Introduction
Lung cancer is the leading cause of cancer-related
death in the world, followed by colorectal, liver and stomach
cancers (1,2), and non-small cell lung cancer (NSCLC)
accounts for ~85% of patients with lung cancer (3,4). Lung
squamous cell carcinoma (LSCC) is the second most common
histological subtype and is associated with poor prognosis
(5). Despite the diagnosis and
treatment of NSCLC have been advanced, the main treatment strategy
for LSCC remains surgical resection or radiation with chemotherapy
(6,7). However, the recurrence rate was high
among the patients who suffer postoperative (8). Previously, PD1/PD-L1 immunotherapies
have improved the clinical outcome of patients with NSCLC and
demonstrated a notable efficacy (9–11), and
targeted therapies have improved outcomes for certain subtypes, but
patients with LSCC are not likely to benefit from these treatments
(12). Thus, it is imperative to
search for new drugs and effective therapeutic strategies against
LSCC to reduce the mortality rate.
Nowadays, numerous natural medicines have been
proved to inhibit or reverse the progression of several cancers and
appeared to be promising novel anticancer drugs due to high
efficiency and low toxicity. Licochalcone A (LCA) is a natural and
useful flavonoid extracted from licorice, which possesses several
pharmacological properties such as antibacterial (13), antitumor (14), anti-inflammatory (15,16)
and antioxidation properties (17).
In recent decades, studies have demonstrated that LCA exhibited
potent antitumor effects in several epithelial carcinoma cells by
regulating numerous signaling pathways, such as the oxidative
stress, the mitogen-activated protein kinase (MAPK) and the
apoptosis pathway (18). For
example, LCA induced apoptosis of gastric cancer cells by
activating the MAPK and PI3K/AKT signaling pathways (17). However, LSCC is a distinct subtype
of NSCLC, and the antitumor effects of LCA on LSCC remain
unknown.
F-box protein 5 (FBXO5) is involved in the
regulation of proliferation, apoptosis, epithelial-mesenchymal
transition and drug resistance (19,20).
FBXO5 promotes proper mitotic entry by blocking the
anaphase-promoting complex/cyclosome activity (21,22).
It functions as a cell cycle regulator and plays a crucial role in
proliferation and tumorigenesis (23,24).
Evidence indicates that FBXO5 is upregulated in various malignant
tumors compared with matched normal tissues. Previous research
indicated that high FBXO5 expression would cause genomic
instability and mitotic catastrophe, contributing to tumorigenesis
of breast cancer (25) and
esophageal squamous cell carcinoma (26). In addition, existing evidence has
suggested that FBXO5 expression level was higher in LSCC compared
with normal tissue, and the overexpressed FBXO5 leads to shorter
overall survival (OS) in patients with LSCC (27). Therefore, FBXO5 may be a potential
oncogene and therapeutic target in LSCC.
The present study aimed to detect the effect of LCA
on cell proliferation, cell cycle, apoptosis and gene expression on
LSCC in vitro and the antitumor activities in vivo,
and further explore the underlying mechanisms.
Materials and methods
Cell culture
The LSCC cells H226 (cat. no. TCHu235), H1703 (cat.
no. SCSP-593) and human bronchial epithelial cells (HBE; cat. no.
CL-0346) were maintained in RPMI-1640 (cat. no. C11875500BT) media
supplemented with 10% fetal bovine serum (cat. no. 10099-141C;
Gibco; Thermo Fisher Scientific, Inc.) and cultured in a 37°C
incubator set with 5% CO2, humidified environment. H226
and H1703 cells were purchased from The Cell Bank of Type Culture
Collection of The Chinese Academy of Sciences. HBE cells were
purchased from Procell Life Science & Technology Co., Ltd.
Chemical reagents and antibodies
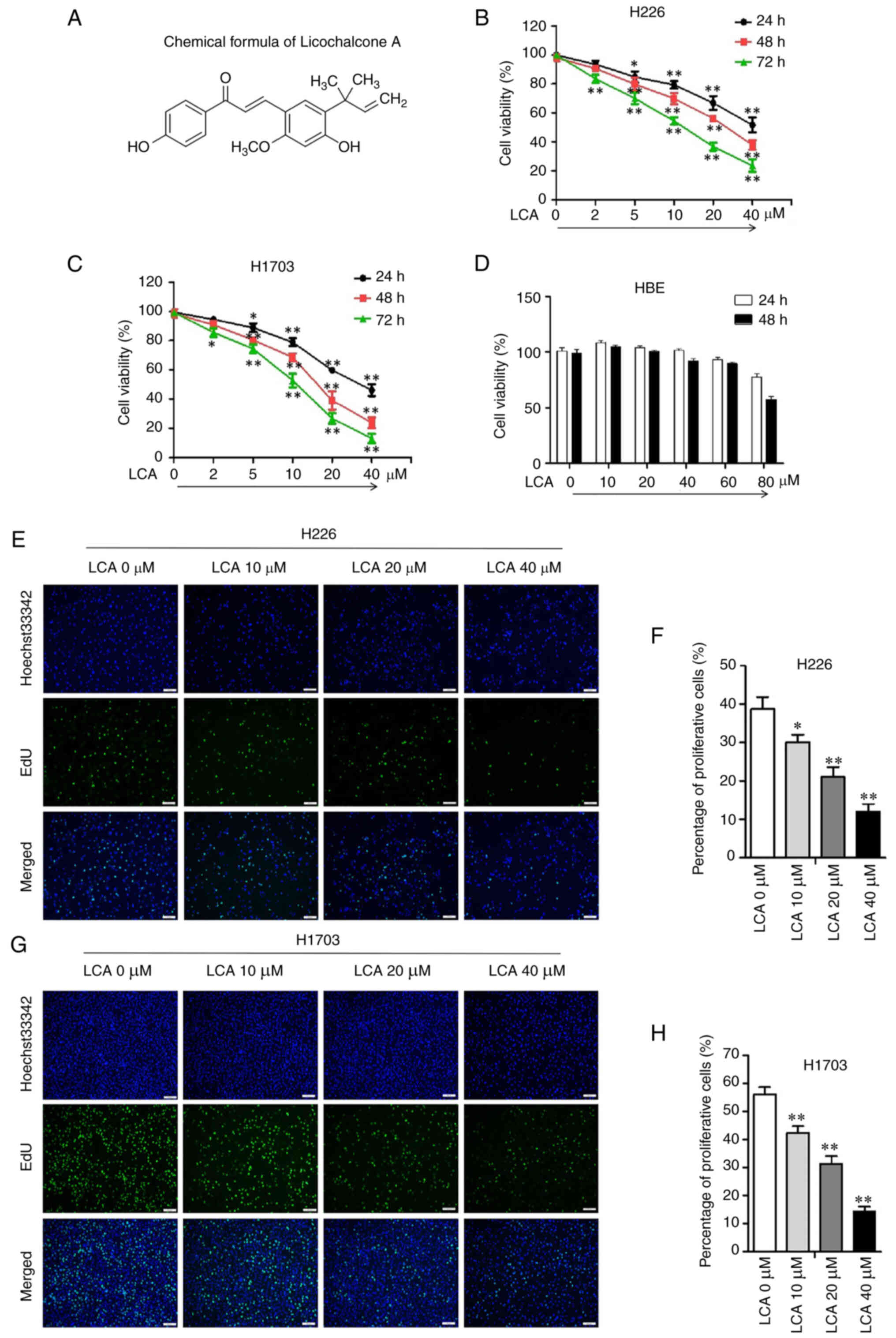
LCA (Cas. no. 58749-22-7; Fig. 1A; cat. no. S01015) was purchased
from Nanjing Dilger Medical Technology Co. Ltd. and dissolved in
dimethyl sulfoxide (DMSO; cat. no. D8371; Beijing Solarbio Science
& Technology Co., Ltd.). FITC Annexin V Apoptosis Detection kit
was acquired from BD (cat. no. 556547, BD Pharmingen; BD
Biosciences). CDK2 (1:1,000; cat. no. ab32147; Abcam), CDK4
(1:1,000; cat. no. ab108357; Abcam), cyclin D1 (1:5,000; cat. no.
ab134175; Abcam), cyclin E (1:1,000; cat. no. WL01072; Wanleibio
Co., Ltd.), cleaved caspase 3 (1:1,000; cat. no. AF7022; Affinity
Biosciences, Ltd.), cleaved poly(ADP-ribose)polymerase-1 (PARP1)
(1:5,000; cat. no. ab32064; Abcam), ERK (1:1,000; cat. no.
ab184699; Abcam), phosphorylated (p-)ERK (1:1,000; cat. no.
ab201015; Abcam), p38 MAPK (1:1,000; cat. no. 8690; Cell Signaling
Technology, Inc.), p-p38 MAPK (1:1,000; cat. no. 4511; Cell
Signaling Technology, Inc.), FBXO5 (1:5,000; cat. no. ab184950;
Abcam), GAPDH (1:5,000; cat. no. 60004-1-IG; Proteintech Group,
Inc.), HRP-goat anti-mouse (1:5,000; cat. no. EM35110-01; Beijing
Emarbio Science & Technology Co., Ltd.) and HRP-goat
anti-rabbit antibody (1:5,000; cat. no. EM35111-01; Beijing Emarbio
Science & Technology Co., Ltd.).
MTT detection
The cells were seeded in 96-well plates at a density
of 5×103 cells per well in a 100-µl medium and incubated
overnight. The H226 and H1703 cells were cultured with LCA at a
dose of 0, 2, 5, 10, 20 and 40 µM for 24, 48 and 72 h. HBE cells
were cultured in the medium with a concentration of 0, 10, 20, 40,
60 and 80 µM LCA for 24 and 48 h. After treatment, 20 µl of 5 mg/ml
MTT (Beijing Solarbio Science & Technology Co., Ltd.) was added
to the well and incubated for at least 3 h in an incubator set at
37°C, then the medium was removed from each well and 200 µl DMSO
was added. The absorbance at the wavelength of 490 nm (OD value)
was measured by Infinite M200 Pro NanoQuant (Tecan Group, Ltd.) and
then the cell viability and IC50 value were measured
according to the OD value. Based on the result of MTT, the
IC50 was calculated for H226 and H1703 cells.
Subsequently, LCA was selected at the dose of 0, 10, 20 and 40 µM
in the following experiments.
5-ethynyl-2′-deoxyuridine (EdU)
staining
Cell proliferation was detected using
BeyoClick™ EdU-488 Assay kit (cat. no. C0071S; Beyotime
Institute of Biotechnology). LSCC cells were plated in six-well
plates and incubated overnight, then treated with different doses
of LCA for 16 h. EdU solution was prepared and final 10 µM EdU
working solution was added to the medium for incubating for 3 h at
37°C. Subsequently, 4% paraformaldehyde was used to fix cells for
15 min and washed off with PBS containing 3% BSA (Beijing Solarbio
Science & Technology Co., Ltd.). Next, the cells were
permeabilized with enhanced immunostaining permeabilization buffer
(cat. no. P0097; Beyotime Institute of Biotechnology) for 15 min
and washed. Then, the EdU detection was performed followed the
manufacturer's protocols and the nuclei were stained with Hoechst
33342 for 10 min while avoiding light at room temperature.
Fluorescence images were obtained by Olympus microscope (Olympus
Corporation). The number of EdU-positive cells was counted and
calculated in five randomly selected fields. Cell proliferation
rate=EdU-positive cells/DAPI-positive cells ×100%. The experiments
were performed in triplicate.
Cell cycle assay
The cell cycle distribution was evaluated by FCM
using propidium iodide (PI; cat. no. P8080; Beijing Solarbio
Science & Technology Co., Ltd.). H226 and H1703 cells were
cultured in 6 cm dishes and treated with LCA (0, 10, 20 and 40 µM)
for 24 h in a 37°C incubator set with 5% CO2, humidified
environment. After treatment, the cells were harvested and
resuspended, and subsequently fixed with 70% cold ethyl alcohol
overnight at 4°C. The next day, cells were centrifuged at 1,000 × g
for 10 min and washed. Following this, the cells were incubated
with RNase A and 500 µl PI in a 37°C water bath for 30 min. The
cell cycle distribution of LSCC was measured on the Accuri C6 flow
cytometer (BD Biosciences) and analyzed with FlowJo 7.6 software
(FlowJo LLC).
Cell apoptosis assay
Cellular apoptosis was evaluated by FCM using
FITC-Annexin V Apoptosis Detection Kit. H226 and H1703 cells were
treated with LCA (0, 10, 20 and 40 µM) for 24 h in a 37°C incubator
set with 5% CO2, humidified environment. After
treatment, the cells were harvested and assayed according to the
manufacturer's instructions. The apoptotic rate was analyzed by the
Accuri C6 flow cytometer (BD Biosciences).
4D-data-independent acquisition
(4D-DIA) proteomics
H1703 cells were cultured overnight in 100-mm
culture dishes until the density reached 40–50%. Subsequently, the
cells were treated with or without 40 µM LCA for 24 h in a 37°C
incubator set with 5% CO2, humidified environment. The
medium was aspirated, and the cells were washed with pre-cooled PBS
solution twice. Next, the cells were gently scraped off with cell
scraper and transferred to a centrifuge tube, centrifuged at 4°C,
300–500 × g for 5 min, the supernatant was discarded and washed
once with PBS. PBS was added to make cell suspensions and
1×107 cells were counted in each sample by a cell
counter. The cells were immediately frozen with liquid nitrogen for
5–10 min, then stored in a −80°C refrigerator for proteomics assay.
Before the differential protein expression assays, the qualitative
and quantitative protein analysis of the 4D mass spectrometry data
was performed by DIA-NN (v1.8.1) software. The Pearson's
correlation analysis was used to evaluate the correlation among
samples. Principal component analysis (PCA) was used to understand
the variances between the different sample groups.
Network pharmacological analysis
PubChem (https://pubchem.ncbi.nlm.nih.gov/) was used for
searching ‘LCA’ and obtained the SMILES number, the potential
targets genes of LCA were predicted by SwissTarget Prediction
(http://www.swisstargetprediction.ch/). The potential
LSCC therapeutic targets were identified by DisGeNET databases
(https://www.disgenet.org/search). The
Sangerbox (http://sangerbox.com/) was used to
perform the Gene Ontology and Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway enrichment analyses of the differentially
expressed genes (DEGs). The DEGs were imported into the Venny 2.1
online tool (https://bioinfogp.cnb.csic.es/tools/venny/index.html)
to screen the overlapping genes. Protein-protein interaction (PPI)
analysis was performed using the STRINGDB (http://string-db.org/) protein interaction
database.
Bioinformatics analyses
The TIMER2.0 database (http://timer.cistrome.org/) and UALCAN database
(https://ualcan.path.uab.edu/analysis)
were used to study the DEGs between tumor and adjacent normal
tissues based on The Cancer Genome Atlas (TCGA) project. In TIMER
2.0 database, the differential expression between tumor and
adjacent normal tissues for any gene of interest across all TCGA
tumors. Distributions of gene expression levels are displayed using
box plots. The statistical significance computed by the Wilcoxon
test is annotated by the number of stars (*P<0.05, **P<0.01
and ***P<0.001).
Molecular docking
The canonical 2D structure of Licochalcone A (CAS
no. 58749-22-7) was obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov/). ChemOffice
software (https://www.downza.cn/) was used to
optimize and minimize the structure energy of LCA, then convert the
2D structure to 3D. The ID of FBXO5 (Q9UKT4) was obtained from
Uniprot database (https://www.uniprot.org/), and the crystal structure
of the FBXO5 (PDB ID: 2M6N) was obtained from PDB database
(http://www.rcsb.org/). The mol2 files of LCA and
the PDB files of FBXO5 were imported to AutoDocK Tools, converted
to PDBQT format and parameters were set to determine the active
pocket. Molecular docking was performed with AutoDock 1.5.6
software. The docking image was acquired using PyMol software
(https://pymol.org/2/).
Transient transfection of small
interfering siRNA
siRNAs against FBXO5 (si-FBXO5) and the negative
control siRNA (si-NC) were purchased from CENTRAL BIOL (www.generalbiol.com/). The sequences of si-FBXO5 and
si-NC are listed in Table SI.
H1703 cells were transfected with si-FBXO5 and si-NC for 24 h at
37°C using Lipofectamine® 3000 (cat no. L3000-015; Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. The concentration of siRNAs used in the present study
was 50 nM. Subsequent experiments were performed 24 h after siRNA
transfection.
In vivo xenograft experiment
All operation procedures of this animal study were
approved by the Institutional Animal Care and Use Committee of
Guilin Medical University (approval no. GLMC-IACUC-2021016; Guilin,
China). A total of 20 BALB/c-nu mice (male) at the age of 4 weeks
(mean weight, 16±0.5 g) were obtained from Hunan SJA Laboratory
Animal Co., Ltd. The mice were housed in pathogen-free environment
with controlled temperature (20–25°C), humidity (40–70%), and free
access to rodent chow and water. After 5 days of acclimation, equal
volumes (200 µl) of 4×106 H1703 cells resuspended in PBS
(4×106 cells/mouse) were injected into the right flank
of mice. After 7 days, when the tumors were visible, the mice were
divided into 4 groups randomly (n=5): i) The control group (saline
containing 20% SBE-β-CD), ii) the LCA 7.5 mg/kg group, iii) the LCA
15 mg/kg group and iv) the cisplatin (CDDP) 2 mg/kg group. LCA was
dissolved in saline containing 20% SBE-β-CD. The mice were
intraperitoneally injected with 200 µl of drugs once a day for 10
days. The body weight, length and width of the tumor were measured
every day, and the tumor volume was calculated as follows: length ×
width2/2. During the process of the animal experiments,
none of the mice succumbed. After 10 days of LCA administration,
the mice were anesthetized by intraperitoneal injection of sodium
pentobarbital (50 mg/kg) and euthanized by cervical dislocation.
The humane endpoints included the tumor volume in the control group
(if it reached ~1,500 mm3) and the dietary activities
(if they were interfered). The tumors and vital organs were excised
and weighted after the absence of a heartbeat and breath, one
portion of the tumor was frozen with liquid nitrogen immediately
for western blotting, while the other portion was fixed in
paraformaldehyde for histopathological experiments. The vital
organs (heart, liver, spleen, lung and kidney) were fixed in
paraformaldehyde for hematoxylin and eosin (H&E) staining.
Western blot analysis
LSCC cells treated with LCA at the concentration of
0, 10, 20 and 40 µM. After treatment, the cells were harvested and
lysed in cold RIPA lysis buffer (cat. no. P0013B; Beyotime
Institute of Biotechnology) with PMSF and protease inhibitor for 30
min on ice. The supernatant was collected after being centrifuged
and the concentration was measured by BCA kit (Beyotime
Biotechnology). A total of 20 µg of protein was separated by
SDS-PAGE (10–12%) electrophoresis and further transferred onto the
nitrocellulose membrane. Subsequently, the membrane was blocked
with 5% non-fat milk in TBST buffer at room temperature for 2 h and
was then incubated overnight under the primary antibodies at 4°C.
After washed for 3 times with TBST, the membrane was incubated with
secondary antibody for 1 h at room temperature the next day. The
specific bands were visualized by ECL kit (cat. no. P0018S;
Beyotime Institute of Biotechnology) and analyzed by ImageJ (v1.48)
software (National Institutes of Health).
Histopathology
Tumor tissues and vital organs were fixed in
paraformaldehyde (4% PFA) for 24 h at room temperature. A serial
alcohol gradient was used to dehydrate the tissues, after
dehydration, the tissues were embedded on paraffin wax blocks. The
paraffin blocks were cut to 4-µm sections for H&E staining
(hematoxylin for 5 min and eosin for 3 min) at room temperature.
Images were captured using a light microscope (Olympus
Corporation).
Statistical analysis
All data were performed using the GraphPad Prism 5
(Dotmatics) and SPSS software version 23.0 (IBM Corp.). The paired
Student's t-test was used on data between two groups after testing
for normality and homogeneity of variance, and one-way ANOVA was
used for multiple comparisons followed by the post hoc Bonferroni
test. The data was expressed as the mean ± standard deviation (SD),
and P<0.05 was considered to indicate a statistically
significant difference.
Results
LCA inhibits the proliferation in LSCC
cells but not in HBE cells in vitro
MTT assay revealed that LCA significantly inhibited
the cell viability in H226 (Fig.
1B) and H1703 (Fig. 1C) cells.
Furthermore, the inhibitory effect of LCA (5, 10, 20 and 40 µM) on
LSCC cells was in a dose and time-dependent manner. The MTT result
showed that the cell viability of HBE was 93.31±2.98 and
89.66±2.25% when the concentration of LCA was 60 µM at 24 and 48 h
(Fig. 1D). These results indicated
that LCA exhibited no significant cytotoxicity against HBE cells in
a treatment with up to 60 µM of the drug. EdU staining assay
indicated that the proliferation rate was significantly reduced in
H226 (Fig. 1E and F) and H1703
cells (Fig. 1G and H) after LCA
treatment. These results revealed that LCA suppressed the
proliferation capacity of LSCC cells.
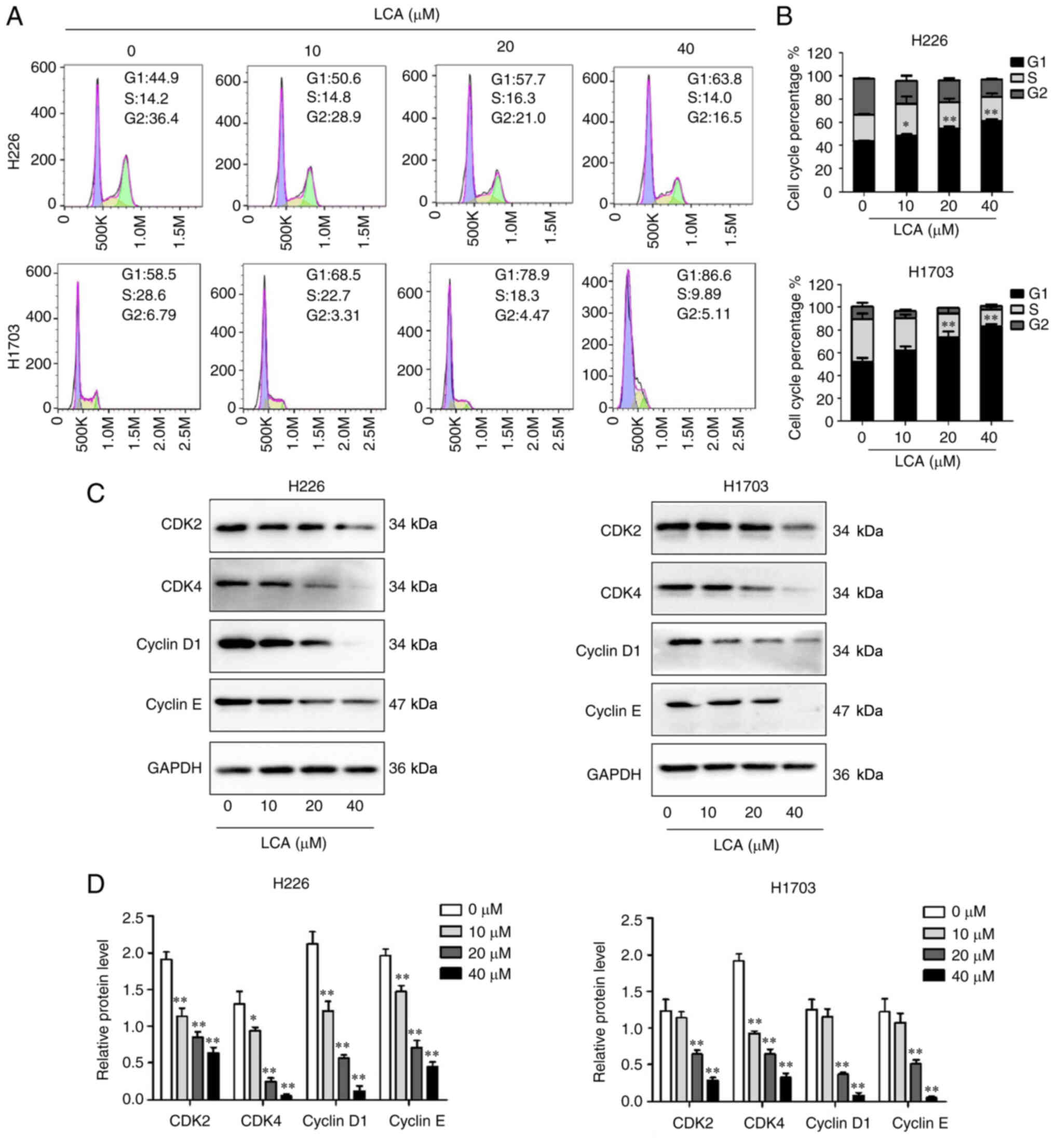
LCA induces cell cycle arrest in LSCC
cells
To assess the effect of LCA on cell cycle, the
distribution of cell cycle in H226 and H1703 cells after treatment
with different concentrations of LCA was investigated by FCM. As
illustrated in Fig. 2A and B, the
proportion of G1 phase at concentrations of 0, 10, 20 and 40 µM in
H226 cells were 43.70±0.78, 48.40±2.03, 54.67±2.71 and 61.33±2.19%,
respectively, and the ratio of cells at G2 phase was decreased. LCA
significantly induced G1 phase arrest in a dose-dependent manner
(10, 20 or 40 vs. control: P<0.05, P<0.01 or P<0.01) in
H226 cells. Similarly, in H1703 cells, the G1 phase proportion at
concentrations of 0, 10, 20 and 40 µM were 52.03±6.09, 61.93±5.92,
73.47±9.07 and 83.40±3.15%, respectively, and the ratio of cells at
S phase decreased. LCA at concentrations of 20 and 40 µM could
significantly increase the G1 phase proportion (20 or 40 vs.
control: P<0.01 or P<0.01) in H1703 cells. These results
indicated that LCA treatment significantly increased the
distribution of G1 phase in a dose-dependent manner in a certain
range of concentrations. According to the results, the expression
level of proteins related to G1 phase was further explored. Western
blot analysis results (Fig. 2C and
D) indicated that LCA decreased the protein levels of cyclin
D1, cyclin E, CDK2 and CDK4 in H226 cells and H1703 cells. These
results suggested that LCA caused G1 phase arrest by regulating the
proteins related to G1 phase.
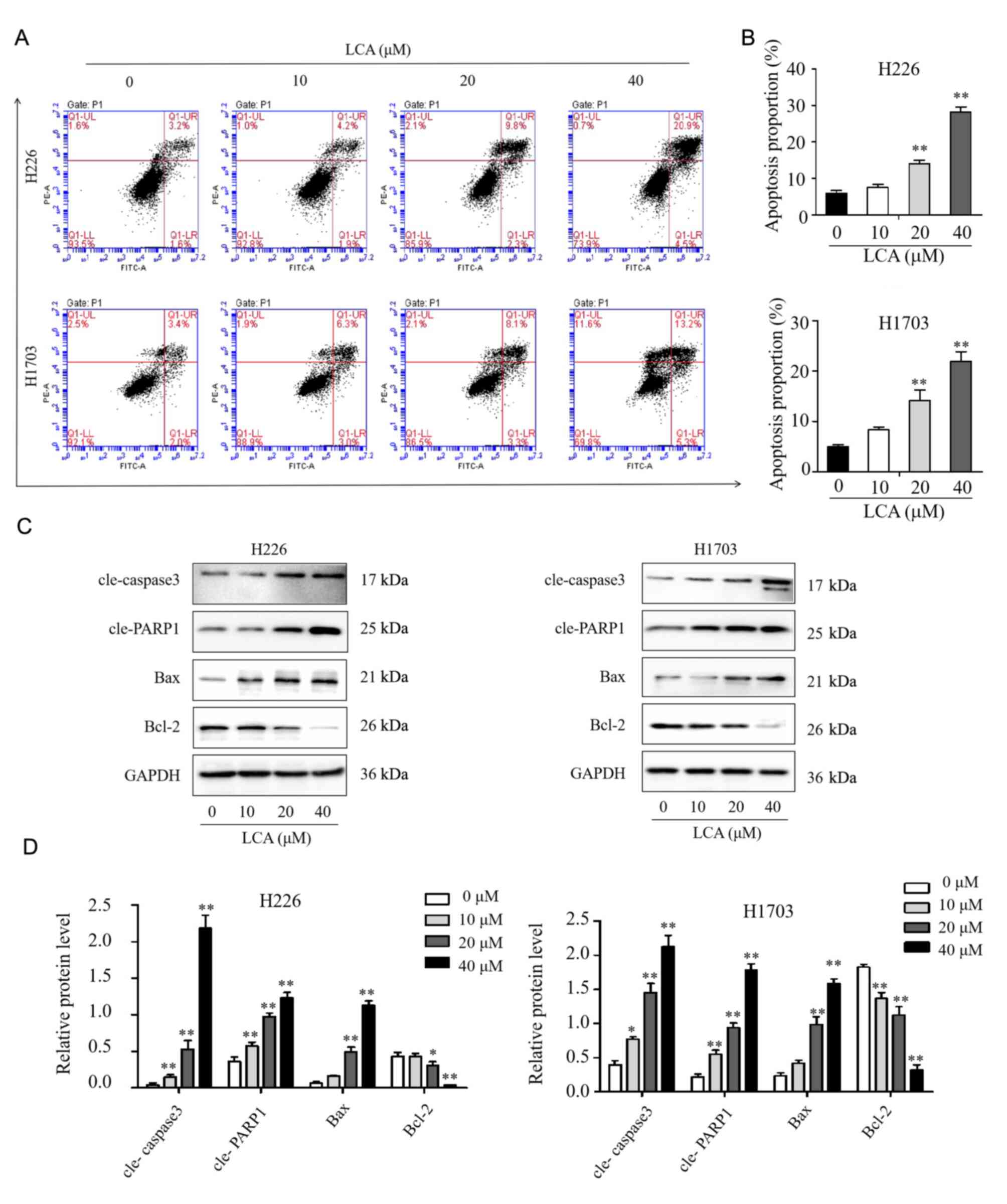
LCA induces apoptosis in LSCC
cells
The cell apoptosis of LSCC induced by LCA were
performed through FCM with the annexin V/PI double staining method.
After LCA treated for 24 h, the apoptotic rates of H226 cells at
concentrations of 0, 10, 20, and 40 µM were 6.13±1.16, 7.67±1.37,
14.07±1.70 and 28.20±2.47%, respectively. Similarly, in H1703
cells, the apoptotic rates were 5.03±0.64, 8.37±0.95, 14.17±3.65
and 21.93±3.35%, respectively (Fig. 3A
and B). At the concentration of 20 and 40 µM, the apoptotic
rates were significantly increased in both H226 and H1703 cells
compared with control (P<0.01). These results demonstrated that
LCA could induce apoptosis in dose-dependent manner within the
range of certain concentrations. As demonstrated in Fig. 3C and D, western blot analysis
results indicated that LCA significantly increased the expression
level of Bax, cleaved PARP1 and cleaved caspase 3 in H226 and H1703
cells, while decreased the level of Bcl-2. Overall, these results
indicated that LCA induced cell apoptosis by the
mitochondrial-mediated pathway in LSCC cells.
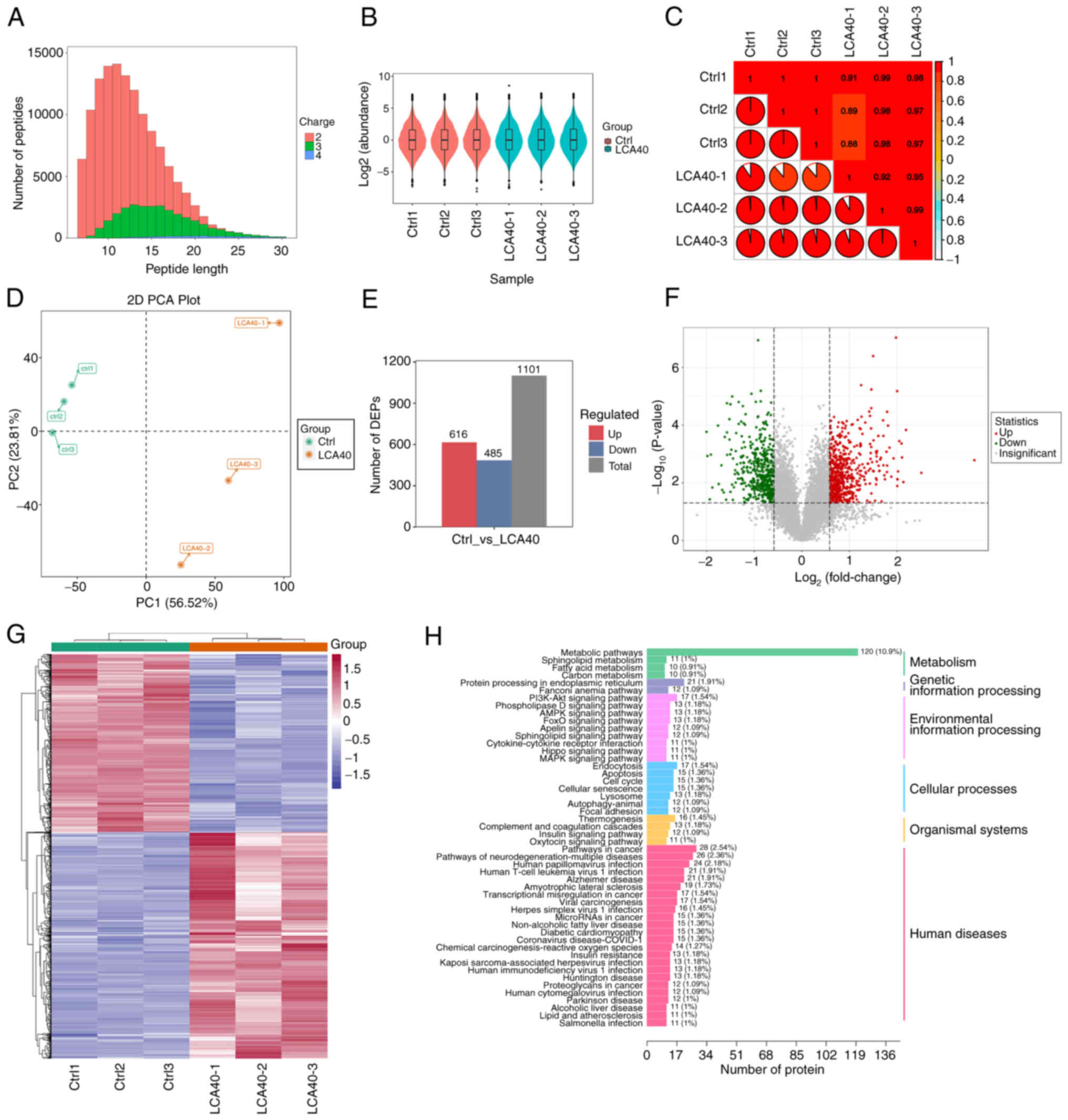
Proteomic analysis
4D-DIA quantitative proteomic technique was used to
identify the differential proteins and potential molecular
mechanisms after LCA treatment in LSCC cells. Most of the peptides
were distributed at 7–20 amino acids and the distribution of
peptide length met the quality control requirements (Fig. 4A). Box plots and violin plots
(Fig. 4B) indicated favorable
consistency within the sample group. Pearson's correlation
coefficients indicated high correlation in the expression patterns
between the samples (Fig. 4C). PCA
revealed satisfactory repeatability in the samples of control and
LCA treatment group (Fig. 4D).
Histogram (Fig. 4E) and Volcano
plots (Fig. 4F) demonstrated a
total of 1101 differentially expressed proteins (DEPs) after LCA
treated, including 616 upregulated and 485 downregulated DEPs. The
heatmap revealed the clustering of DEGs between the control and
LCA-treatment group (Fig. 4G).
Enrichment analyses of these DEPs based on KEGG databases revealed
that PI3K/Akt signaling pathway, AMPK signaling pathway, Hippo
signaling pathway, FoxO signaling pathway and MAPK signaling
pathway were enriched (Fig.
4H).
Network pharmacological analysis
The SwissTarget Prediction database search revealed
that 101 relevant targets for LCA were obtained. Disgenet databases
identified 671 potential therapeutic targets related to LSCC.
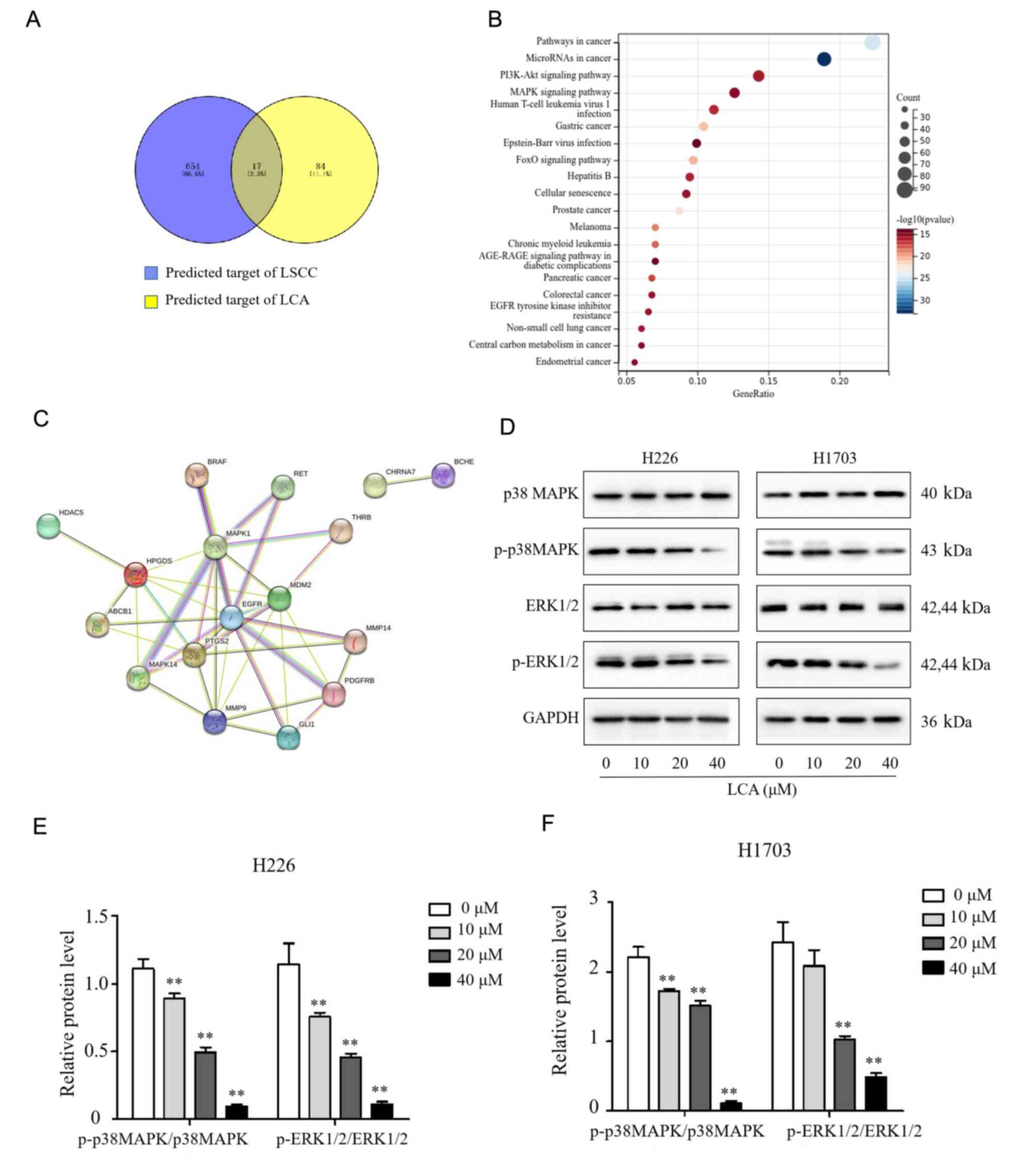
Comparative analysis of the targets obtained 17 overlapping genes
(Fig. 5A). Moreover, the KEGG
Pathway enrichment results detected that the DEPs mainly involved
in pathways in cancer, microRNAs in cancer, PI3K/Akt signaling
pathway and MAPK signaling pathway (Fig. 5B). These signaling pathways may be
closely related to the effects of LCA treatment in LSCC. A PPI
network was constructed among the 17 differentially expressed
proteins. The interacting proteins were MAPK1 (ERK2), MAPK14 (p38
MAPK), EGFR, PTGS2, PDGFRB, MMP9, MMP14, GLI1, MDM2, HPGDS, ABCB1,
RET and THRB (Fig. 5C); and the
MAPK1 (ERK2), MAPK14 (p38 MAPK), EGFR, PTGS2, PDGFRB were the core
targets. Combined with the results of proteomics, it was
hypothesized that LCA inhibits proliferation of LSCC via
suppression of the MAPK signaling pathways. Subsequently, western
blotting results demonstrated that LCA treatment inhibited the
expression of p-p38 MAPK and p-ERK1/2 in a dose-dependent manner
while the total p38 MAPK and ERK1/2 were not changed in both H1703
and H226 cells (Fig. 5D-F).
Bioinformatics analysis
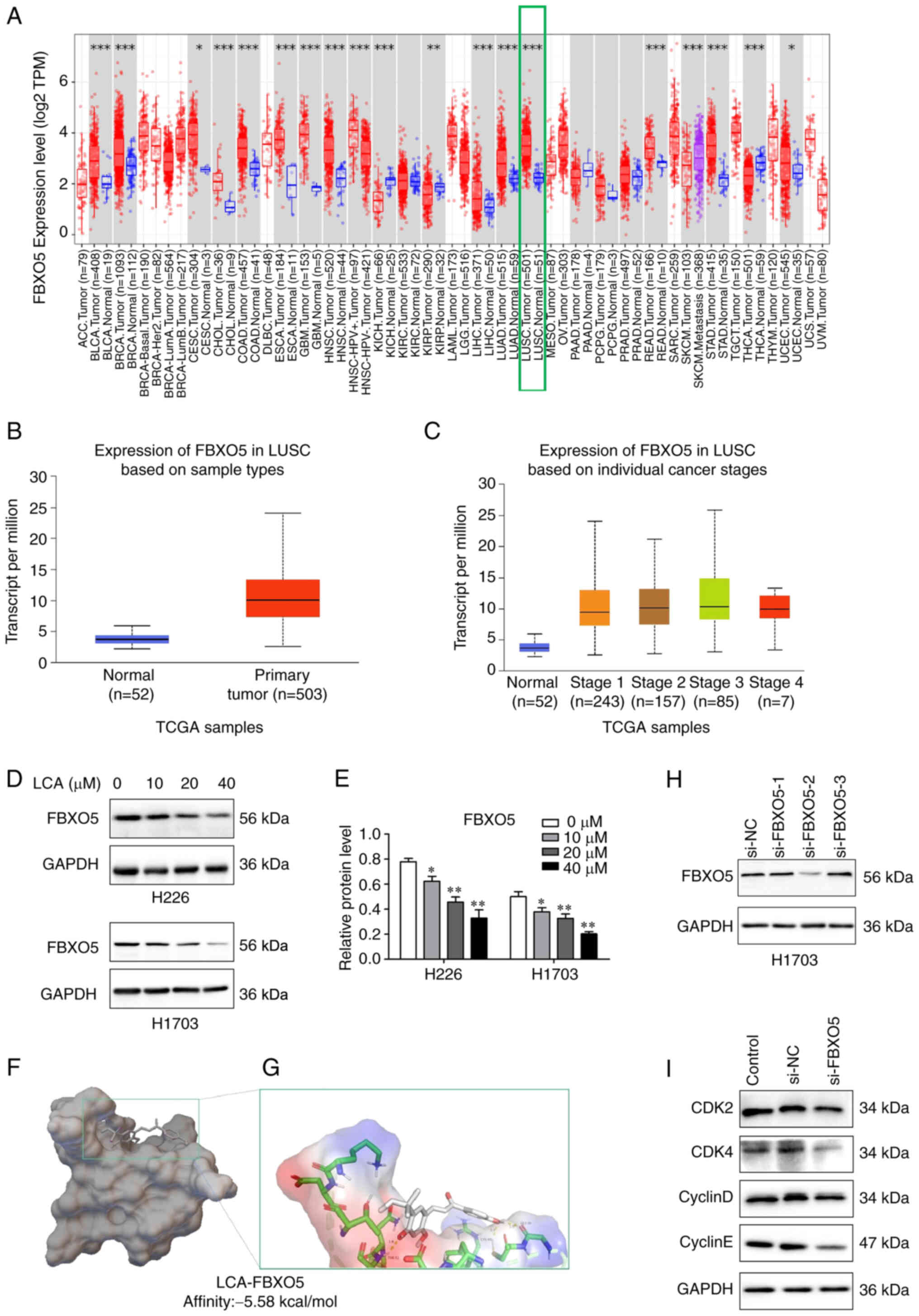
The top 5 upregulated and downregulated DEPs
obtained from proteomic analysis were presented in Table I. Bioinformatics analysis of these
10 DEPs through TIMER databases demonstrated that FBXO5 level was
higher in 14 types of cancer, such as LIHC, lung adenocarcinoma,
LSCC, bladder and breast cancer (Fig.
6A). The FBXO5 expression was significantly elevated in LSCC
compared with the corresponding normal tissues through TCGA data
(Fig. 6B). Moreover, the expression
of FBXO5 was significantly elevated in each pathological stage
(Fig. 6C). In addition, the result
of western blot analysis demonstrated that FBXO5 expression
decreased significantly in a concentration-dependent manner after
LCA treatment (Fig. 6D and E).
Therefore, it was perceived that FBXO5 may be a potential target of
LCA in the LSCC cells. Molecular docking has been widely used in
prediction of the interactions between a target protein (enzyme)
and drug molecules (ligands) at a molecular level (28). In the present study, the result
revealed that LCA is tightly bound to the active site of FBXO5 with
a binding energy of-5.58 kcal/mol, in which the hydroxyl group of
the LCA molecule forms three hydrogen bonds with the TYR52, CYS40
and GLY 39 (Fig. 6F and G). To
identify the effect of FBXO5 in LSCC cells, H1703 cells were
transfected with FBXO5 siRNA to knockdown the FBXO5. Si-FBXO5-2 was
chosen for its significant inhibition on the expression of FBXO5 in
H1703 cells (Fig. 6H). MTT
experiment exhibited that the cell viability of H1703 cells treated
with FBXO5-2 was lower compared with the negative control group
(Fig. S1). Moreover, FBXO5
silencing markedly reduced the expression levels of CDK2, CDK4,
cyclin D1 and cyclin E (Fig. 6I).
All the aforementioned results indicated that FBXO5 may be a
potential target of LCA.
 | Table I.Top 10 differentially expressed
proteins (LCA vs. Control). |
Table I.
Top 10 differentially expressed
proteins (LCA vs. Control).
|
|
| LCA treatment vs.
Control |
|---|
|
|
|
|
|---|
| Protein | Gene | Fold change | P-value |
|---|
| P01033 | TIMP1 |
12.34 | 0.0016 |
| Q96PD7 | DGAT2 |
5.71 | 0.0045 |
| P08138 | NGFR |
4.5617 | 0.0001 |
| Q6ZNF0 | ACP7 |
4.3918 | 0.0095 |
| P00740 | F9 |
4.3059 | 0.0013 |
| Q6PCB0 | VWA1 |
0.2497 | 0.0002 |
| P15104 | GLUL |
0.2529 | 0.0012 |
| Q6EMK4 | VASN |
0.2616 | 0.0376 |
| Q8TDB6 | DTX3L |
0.2661 | 0.0011 |
| Q9UKT4 | FBXO5 |
0.2716 | 0.0146 |
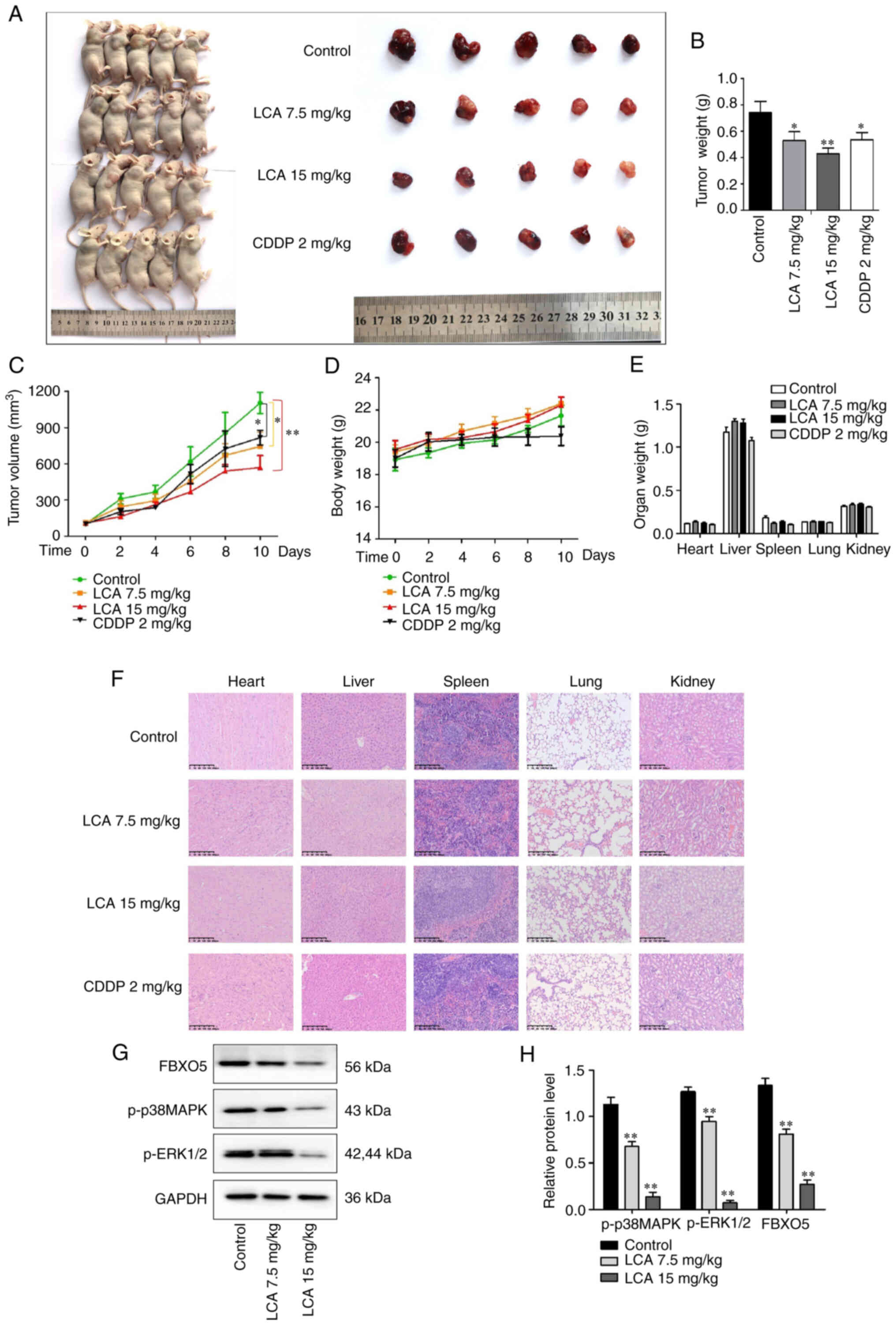
LCA inhibits the growth of LSCC in
vivo
Tumor-transplanted mouse models of H1703 cells were
used to evaluate the antitumor effect of LCA in vivo. As
revealed in Fig. 7A, images of the
mice and tumors of each group were captured. The maximum tumor
volume was 1,418.48 mm3 and the weight was 0.9 g in the
control group. Compared with control, the weights and volumes of
the tumor in LCA treatment groups were significantly decreased
(Fig. 7B and C). As shown in
Fig. 7D, no significant changes in
body weight were detected among groups. The body weight was
decreased in the CDDP group compared with control, but the loss was
not significant (P>0.05). In toxicological experiments, the
changes of organ weight and histological changes are important
indexes to evaluate the toxicity of drugs or compounds (29). To detect the possible potential
toxicity to organs, the organ weight was measured and histological
examination was performed. No significant changes in organ weights
were revealed among groups (Fig.
7E). H&E staining results revealed that no significant
toxicities were observed in the vital organs of mice (Fig. 7F). Western blot analysis
demonstrated that LCA treatment decreased the expression of FBXO5,
p-ERK1/2 and p-p38MAPK (Fig. 7G and
H) in vivo, which was consistent with the suppression
effect of LCA on the MAPK pathway and the expression of FBXO5 in
vitro. The aforementioned data indicated that LCA exhibited
obvious antitumor activity with low toxicity on LSCC in
vivo.
Discussion
Despite advancements in diagnosis and therapeutic
techniques, the survival rates of lung cancer remain low in the
world. LSCC is a highly heterogeneous malignancy with high tumor
mutational burden. The molecular profile of LSCC was significantly
different from that of adenocarcinoma (30,31).
PD1/PD-L1 immune checkpoint blockade and targeted therapies
improved the outcome of patients with lung adenocarcinoma, while
these therapies were not available for patients with LSCC.
Therefore, it is essential to find novel drugs or therapeutics
which are effective and have low side effects. Previous studies
demonstrated that Chinese traditional herbal medicine licorice
displays anti-inflammatory, antitussive and antitumor activities
(32–34). LCA is extracted from licorice and
possess antitumor properties in several cancers (35–37).
In the present study, LCA effectively inhibited the
proliferation of H226 and H1703 cells, while exhibiting no
significant cytotoxicity against HBE cells even at a high
concentration. The antitumor effect of LCA was also investigated
via subcutaneous xenograft models and it was revealed that LCA
suppressed xenograft tumor growth in mice. These results indicated
that LCA effectively inhibited cell proliferation in vitro
and xenograft tumor growth in vivo. Moreover, H&E
staining of vital organs indicated that the application of LCA was
safe within a certain concentration range in vivo.
Cell cycle regulation is closely related to
proliferation of tumors. Cell cycle is strictly regulated by
cyclins and their associated specific CDKs (38), such as cyclin D1, CDK4, cyclin E and
CDK2, which are of importance for the transition from G1 to S phase
(39). In the present study, it was
revealed that LCA increased the cell number in G1 phase in LSCC
cells and decreased the protein levels of cyclin D1, cyclin E, CDK2
and CDK4. These findings were consistent with the previous studies
that LCA induced G1 phase arrest in MCF-7 (14) and HCT-116 (40) cells. The aforementioned data
suggested that LCA induced G1 phase arrest and downregulated the
expression level of cell cycle-related protein.
Apoptosis is a programmed cellular process that
occurs under either physiological or pathological conditions
(41). Apoptosis exerts a vital
role in the treatment of cancer, and induction of apoptosis is the
eventual aim of numerous cancer therapies (42). The findings of the present study
displayed that LCA induced apoptosis in LSCC cells and upregulated
the level of cleaved caspase-3, cleaved PARP1 and Bax, meanwhile,
LCA downregulated the level of Bcl-2. Therefore, LCA could induce
cell apoptosis by the mitochondrial-mediated intrinsic pathway in
LSCC cells. In the present study, the observed inhibition of cell
viability and proliferation was not only dependent on increased
cell apoptosis resulting from LCA treatment, but also in the cell
cycle arrest. Apoptotic rates reflect part of proliferation
inhibition.
Proteomics is widely used in drug development and
cancer research (43). Proteomics
combination with bioinformatics can analyze the mechanisms and
biochemical processes of numerous complex diseases, such as
diabetes and cancers (44). KEGG
analyses of proteomic results revealed that the DEPs regulated by
LCA were associated with the MAPK signaling pathway. And the
network pharmacological analysis indicated that the MAPK1 (ERK2)
and MAPK14 (p38 MAPK) were the core targets of LCA against LSCC.
MAPK signaling pathways are involved in several biological
processes and regulate cell proliferation, apoptosis and immune
escape of several cancers (45).
The ERK1/2 and p38 MAPK signaling pathways have been found to be
activated and overexpressed in lung cancer (46,47).
In the present study, LCA significantly suppressed ERK1/2
phosphorylation and p38 MAPK phosphorylation in vivo and
in vitro.
Moreover, 4D-DIA proteomics also identified that
FBXO5 protein expression was decreased after LCA treatment in LSCC
cells. FBXO5 is vital in regulation of cycle proliferation and
tumorigenesis. FBXO5 promotes cell proliferation and inhibits DNA
re-replication via accumulation of cyclin A2 and geminin (48). FBXO5 was upregulated in various
malignant tumors compared with matched normal tissues (25,49). A
previous study has demonstrated that targeting FBXO5 could
eliminate cutaneous squamous cell carcinoma cells with no effect on
normal skin cells (50). Existing
evidence has suggested that FBXO5 was upregulated in LSCC compared
with normal tissue and associated with shorter OS in patients with
LSCC (27). And FBXO5 may function
as an oncogene and may be a novel therapeutic target in various
malignancies (51). In the present
study, compared with the adjacent normal tissue, FBXO5 was highly
expressed in LSCC tissues based on the TCGA data, and FBXO5 level
was significantly elevated in each pathological stage. The
aforementioned results suggested that FBXO5 played an important
role in LSCC and may be a therapeutic target for LSCC treatment.
Western blot analysis revealed that LCA significantly decreased the
FBXO5 protein expression. Furthermore, in order to ascertain the
effect of FBXO5, MTT experiments were used to detect the cell
viability in FBXO5-silenced H1703 cells. FBXO5 knockdown inhibited
the cell viability and decreased the expression levels of CDK2,
CDK4, cyclin D1 and cyclin E in H1703 cells. These findings
indicated that FBXO5 protein knockdown inhibit the proliferation of
H1703 cells.
There are certain limitations to the present study.
In order to ascertain whether FBXO5 was the therapeutic target for
antitumor effects of LCA, rescue experiments should be performed in
the FBXO5-silenced H1703 cells in the future.
In summary, LCA inhibited the proliferation of LSCC
cells, induced cell cycle arrest and apoptosis in vitro and
significantly inhibited the tumorigenesis with few adverse effects
in vivo. In addition, LCA inhibited ERK1/2 phosphorylation
and p38 MAPK phosphorylation, and suppressed the expression of
FBXO5 in vivo and in vitro. In conclusion, LCA may be
a potential therapeutic candidate of LSCC.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant no. 82160615).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XF, LW, JW and XD designed the experiments. XF and
JW performed the experiments and data collection. XF wrote the
original draft and revised the manuscript. GG and MJ analyzed the
data. LW and JW critically reviewed and edited the manuscript. All
authors read and approved the final version of the manuscript. XF
and XD confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Animal Care and Use Committee of Guilin Medical University
(approval no. GLMC-IACUC-2021016; Guilin, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
LSCC
|
lung squamous cell carcinoma
|
|
LCA
|
licochalcone A
|
|
FCM
|
flow cytometry
|
|
4D-DIA proteomics
|
4D-data-independent acquisition
proteomics
|
|
MAPK
|
mitogen-activated protein kinase
|
|
ERK1/2
|
extracellular regulated protein
kinases 1 and 2
|
|
CDK2
|
cyclin dependent kinase 2
|
|
CDK4
|
cyclin dependent kinase 4
|
|
PARP1
|
poly(ADP-ribose)polymerase-1
|
|
FBXO5
|
F-box protein 5
|
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cheng TY, Cramb SM, Baade PD, Youlden DR,
Nwogu C and Reid ME: The international epidemiology of lung cancer:
Latest trends, disparities, and tumor characteristics. J Thorac
Oncol. 11:1653–1671. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Herbst RS, Morgensztern D and Boshoff C:
The biology and management of non-small cell lung cancer. Nature.
553:446–454. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Derman BA, Mileham KF, Bonomi PD, Batus M
and Fidler MJ: Treatment of advanced squamous cell carcinoma of the
lung: A review. Transl Lung Cancer Res. 4:524–532. 2015.PubMed/NCBI
|
|
6
|
Rittmeyer A, Barlesi F, Waterkamp D, Park
K, Ciardiello F, von Pawel J, Gadgeel SM, Hida T, Kowalski DM, Dols
MC, et al: Atezolizumab versus docetaxel in patients with
previously treated non-small-cell lung cancer (OAK): A phase 3,
open-label, multicentre randomised controlled trial. Lancet.
389:255–265. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gandara DR. Hammerman PS, Sos ML, Lara PN
Jr and Hirsch FR: Squamous cell lung cancer: From tumor genomics to
cancer therapeutics. Clin Cancer Res. 21:2236–2243. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kennedy GT, Azari FS, Bernstein E, Nadeem
B, Chang AE, Segil A, Sullivan N, Marfatia I, Din A, Desphande C,
et al: A prostate-specific membrane antigen-targeted near-infrared
conjugate for identifying pulmonary squamous cell carcinoma during
resection. Mol Cancer Ther. 21:546–554. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ,
Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, et al:
Safety and activity of anti-PD-L1 antibody in patients with
advanced cancer. N Engl J Med. 366:2455–2465. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Topalian SL, Hodi FS, Brahmer JR,
Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD,
Sosman JA, Atkins MB, et al: Safety, activity, and immune
correlates of anti-PD-1 antibody in cancer. N Engl J Med.
366:2443–2454. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ferris RL, Blumenschein G Jr, Fayette J,
Guigay J, Colevas AD, Licitra L, Harrington K, Kasper S, Vokes EE,
Even C, et al: Nivolumab for recurrent squamous-cell carcinoma of
the head and neck. N Engl J Med. 375:1856–1867. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xia L, Liu Y and Wang Y: PD-1/PD-L1
blockade therapy in advanced non-small-cell lung cancer: Current
status and future directions. Oncologist. 24 (Suppl 1):S31–S41.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang ZF, Liu J, Yang YA and Zhu HL: A
review: The anti-inflammatory, anticancer and antibacterial
properties of four kinds of licorice flavonoids isolated from
licorice. Curr Med Chem. 27:1997–2011. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bortolotto LF, Barbosa FR, Silva G,
Bitencourt TA, Beleboni RO, Baek SJ, Marins M and Fachin AL:
Cytotoxicity of trans-chalcone and licochalcone A against breast
cancer cells is due to apoptosis induction and cell cycle arrest.
Biomed Pharmacother. 85:425–433. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lv H, Ren H, Wang L, Chen W and Ci X: Lico
A enhances Nrf2-mediated defense mechanisms against t-BHP-induced
oxidative stress and cell death via Akt and ERK activation in RAW
264.7 cells. Oxid Med Cell Longev. 2015:7098452015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
de Freitas KS, Squarisi IS, Acesio NO,
Nicolella HD, Ozelin SD, Reis Santos de Melo M, Guissone APP,
Fernandes G, Silva LM, da Silva Filho AA and Tavares DC:
Licochalcone A, a licorice flavonoid: Antioxidant, cytotoxic,
genotoxic, and chemopreventive potential. J Toxicol Environ Health
A. 83:673–686. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hao W, Yuan X, Yu L, Gao C, Sun X, Wang D
and Zheng Q: Licochalcone A-induced human gastric cancer BGC-823
cells apoptosis by regulating ROS-mediated MAPKs and PI3K/AKT
signaling pathways. Sci Rep. 5:103362015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li MT, Xie L, Jiang HM, Huang Q, Tong RS,
Li X, Xie X and Liu HM: Role of licochalcone A in potential
pharmacological therapy: A review. Front Pharmacol. 13:8787762022.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lin M, Xu Y, Gao Y, Pan C, Zhu X and Wang
ZW: Regulation of F-box proteins by noncoding RNAs in human
cancers. Cancer Lett. 466:61–70. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Song Y, Lin M, Liu Y, Wang ZW and Zhu X:
Emerging role of F-box proteins in the regulation of
epithelial-mesenchymal transition and stem cells in human cancers.
Stem Cell Res Ther. 10:1242019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Reimann JD, Gardner BE, Margottin-Goguet F
and Jackson PK: Emi1 regulates the anaphase-promoting complex by a
different mechanism than Mad2 proteins. Genes Dev. 15:3278–3285.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Miller JJ, Summers MK, Hansen DV, Nachury
MV, Lehman NL, Loktev A and Jackson PK: Emi1 stably binds and
inhibits the anaphase-promoting complex/cyclosome as a
pseudosubstrate inhibitor. Genes Dev. 20:2410–2420. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Di Fiore B and Pines J: Defining the role
of Emi1 in the DNA replication-segregation cycle. Chromosoma.
117:333–338. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vaidyanathan S, Cato K, Tang L, Pavey S,
Haass NK, Gabrielli BG and Duijf PHG: In vivo overexpression of
Emi1 promotes chromosome instability and tumorigenesis. Oncogene.
35:5446–5455. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu X, Wang H, Ma J, Xu J, Sheng C, Yang
S, Sun L and Ni Q: The expression and prognosis of Emi1 and Skp2 in
breast carcinoma: Associated with PI3K/Akt pathway and cell
proliferation. Med Oncol. 30:7352013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guan C, Zhang J, Zhang J, Shi H and Ni R:
Enhanced expression of early mitotic inhibitor-1 predicts a poor
prognosis in esophageal squamous cell carcinoma patients. Oncol
Lett. 12:114–120. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang K, Qu X, Liu S, Yang X, Bie F, Wang
Y, Huang C and Du J: Identification of aberrantly expressed F-box
proteins in squamous-cell lung carcinoma. J Cancer Res Clin Oncol.
144:1509–1521. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pinzi L and Rastelli G: Molecular docking:
Shifting paradigms in drug discovery. Int J Mol Sci. 20:43312019.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li F, Wang L, Cai Y, Luo Y and Shi X:
Safety assessment of desaminotyrosine: Acute, subchronic oral
toxicity, and its effects on intestinal microbiota in rats. Toxicol
Appl Pharmacol. 417:1154642021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Friedlaender A, Banna G, Malapelle U,
Pisapia P and Addeo A: Next generation sequencing and genetic
alterations in squamous cell lung carcinoma: Where are we today?
Front Oncol. 9:1662019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cancer Genome Atlas Research Network, .
Comprehensive molecular profiling of lung adenocarcinoma. Nature.
511:543–550. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hosseinzadeh H and Nassiri-Asl M:
Pharmacological effects of Glycyrrhiza spp. and its bioactive
constituents: Update and review. Phytother Res. 29:1868–1886. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li K, Ji S, Song W, Kuang Y, Lin Y, Tang
S, Cui Z, Qiao X, Yu S and Ye M: Glycybridins A-K, bioactive
phenolic compounds from Glycyrrhiza glabra. J Nat Prod. 80:334–346.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Song W, Si L, Ji S, Wang H, Fang XM, Yu
LY, Li RY, Liang LN, Zhou D and Ye M: Uralsaponins M-Y, antiviral
triterpenoid saponins from the roots of Glycyrrhiza uralensis. J
Nat Prod. 77:1632–1643. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gao F, Li M, Yu X, Liu W, Zhou L and Li W:
Licochalcone A inhibits EGFR signalling and translationally
suppresses survivin expression in human cancer cells. J Cell Mol
Med. 25:813–826. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu KD, Miao Y, Li P, Li PP, Liu J, Li J
and Cao F: Licochalcone A inhibits cell growth through the
downregulation of the Hippo pathway via PES1 in cholangiocarcinoma
cells. Environ Toxicol. 37:564–573. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hu C, Zuo Y, Liu J, Xu H, Liao W, Dang Y,
Luo C, Tang L and Zhang H: Licochalcone A suppresses the
proliferation of sarcoma HT-1080 cells, as a selective R132C mutant
IDH1 inhibitor. Bioorg Med Chem Lett. 30:1268252020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mori M, Bogdan A, Balassa T, Csabai T and
Szekeres-Bartho J: The decidua-the maternal bed embracing the
embryo-maintains the pregnancy. Semin Immunopathol. 38:635–649.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chang Z, Kuang HX, Zhou X, Zhu H, Zhang Y,
Fu Y, Fu Q, Jiang B, Wang W, Jiang S, et al: Temporal changes in
cyclinD-CDK4/CDK6 and cyclinE-CDK2 pathways: Implications for the
mechanism of deficient decidualization in an immune-based mouse
model of unexplained recurrent spontaneous abortion. Mol Med.
28:1002022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu P, Yu T, Wu J and Chen J: Licochalcone
a induces ROS-mediated apoptosis through TrxR1 inactivation in
colorectal cancer cells. Biomed Res Int.
2020:58750742020.PubMed/NCBI
|
|
41
|
Morana O, Wood W and Gregory CD: The
apoptosis paradox in cancer. Int J Mol Sci. 23:13282022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Khodavirdipour A, Piri M, Jabbari S,
Keshavarzi S, Safaralizadeh R and Alikhani MY: Apoptosis detection
methods in diagnosis of cancer and their potential role in
treatment: Advantages and disadvantages: A review. J Gastrointest
Cancer. 52:422–430. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ji Q, Zhu F, Liu X, Li Q and Su SB: Recent
advance in applications of proteomics technologies on traditional
Chinese medicine research. Evid Based Complement Alternat Med.
2015:9831392015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Monti C, Zilocchi M, Colugnat I and
Alberio T: Proteomics turns functional. J Proteomics. 198:36–44.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yong HY, Koh MS and Moon A: The p38 MAPK
inhibitors for the treatment of inflammatory diseases and cancer.
Expert Opin Investig Drugs. 18:1893–1905. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Greenberg AK, Basu S, Hu J, Yie TA,
Tchou-Wong KM, Rom WN and Lee TC: Selective p38 activation in human
non-small cell lung cancer. Am J Respir Cell Mol Biol. 26:558–564.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Sugiura R, Satoh R and Takasaki T: ERK: A
double-edged sword in cancer. ERK-dependent apoptosis as a
potential therapeutic strategy for cancer. Cells. 10:25092021.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Marzio A, Puccini J, Kwon Y, Maverakis NK,
Arbini A, Sung P, Bar-Sagi D and Pagano M: The F-box
domain-dependent activity of EMI1 regulates PARPi sensitivity in
triple-negative breast cancers. Mol Cell. 73:224–237.e6. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhao Y, Tang Q, Ni R, Huang X, Wang Y, Lu
C, Shen A, Wang Y, Li C, Yuan Q, et al: Early mitotic inhibitor-1,
an anaphase-promoting complex/cyclosome inhibitor, can control
tumor cell proliferation in hepatocellular carcinoma: Correlation
with Skp2 stability and degradation of p27(Kip1). Hum Pathol.
44:365–373. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
McHugh A, Fernandes K, Chinner N, Ibrahim
AFM, Garg AK, Boag G, Hepburn LA, Proby CM, Leigh IM and Saville
MK: The identification of potential therapeutic targets for
cutaneous squamous cell carcinoma. J Invest Dermatol.
140:1154–1165.e5. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liu P, Wang X, Pan L, Han B and He Z:
Prognostic significance and immunological role of FBXO5 in human
cancers: A systematic pan-cancer analysis. Front Immunol.
13:9017842022. View Article : Google Scholar : PubMed/NCBI
|