC-1 cells decreased the inhibitory effects of miR-7515
overexpression on cell proliferation (Fig. 8A). Similarly, the wound healing assay
results demonstrated that the migratory rate of SW1990 and PANC-1
cells co-transfected with the miR-7515 overexpression lentivirus
and IGF-1 overexpression plasmid was increased compared with cells
only transfected with miR-7515 overexpression lentivirus (Fig. 8B). Furthermore, the overexpression of
IGF-1 in miR-7515 overexpression cells upregulated the expression
levels of cyclin D1 (G1 phase marker), CDK2
(G1 phase marker), MMP-2 (metastasis marker) and MMP-9
(metastasis marker) (Fig. 8C). These
results indicated that IGF-1 may regulate the proliferation
migration and invasion of PC cells via targeting the IGF-1-induced
Ras/Raf/MEK/ERK signaling pathway.
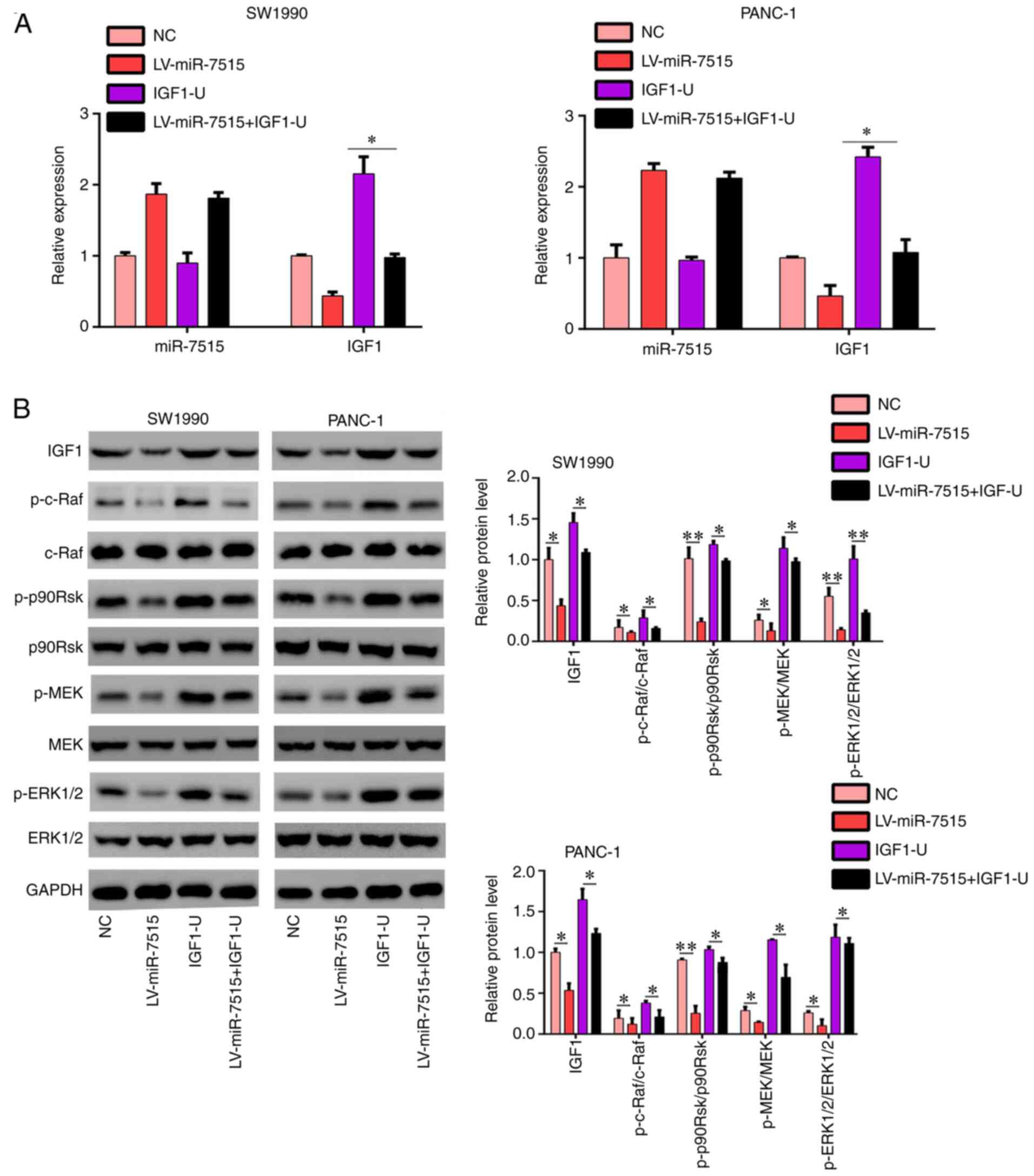 | Figure 7.IGF-1 overexpression reverses the
regulatory effects of miR-7515 overexpression on the Ras/Raf/MEK/
ERK signaling pathway. PANC-1 and SW1990 cells were transfected
with miR-7515 overexpression lentivirus, IGF-1 overexpression
plasmid, miR-7515 overexpression lentivirus + IGF-1 overexpression
plasmid and corresponding negative control. (A) Reverse
transcription-quantitative PCR was used to analyze the expression
levels of IGF-1 and miR-7515 in each group. (B) Western blotting
was used to analyze the expression levels of IGF-1, p-c-Raf,
p-p90RSK, p-MEK, MEK, p-ERK1/2 and ERK1/2 in each group of cells.
*P<0.05 and **P<0.01. IGF-1, insulin like growth factor 1;
miR, microRNA; p-, phosphorylated; p90RSK, ribosomal protein S6
kinase α-1; NC, negative control; LV, lentivirus. |
MEK inhibitors block the effects of
miR-7515 knockdown on proliferation invasion and migration of PC
cells
To determine whether miR-7515 regulated the
proliferation invasion and migration of PC cells via regulating
IGF-1 expression, AsPC-1 and BXPC-3 cells were transfected with the
miR-7515 inhibitor and treated with the MEK inhibitor PD98059. The
results of the CCK-8 assays demonstrated that the treatment of
AsPC-1 and BXPC-3 cells with the MEK inhibitor decreased the
promoting effects of the miR-7515 inhibitor on cell proliferation
(Fig. 9A). The results of the wound
healing assay revealed that the migration rate in AsPC-1 and BXPC-3
cells transfected with the miR-7515 inhibitor and treated with the
MEK inhibitor, PD98059, was decreased compared with cells only
transfected with the miR-7515 inhibitor (Fig. 9B). Furthermore, compared to the group
only transfected with the MEK inhibitor, the inhibition of MEK in
miR-7515 inhibitor-transfected cells upregulated the expression
levels of cyclin D1, CDK2, MMP-2 and MMP-9 (Fig. 9C). These findings indicated that IGF-1
may regulate the proliferation, migration and invasion of PC cells
via targeting the IGF-1-induced Ras/Raf/MEK/ERK signaling
pathway.
Discussion
PC is a lethal malignant tumor of the digestive
system that is accompanied by high mortality rates. At present, the
successful treatment for PC remains a significant challenge
(2,21). Previous studies have reported the role
of numerous miRNAs in the occurrence and the development of PC
(22–24). However, to the best of our knowledge,
the molecular mechanisms underlying the role of miRNAs in PC remain
largely unknown.
miR-7515 is a novel miRNA identified by Lee et
al in 2013 (25). A previous
study demonstrated that miR-7515 expression levels were
downregulated in lung cancer cells, which inhibited lung cancer
cell proliferation via targeting c-Met (25). Similarly, Chong et al (26) revealed that the expression levels of
miR-7515 were downregulated in recurrent ovarian cancer compared
with primary ovarian cancer. However, to the best of our knowledge,
the role of miR-7515 in PC remains unknown. The results of the
present study revealed that the expression levels of miR-7515 were
downregulated in PC tissues and cell lines, and the low expression
of miR-7515 predicted a poor clinical outcome. The overexpression
of miR-7515 in PC cells inhibited cell proliferation, migration and
invasion both in vitro and in vivo, while miR-7515
knockdown increased cell proliferation, invasion and migration
in vitro. To the best of our knowledge, this is the first
evidence to suggest that miR-7515 may play a tumor-suppressive role
in PC by inhibiting the proliferation, migration and invasion of PC
cells.
Accumulating evidence indicates that IGF-related
signaling pathways promote the progression of PC to an advanced
stage via stimulating the proliferation, metastasis and
chemoresistance of PC cells (27).
IGF-1 is a member of IGF family, and previous studies have
demonstrated that the expression levels of IGF-1 were upregulated
in PC tissues, which indicates its potential as a diagnostic
biomarker in PC (28,29). The upregulated expression of IGF-1
also predicted a poor clinical outcome in patients with PC.
Moreover, the IGF-1 receptor (IGF-1R) was revealed to have tyrosine
kinase activity (30). Upon IGF-1R
binding with IGF-1, the receptor is activated and increases the
phosphorylation levels of AKT and MEK proteins, leading to the
activation of intracellular signaling pathways, such as the
PI3K/Akt/mTOR pathway and RAS/RAF/MEK/ERK signaling pathway
(31). Therefore, the knockdown of
IGF-1 expression may represent a potential strategy for the
treatment of PC. In the present study, the target genes of miR-7515
were demonstrated to be most enriched in the Ras signaling pathway.
In particular, IGF-1 was identified as a target gene of miR-7515,
and was revealed to have a high binding score and to be involved in
the Ras signaling pathway. Thus, it was hypothesized that miR-7515
may play a tumor-suppressive role in PC via targeting IGF-1
expression, as well as the Ras/RAF/MEK/ERK signaling pathway.
Consistent with this hypothesis, the results of the present study
revealed that miR-7515 directly bound with IGF-1 and regulated its
expression, in addition to regulating the Ras/Raf/MEK/ERK signaling
pathway. However, the exact mechanism whereby miRNA-7515 inhibits
the Ras/RAF/MEK/ERK signaling pathway requires further
investigation. IGF-1 expression levels were revealed to be
upregulated in PC tissues and negatively co-expressed with miR-7515
expression. In addition, the overexpression of IGF-1 blocked the
inhibitory effects of miR-7515 overexpression on proliferation,
migration and invasion of PC cells. Treatment with the MEK
inhibitor, PD98059, also blocked the effects of knockdown of
miR-7515 on proliferation, migration and invasion of PC cells. To
the best of our knowledge, the present study was the first to
determine the potential molecular mechanism of miR-7515 in PC. In
fact, miRNAs could exhibit powerful effects on the development of
malignancy due to their potential to control numerous target genes
(32). However, this phenomenon is
also a limitation in miRNA research, and future studies are needed
to further explore whether there are other pathways involved in PC
progression.
In conclusion, the findings of the present study
indicated that miR-7515 may be a novel tumor suppressor in PC, and
it may suppress proliferation, migration and invasion of PC cells
by targeting the IGF-1-induced Ras/Raf/MEK/ERK signaling pathway.
Although miRNA-based therapeutics are still under development, our
results are encouraging, and suggest that miR-7515 and IGF-1 could
serve as clinical targets for the treatment of PC or other tumor
types in the future.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant. no. 81960433) and the
National Natural Science Foundation of Guizhou Medical University
(grant. no. 19NSP034).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
SL and ZZ confirmed the authenticity of all the raw
data. WC and ZZ designed the study. WC, ZZ and ZH performed the
experiments and analyzed the data. ZZ and WC wrote the manuscript
and were responsible for language revisions. SL made substantial
contributions to conception and design, acquisition of data, and
analysis and interpretation of the data. All the authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all
patients prior to participation and the patient experimental
protocol was approved by the Human Research Ethics Review Committee
of Guizhou Medical University (Guiyang, China). All animal
experiments were performed in strict accordance with the Guide for
the Care and Use of Laboratory Animals and the Principles for the
Utilization and Care of Vertebrate Animals. The animal experimental
protocols were approved (approval. no. 2000025) by the Ethics
Committee of Guizhou Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Riquelme E, Zhang Y, Zhang L, Montiel M,
Zoltan M, Dong W, Quesada P, Sahin I, Chandra V, Lucas AS, et al:
Tumor microbiome diversity and composition influence pancreatic
cancer outcomes. Cell. 178:795–806. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
McGuigan A, Kelly P, Turkington RC, Jones
C, Coleman HG and McCain RS: Pancreatic cancer: A review of
clinical diagnosis, epidemiology, treatment and outcomes. World J
Gastroenterol. 24:4846–4861. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mendt M, Kamerkar S, Sugimoto H, McAndrews
KM, Wu CC, Gagea M, Yang S, Blanko EV, Peng Q, Ma X, et al:
Generation and testing of clinical-grade exosomes for pancreatic
cancer. JCI Insight. 19:e992632018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu GF, Li GJ and Zhao H: Efficacy and
toxicity of different chemotherapy regimens in the treatment of
advanced or metastatic pancreatic cancer: A network meta-analysis.
J Cell Biochem. 119:511–523. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Srikok S, Patchanee P, Boonyayatra S and
Chuammitri P: Potential role of MicroRNA as a diagnostic tool in
the detection of bovine mastitis. Prev Vet Med. 182:1051012020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kagiya T: MicroRNAs: Potential biomarkers
and therapeutic targets for alveolar bone loss in periodontal
disease. Int J Mol Sci. 17:13172016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Aslan M, Shahbazi R, Ulubayram K and
Ozpolat B: Targeted therapies for pancreatic cancer and hurdles
ahead. Anticancer Res. 38:6591–6606. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Qu K, Zhang X, Lin T, Liu T, Wang Z, Liu
S, Zhou L, Wei J, Chang H, Li K, et al: Circulating miRNA-21-5p as
a diagnostic biomarker for pancreatic cancer: Evidence from
comprehensive miRNA expression profiling analysis and clinical
validation. Sci Rep. 7:16922017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tu MJ, Ho PY, Zhang QY, Jian C, Qiu JX,
Kim EJ, Bold RJ, Gonzalez FJ, Bi H and Yu AM: Bioengineered
miRNA-1291 prodrug therapy in pancreatic cancer cells and
patient-derived xenograft mouse models. Cancer Lett. 442:82–90.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu Z, Zhao S, Wang L, Wang J and Zhou J:
miRNA-339-5p plays an important role in invasion and migration of
pancreatic cancer cells. Med Sci Monit. 25:7509–7517. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang Y, Sun Y, Wang H, Li H, Zhang M, Zhou
L, Meng X, Wu Y, Liu P, Liu X, et al: MicroRNA-221 induces
autophagy through suppressing HDAC6 expression and promoting
apoptosis in pancreatic cancer. Oncol Lett. 16:7295–7301.
2018.PubMed/NCBI
|
|
12
|
Lv F, Zheng K, Yu J and Huang Z:
MicroRNA-661 expression is upregulated in pancreatic ductal
adenocarcinoma and promotes cell proliferation. Oncol Lett.
16:6293–6298. 2018.PubMed/NCBI
|
|
13
|
Zhang X, Shi H, Lin S, Ba M and Cui S:
MicroRNA-216a enhances the radiosensitivity of pancreatic cancer
cells by inhibiting beclin-1-mediated autophagy. Oncol Rep.
34:1557–1564. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sun Y, Zhu Q, Zhou M, Yang W, Shi H, Shan
Y, Zhang Q and Yu F: Restoration of miRNA-148a in pancreatic cancer
reduces invasion and metastasis by inhibiting the wnt/β-catenin
signaling pathway via downregulating maternally expressed gene-3.
Exp Ther Med. 17:639–648. 2019.PubMed/NCBI
|
|
15
|
Mutgan AC, Besikcioglu HE, Wang S, Friess
H, Ceyhan GO and Demir IE: Insulin/IGF-driven cancer cell-stroma
crosstalk as a novel therapeutic target in pancreatic cancer. Mol
Cancer. 17:662018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rozengurt E: Mechanistic target of
rapamycin (mTOR): A point of convergence in the action of
insulin/IGF-1 and G protein-coupled receptor agonists in pancreatic
cancer cells. Front Physiol. 5:3572014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yakovenko A, Cameron M and Trevino JG:
Molecular therapeutic strategies targeting pancreatic cancer
induced cachexia. World J Gastrointest Surg. 10:95–106. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Takahashi T, Ichikawa H, Morimoto Y,
Tsuneyama K and Hijikata T: Inhibition of EP2/EP4 prostanoid
receptor-mediated signaling suppresses IGF-1-induced proliferation
of pancreatic cancer BxPC-3cells via upregulating γ-glutamyl
cyclotransferase expression. Biochem Biophys Res Commun.
516:388–396. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
National Research Council (US) Committee
for the Update of the Guide for the Care and Use of Laboratory
Animals, . Guide for the Care and Use of Laboratory Animals.
National Academies Press (US); Washington, DC: 2011
|
|
21
|
Lei S, He Z, Chen T, Guo X, Zeng Z, Shen Y
and Jiang J: Long noncoding RNA 00976 promotes pancreatic cancer
progression through OTUD7B by sponging miR-137 involving EGFR/MAPK
pathway. J Exp Clin Cancer Res. 38:4702019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nagathihalli NS, Castellanos JA,
Lamichhane P, Messaggio F, Shi C, Dai X, Rai P, Chen X, Vansaun MN
and Merchant NB: Inverse correlation of STAT3 and MEK signaling
mediates resistance to RAS pathway inhibition in pancreatic cancer.
Cancer Res. 78:6235–6246. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu SZ, Xu HC, Wu XL, Liu P, Shi YC, Pang
P, Deng L, Zhou GX and Chen XY: Dihydrosanguinarine suppresses
pancreatic cancer cells via regulation of mut-p53/WT-p53 and the
Ras/Raf/Mek/Erk pathway. Phytomedicine. 59:1528952019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu L, Yuan X, Ni J, Shen L, Cai M and
Jiang D: Gain of microRNA-103 triggers metastatic behavior by
targeting ubiquitin specific peptidase 10 in pancreatic cancer. Int
J Clin Exp Pathol. 12:1214–1223. 2019.PubMed/NCBI
|
|
25
|
Lee JM, Yoo JK, Yoo H, Jung HY, Lee DR,
Jeong HC, Oh SH, Chung HM and Kim JK: The novel miR-7515 decreases
the proliferation and migration of human lung cancer cells by
targeting c-Met. Mol Cancer Res. 11:43–53. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chong GO, Jeon HS, Han HS, Son JW, Lee YH,
Hong DG, Lee YS and Cho YL: Differential MicroRNA expression
profiles in primary and recurrent epithelial ovarian cancer.
Anticancer Res. 35:2611–2617. 2015.PubMed/NCBI
|
|
27
|
Dey S, Liu S, Factora TD, Taleb S,
Riverahernandez P, Udari L, Zhong X, Wan J and Kota J: Global
targetome analysis reveals critical role of miR-29a in pancreatic
stellate cell mediated regulation of PDAC tumor microenvironment.
BMC Cancer. 20:6512020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang SY, Miah A, Pabari A and Winslet M:
Growth factors and their receptors in cancer metastases. Front
Biosci (Landmark Ed). 16:531–538. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hwang HJ, Oh MS, Lee DW and Kuh HJ:
Multiplex quantitative analysis of stroma-mediated cancer cell
invasion, matrix remodeling, and drug response in a 3D co-culture
model of pancreatic tumor spheroids and stellate cells. J Exp Clin
Cancer Res. 38:2582019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kopantzev EP, Kopantseva MR, Grankina EV,
Mikaelyan A, Egorov VI and Sverdlov ED: Activation of IGF/IGF-IR
signaling pathway fails to induce epithelial-mesenchymal transition
in pancreatic cancer cells. Pancreatology. 19:390–396. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bertrand FE, Steelman LS, Chappell WH,
Abrams SL, Shelton JG, White ER, Ludwig DL and McCubrey JA: Synergy
between an IGF-1R antibody and Raf/MEK/ERK and PI3K/Akt/mTOR
pathway inhibitors in suppressing IGF-1R-mediated growth in
hematopoietic cells. Leukemia. 20:1254–1260. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Catalanotto C, Cogoni C and Zardo G:
MicroRNA in control of gene expression: An overview of nuclear
functions. Int J Mol Sci. 17:17122016. View Article : Google Scholar : PubMed/NCBI
|