The hedgehog (HH) signaling pathway was first
discovered by Nusslein-Volhard and Wieschaus in the 1980s when
screening for genes that affect Drosophila embryonic
development. In the following decades, the HH pathway was confirmed
to be a highly conserved signaling mechanism, widely related to
human physiology and pathology (1).
Through highly regulated activation, HH coordinates the development
of multiple central systems and limb formation in embryos. In
adults, HH is mainly involved in stem cell renewal, wound healing,
organ homeostasis, tissue repair and tumorigenesis (2,3).
The role of HH is mainly dependent on the signal
transduction of the following molecules: Ligand family member
protein, HH ligands; two receptors, patched (PTCH) and smoothened
(SMO); the nuclear factors, kinesin family member 7, casein kinase
1, suppressor of fused protein (SUFU) and glioma-associated
oncogene (GLI); and related target genes (4).
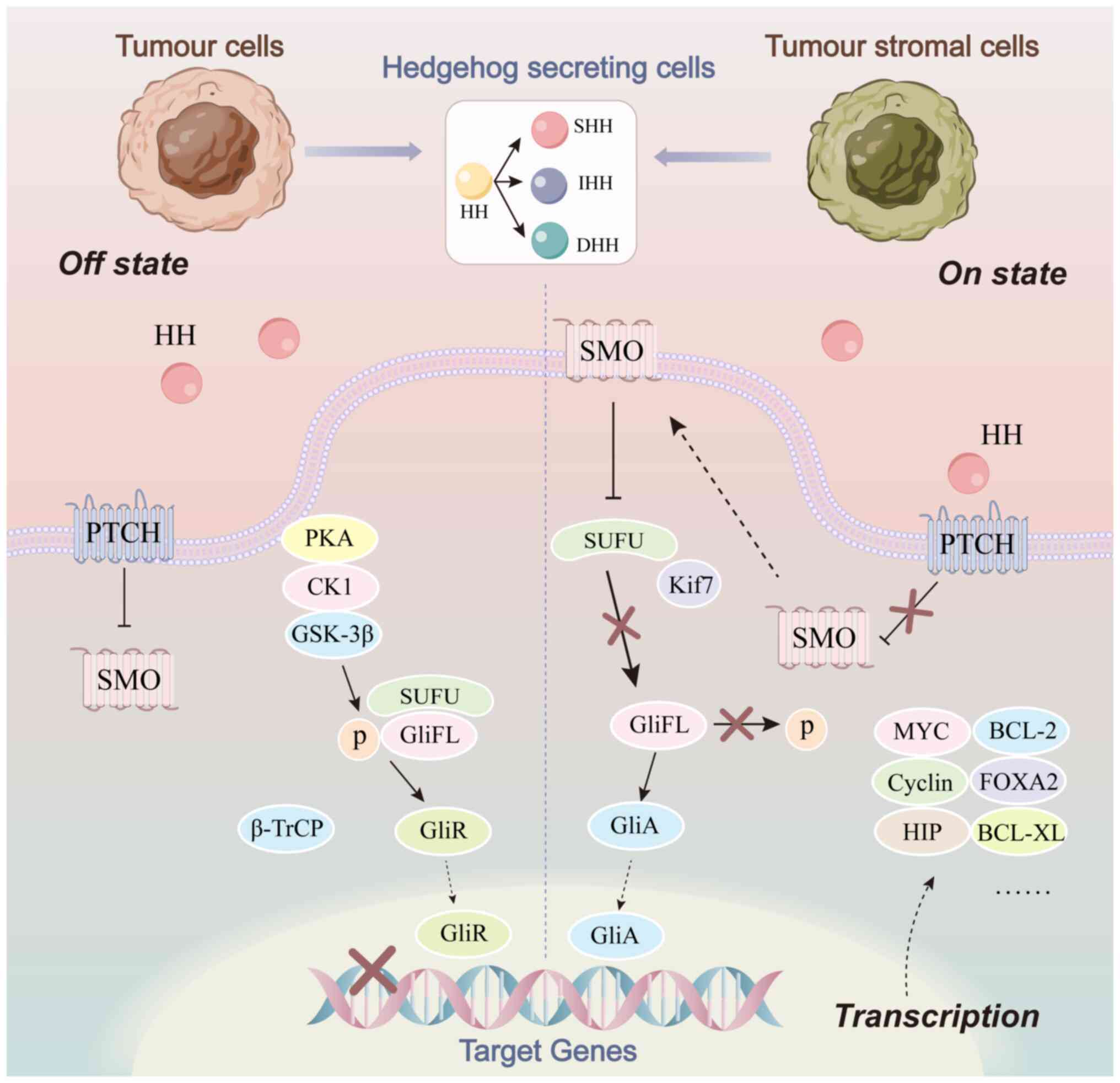
As with several classical signaling pathways, the
classical HH pathway is initiated by the binding of ligands to
receptors. HH ligands is currently known to have three subtypes in
mammals, including sonic HH (SHH), Indian HH (IHH) and desert HH
(DHH), of which SHH is the most widely expressed and most active.
When ligand is not present (off-state), the coupling receptor, SMO,
on intracellular vesicles is inhibited by PTCH, which is located on
primary cilia. In the absence of accumulation, SMO cannot transmit
HH signal to downstream SUFU and therefore GLI activator (GLI-A) is
not released. Instead, a GLI transcriptional repressor is formed to
inhibit the expression of the target genes (5). When ligand is available (on-state),
ligand binds to and internalizes PTCH, relieving the inhibition of
SMO. As a result, full-length GLI receives the relevant signals to
form GLI-A and promotes the transcription of HH target genes
(4,5). Activation of the classical HH pathway
can also be divided into autocrine, paracrine and reverse secretory
modes, according to the source of HH ligand. The interaction
between tumor cells and tumor stromal cells is realized by such
pathways (Fig. 1).
The specific mechanism of action of the
non-classical HH pathway has not yet been fully understood.
However, it appears that when the classical pathway cannot function
in a cytotoxic or stressful state, the non-classical pathway is an
alternative activation pathway for transmitting HH signals
(6). In the PTCH-SMO-GLI axis, the
key proteins of HH signal transmission can alter their original
conformation by coupling with other molecules due to their unique
structure, thus regulating target genes without being constrained
by upstream/downstream signals, such as apoptosis factors gathering
in the C-terminal tail of PTCH or formation of the SMO-Gi (one
family of G proteins) protein complex (7). GLI-related non-classical pathways are
becoming increasingly prominent in cancer development due to their
involvement in a number of signaling pathways known to be related
to cancer (8).
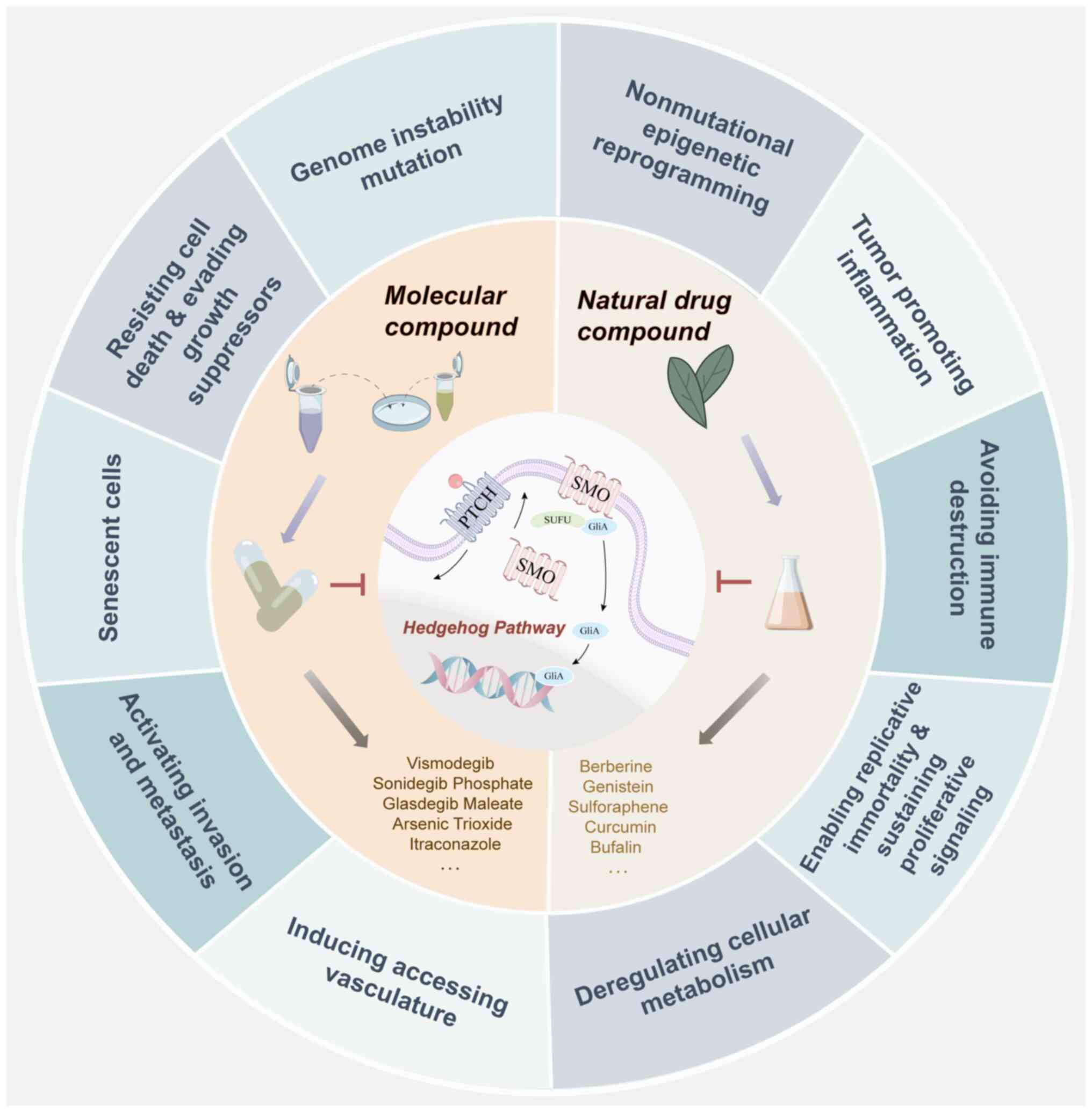
Following the research progress on tumorigenesis and
development, oncologists have summarized the characteristic
parameters of tumors into 14 types (9), which has provided a logical framework
for understanding the notable diversity of tumors. There is
sufficient evidence to support that abnormal expression of the HH
pathway manipulates tumor cell growth, proliferation, survival,
angiogenesis and metastasis and the metabolic reprogramming of
tumor cells and the microenvironment (10,11).
Based on the relationship between the HH pathway and
cancer phenotype, the present review mainly discusses the molecular
mechanisms identified in the latest discoveries of the HH pathway
in tumorigenesis and development, as well as the related factors
regulating these phenotypic changes. Several molecular synthetic
and natural drugs that have been revealed to inhibit abnormal
activation of HH are also summarized, for exploring the possibility
of treating related cancer types (Fig.
2).
According to the current understanding, cancer tends
to be defined as a group of malignant diseases with multiple
genetic origins. Therefore, gene mutations cannot be ignored in the
occurrence of cancer. In normal tissues and organs, the HH pathway
is a crucial pathway for development. For instance, the HH pathway
controls the migration of granular neuronal precursors to the
correct location in the brain (12). Once mutation of HH occurs, the
opportunity for tumorigenesis occurs. According to the 2021 World
Health Organization guidelines, medulloblastomas can be divided
into four molecular subgroups, of which the evaluation is partially
based on HH genotyping (13).
Gorlin syndrome (GS), a rare autosomal dominant
disease, is caused by mutations in the HH pathway. The typical
mutations observed in GS cause pathogenic variations of PTCH1 and
SUFU. In the differential diagnosis of GS, it has been suggested
that PTCH2 may be a candidate gene for the prediction of
susceptibility, and mutation of PTCH2 has been reported in clinical
practice (14). However, a recent
study asserted that PTCH2 should not be included in the genetic
diagnosis of GS (15). In fact, the
PTCH2 gene has different functional characteristics to PTCH1. PTCH2
is considered to coordinate PTCH1 to alter SMO localization and
plays an auxiliary role in regulating the HH pathway (12). This may explain the absence of a
statistically significant detection of PTCH2 mutations in the
clinic.
A recent study indicated that 68% of SUFU pathogenic
variation carriers had at least one type of tumor, and the
incidence of tumors in their relatives reached 44.1% in individuals
up to 50 years-old (16).
SMO mutations have also attracted wide attention due
to resistance mutations that prevent effective drug binding
(discussed later). In addition, carcinogenic mutation of SMO is
widespread. In this instance, the mutation stabilizes the active
form of SMO and releases it from the conformational restriction of
the inactive state, and thus may induce clonal expansion together
with tumor initiation and invasion (17). SMO multi-site mutations have been
revealed to be a carcinogenic driver for a variety of
epithelial-derived tumors and brain tumors (18).
GLI mutations have also been reported in the clinic.
In addition, a study has suggested that upregulation of a GLI2-β
subtype lacking the N-terminal repressor domain induced chromosome
number and structure aberrations, which disrupt genomic stability
(19).
In addition to single-gene mutations, multi-gene
fusion has also received attention in research. Common gene fusions
occur between GLI1 and other genes (Table SII). At present, the MALAT1-GLI1
gene fusion has been identified as one of the diagnostic criteria
for gastroblastoma (20).
Regardless of the concrete types of multi-gene fusion, inhibition
of HH can be used as a general treatment method. Furthermore, the
upregulation of GLI1 has been defined as an alternative genetic
mechanism for GLI1 fusions, with the characteristics of
co-amplification of the cyclin-dependent kinase (CDK) 4 and MDM2
genes (21,22).
In addition to altering the DNA sequence, DNA can
also be chemically modified by methylation and hydroxy-methylation
to regulate gene expression (23,24).
By querying the MethMarkerDB database (https://methmarkerdb.hzau.edu.cn/) (25), the overall degree of methylation of
HH pathway-related genes was determined and the potential of
differentially methylated regions for the diagnosis (Table SIII) and prognosis (Table SIV) of some cancer types was
underscored. Among them, the low methylation level and high
expression of GLI1 in melanoma, GLI2 in ocular melanomas and GLI3
in stomach cancer suggested an improved patient prognosis. PTCH1 in
kidney clear cell carcinoma and GLI3 in colon and prostate cancer
were also considered to be robust diagnostic biomarkers. In
addition, clinical data supports the suggestion that promoter
methylation is a critical regulatory mechanism of SHH (26), PTCH, GLI (27) and SMO (28) expression. Accordingly, folic acid
(29) and DNA methyltransferase
inhibitors (27) can effectively
block the HH pathway for the treatment of cancer.
Notable heterogeneity in methylation and gene
expression levels were also observed in the MethMarkerDB data
(Tables SIII and SIV). This heterogeneity may be ascribed
to the limited sample size. However, abnormal epigenetic
characteristics of cancer cells can also be induced by
phosphorylation, acetylation and other acylation modifications of
histones in cellular chromatin (23,24).
The phosphorylation of GLI1 can be disrupted by mutations in
AMP-activated protein kinase (AMPK), which increases carcinogenic
potency. As acetylated proteins, GLI1 and GLI2 rely on histone
deacetylase (HDAC)-mediated deacetylation to promote
transcriptional activation. HDAC-related changes also directly lead
to changes in GLI. Therefore, HH and HDAC inhibitors may exert a
synergistic antitumor effect (30).
Additional modifications include: Sufu negating protein 1
ubiquitinates SUFU via ligand of numb-protein X 1 (31), runt-related transcription factor 3
promotes GLI ubiquitination (10)
through the E3 ubiquitin ligase family (32) and protein phosphatase 4 regulatory
subunit 2 promotes SUFU dephosphorylation (33).
Recent developments in the research of ncRNA have
attracted marked attention, particularly microRNA (miRNA), which
plays a regulatory role by directly cutting or preventing the
translation of mRNA. In addition, circular RNA, rich with miRNA
binding sites, can sponge miRNA to relieve the inhibition of mRNA.
The binding of long ncRNAs to RNA-binding proteins confers a
variety of regulatory functions, including managing genomic
instability. The specific regulation of key molecules of the HH
pathway by ncRNAs is summarized in Table II (11,34–127).
As summarized in Table SV, the HH
pathway can also affect the expression of other pathway molecules
by regulating ncRNA to regulate tumor phenotypes.
The HH pathway mechanism in tumor-promoting
inflammation can be exemplified by Hp infection, which has been
proven to be associated with gastric cancer. Once infected, Hp
injects cytotoxic associated gene A into gastric cells, resulting
in the accumulation of bone marrow cells in the stomach following
the secretion of SHH ligands by parietal cells. Then, these cells
differentiate and mature under the induction of factors such as
IFN-α, express GLI1-dependent schlafen 4 and secrete interleukin
(IL)-1β to activate the IL-6/phosphorylated signal transducer and
activator of transcription (STAT)-3 pathway. The activation of SHH
during Hp infection was closely related to the expression of
programmed death-ligand 1 (PD-L1) (128), as well as the emergence of
myeloid-derived suppressor cells (MDSCs), intestinal metaplasia and
soluble polypeptide expression metaplasia (129).
The relationship between hepatocellular carcinoma
and the HH pathway activated by chronic liver injury has also been
clarified. Specifically, hepatitis B virus X protein has been
revealed to directly interact with GLI1 and promote disease
progression when GLI2 is not inhibited by Sestrin 3 (130).
In pan-cancer, activation of the transcriptional
programs of SHH in the non-T cell-inflamed tumor microenvironment
was discovered in a study by Bao et al (131), after performing an unbiased
genome-wide pathway discovery. Furthermore, non-classical HH
activation can promote tumor development by generating
pro-inflammatory cytokines, such as tumor necrosis factor (TNF)-α,
IL-1β (132) and TGF-β (133), in hypoxic or inflammatory
environments. In addition, hypoxia also contributes to the
enrichment of immunosuppressive cells (134), as well as the excessive activation
of the Notch/HH axis (135).
Notably, STAT is one of the main factors of pro-inflammatory signal
transduction. The interaction between STAT and HH is an important
component of inflammation and tumor promotion mechanisms (136). In basal cell carcinoma, IL-6 has
been revealed to cooperate with carcinogenic HH/GLI signaling via
the IL-6R/Janus kinase 2/STAT3 pathway (137). In addition, the antitumor effect
of IFN-γ/STAT1 can be antagonized by the activation of suppressor
of cytokine signaling 1 by GLI1 and GLI2 (138).
By contrast, the anti-inflammatory effect of HH
pathway activation is also a protective mechanism, particularly in
acute inflammation. Increased expression of the anti-inflammatory
cytokine, IL-10, in HH pathway-responsive stromal cells and
concomitant increases in CD4 forkhead box (Fox)p3 regulatory T
cells (Tregs) reduces the tumor burden of colitis-associated colon
cancer (139). Similarly, the
pancreatic gland is also protected from acute pancreatitis by the
SHH/GLI1/IL-10 axis (140). When
considering that the HH pathway is also involved in the
interference of cell differentiation and maturation during tissue
repair following inflammation (141), whether to administer pathway
inhibitors in the early stages of inflammation and the risk of
tumor development remain to be considered.
However, while activation of the HH pathway induces
anti-inflammatory effects, another real problem emerges: Tumor
immune escape (142). In fact,
there is a complex crosstalk between immune cells, cancer cells and
inflammation.
In most tumors, fibroblasts, endothelial cells and
macrophages exhibit strong positive connections with GLI (143). In previous studies, TAMs derived
from MDSCs were revealed to secrete a variety of anti-inflammatory
cytokines and express immune checkpoint ligands that inhibit
effector T cells and recruit Tregs (144,145). Research thus far has mainly
focused on the association of TAM aggregation with abnormal
expression of SHH and activation of HH/GLI (146–148). With the participation of
krüppel-like factor 4 SHH-derived TAM M2 polarization (149), and GLI1 can directly affect the M2
activation state by activating the feedforward loop of STAT6-IL-4ra
(150). In addition, the STAT3
pathway has also been revealed to be involved in the regulation of
PD-L1 expression in TAMs by tumor-derived SHH ligand (151,152). The carcinoma cell-TAM-carcinoma
cell loop may produce a hierarchical amplification effect via the
SHH/GLI2-TGF-β1 loop, promoting tumor growth (153). By targeting tumor-supportive
M2-like TAMs, a synergistic effect of peroxisome proliferator
activated receptor γ with SMO has also been proven (146).
In an infectious state, GLI1 directly increases the
transcription of cyclooxygenase-2/prostaglandin E2 and regulates
miR-324-5p and miR-338-5p. These miRNAs target PD-L1 to increase
its expression, finally realizing the SHH/phosphoinositide-3 kinase
(PI3K)/mTOR/NF-κB signal transduction of dendritic cells and
activating Treg amplification (154). In gastric cancer, this process was
discovered to be mediated by the mTOR signaling pathway, involving
the SMO-independent HH pathway (145). In addition, blocking HH
reprogramed Treg trans-differentiation into inflammatory Th17
cells, which enhanced the recruitment of cytotoxic CD8+
T cells into tumors (155).
HH activity detection can be used to predict the
efficacy of immune checkpoint inhibitors, with the support of
clinical data (156,157). The expression of programmed death
protein-1 (PD-1)/PD-L1 induced by various pro-inflammatory factors
changes under the crosstalk of HH and other pathways (147,158,159). The high prevalence of Tregs within
the tumor microenvironment and induction of PD-1/PD-L1 is likely to
constitute a major mechanism of immunosuppression by HH/GLI
signaling in cancer (147). These
findings increase the therapeutic opportunities for combination
treatments with HH and immune checkpoint inhibitors.
In the unlimited proliferation of tumors, telomerase
determines the replication potential of cancer cells. Research has
revealed that hTERT is a direct target of HH/GLI, and the effect on
hTERT activity is related to differentiating benign and malignant
tumors (160). Reduced cell
proliferation and GLI1 levels have also been observed in cancer
cells with long-term use of telomerase inhibitors (161). However, it cannot be concluded
that TERT forms a feedback loop with HH, since the current
suggestion is that TERT upstream of HH is more likely to activate
the HH pathway by regulating miRNA (162) and recruiting pro-oncogenic
transcription factors (163).
These effects contribute to increasing invasion and are independent
of changes in telomerase activity.
Regulation of the tumor cell cycle is mainly
dependent on two important pathways: The RB transcriptional
corepressor 1 and p53 pathways. Furthermore, mutations in these two
pathways lead to the formation of primary cilia, abnormal elevation
of Hedgehog ligands (164,165) and direct regulation of downstream
cell cycle regulators, such as cyclin (166) and CDKs (167). It mediates HH-induced DNA
replication. This was also partially dependent on the PI3K/AKT
pathway (167). The HH pathway has
also been revealed to block its inhibition of cyclin regulation by
affecting stem cell-related factors, such as BMI-1, to inhibit the
downstream p14 and p16 proteins (168). Another well-known cell cycle
regulator is FoxM1. GLI1 has been demonstrated to bind to FoxM1 and
initiate the effect of Xenopus kinesin-like protein 2 on cell
division (169). The crosstalk
between the Notch pathway as a proliferation-related pathway and
the HH pathway also decreases G1/G0 cycle retardation (170).
The HH pathway has been implicated in regulating
apoptosis. TRAIL has become a new target for cancer treatment due
to reduced toxicity to normal tissues and specificity for tumor
cell apoptosis. The positive crosstalk between NF-κB and the HH
pathway amplifies the effect of resistance to TRAIL-related
apoptosis (171,172). However, blockade of the GLI family
increases TRAIL sensitivity (173).
A considerable number of studies have revealed that
the Bcl-2 family are target genes of the HH pathway, including
BCL-2 (174), myeloid cell
leukemia-1 (174,175), BCL-XL (174) and NOXA (176), which play an anti-apoptotic role
by blocking the activity of the caspase family, leading to TRAIL
resistance. Bcl-2 proteins also form a feedforward signal with SUFU
to further induce the expression of target genes in the HH pathway
(174). Accordingly, inhibition of
the HH pathway provides a new avenue for the treatment of
TRAIL-resistant tumors.
Among the apoptotic pathways, the MYC family
(oncogenes related to apoptosis) also closely interact with HH.
Upregulation of the family member protein, MYCN, in basal cell
carcinomas is a key factor that re-activates dormant HH signals and
induces tumor progression (177).
Similarly, MYCN has been identified as an invasive marker in
neuroblastoma, which was associated with HH signaling, for
determining prognosis. Although the positive or negative
correlation between GLI1 expression and prognosis in neuroblastoma
remains controversial, most evidence suggests that high GLI1
expression indicates an improved prognosis in MYCN-amplified
neuroblastoma (178). An
explanation for this is that protein kinase-like endoplasmic
reticulum kinase-EIF2α pathway is an important mediator in
Hh-dependent autophagy on MYCN-amplified neuroblastoma (179).
In addition to TRAIL and the MYC family, the
anti-apoptotic effects of other apoptosis-related factors also
depended on HH. For instance, TNF-α induces the expression of
activator protein 1 family members and regulates apoptosis through
the HH pathway (180,181). Survivin, another inhibitor of
apoptosis family, has been revealed to be a transcriptional target
of GLI (182). HH/GLI1 signaling
mediates the RNA polymerase III signaling pathway and tRNA
synthesis to regulate the cell cycle and death receptor binding
(183).
It is worth noting that the effect of HH-induced
autophagy on apoptosis does not act alone in various cancer types.
On the one hand, HH-induced autophagy leads to cell death and
affects a variety of cancer types by regulating targets such as
BCL-2 interacting protein 3 and LC3 II (184,185). Correspondingly, certain drugs have
been revealed to induce autophagy and cell death by blocking HH
signaling (186,187). On the other hand, HH-induced
autophagy can also promote tumors by providing energy for tumor
development. SHH inhibition in thyroid cancer activates TAK1,
phosphorylates JNK/AMPK and induces autophagy (181). Therefore, acknowledging the effect
of autophagy on apoptosis caused by inhibition of HH can guide the
usage of drug combinations more reasonably.
In addition to systematic apoptosis pathways,
another phenomenon, dependence receptors triggering apoptosis
signals without ligands, have been gradually recognized. Autocrine
SHH interference in colon, pancreatic and lung cell lines triggers
cell death via PTCH proapoptotic signaling (188). The cell-adhesion
molecule-related/downregulated by oncogenes protein and its ligand,
SHH, perform identically (189).
However, very little optimization work has been conducted on this
finding.
In addition to interfering with apoptosis, HH has
also been implicated in resistance to cell death induced by
physical and chemical factors (166). Under radiation, HH is upregulated,
which may be driven by TGF-β and TNF-α (190) and mediated by mTOR/ribosomal
protein S6 kinase β-1 (191).
Atypical protein kinase Cι/λ and GLI can also form a positive
feedback loop under high levels of radiation to change the
radiosensitivity of tumors (192).
In addition, the HH pathway activated by chemoradiotherapy also
increases the rate of tumor proliferation by upregulating the
G1/cyclin/Rb axis (193). In
response to irradiation, GLI1 activates RNA polymerase I, which
synthesizes ribosomal RNA for accommodating cell proliferation and
division (194). Thus, blocking HH
signaling was demonstrated to be an effective method for inhibiting
accelerated tumor repopulation following therapy.
The HH pathway is also associated with cell
senescence-related diseases. For instance, IHH protects bone
marrow-derived mesenchymal stem cells from senescence-associated
secretory phenotype (SASP)-induced senescence by downregulating the
ROS/mTOR pathway during oxidative stress (195). However, recent studies have
gradually recognized that SASP has a bidirectional regulatory
effect on tumors and may be related to the degree of aging load
(196). In medulloblastoma, a
PTCH1 loss of heterozygosity was discovered to be associated with
high levels of cellular senescence before tumor occurrence.
However, other subsequent spontaneous site mutations, such as in
p53, stimulate inhibition of senescence and promote tumor
development by modulating CDKs (197).
Hypoxia is a mechanism underlying tumor invasion and
metastasis that is partially achieved by activating the HH pathway.
Hypoxia is often accompanied by TNF, NOX4 and other products. TNF-α
can upregulate the expression of GLI1 via NF-κB transcription to
achieve EMT and drug resistance (132). Furthermore, NOX4 expression
triggers the reactive oxygen species-mediated non-canonical HH
pathway in the initiation of EMT (198). Hypoxia inducible factor (HIF) is
also a widely studied regulator. The upregulation of HIF-1α was
revealed to be associated with the upregulation of SHH ligand
secretion and GLI1 expression, thereby increasing metalloproteinase
(MMP) expression and EMT progression (199,200). It should be noted that activation
of the HH pathway during hypoxia leads to an increase in stromal
fibroblasts, then the deposition of fibrous tissue and ultimately
the aggravation of hypoxia (201).
Hypoxia can also provoke HIF-2α to induce GLI1 activation through a
SMO-independent pathway (202),
which can be ablated by PI3K inhibitor or MEK inhibitor (202). This finding also suggests a
complex connection between these pathways.
The crosstalk between the HH, PI3K and MEK pathways
has been gradually clarified. Previously, there was evidence that
fibroblast metastasis required the stimulation of Ras homolog
family member (Rho)A by SMO, which can be realized by a
heterotrimeric Gi proteins/PI3K/Rac1 series of activations
(7). Subsequently, it was revealed
that the PI3K/AKT/mTOR pathway plays an important role in SHH
signaling to promote metastasis (203). More specifically, AKT/GSK3β
signaling mediates the upregulation of GLI1 expression, thereby
obtaining epithelial mesenchymal plasticity (204). In addition, astrocyte elevated
gene-1 protein is induced by PI3K/AKT signaling and is vital in the
metastasis and development of various types of cancer. Therefore,
it was not surprising that PI3K pathway is involved in the
crosstalk process (205).
In the MEK pathway, GLI1 directly binds to the
promoter region of the CXCR4 gene and participates in stimulated
signal transduction of CXCL12 to stimulate the phosphorylation of
ERK (206). This is consistent
with the observation that HH upregulates MMP-9 expression via the
ERK pathway (207), while MEK/ERK
signaling is involved in the regulation of GLI1 activity in turn
(208,209). Furthermore, the MEK/AKT pathway
could be regulated by RAS and others. The widespread occurrence of
this regulation in cancer cells rectifies the singleness of the HH
pathway, which is mainly activated in the microenvironment but not
tumor cells (208,209). The close relationship between
these three pathways supports the notion that the synergistic role
could be amplified by blocking the pathways simultaneously
(210).
The HH and Wnt/β-catenin pathways have also been
demonstrated to impose a synchronized regulation on tumor
metastasis. In fact, a crosstalk between these pathways does exist
(89,211). During the development of cancer
associated fibroblasts (CAFs), two pathways regulate the target
gene of TGF-β/SMAD3 to participate in the EMT process (212). Although uncertainty remains as to
the specific mechanism, it is not difficult to observe that tumor
invasion and metastasis are the ultimate outcome of multiple
pathways.
In the previous section, the upstream and downstream
molecular mechanisms of GLI1 were briefly mentioned. In gastric
cancer, galectin-1 from CAFs binds to β1 integrin and targets GLI1
to promote both EMT and vasculogenic mimicry (213). Signal peptide CUB EGF-like
domain-containing protein 2 (214)
and FoxF1 (215) both regulate
tumor metastasis via GLI1. In addition, S100A4 is a newly
discovered downstream target gene of GLI1 (216). However, a new theory suggests that
tumor progression is dynamically regulated between proliferation
and metastasis through up and downregulated GLI1 levels, rather
than just upregulated GLI1 alone. Moreover, endogenous GLI1 can
directly bind to the promoter of the E-cadherin gene, termed CDH1,
and resist EMT (217).
Compared with GLI1, truncated GLI1 (tGLI1)
demonstrated a stronger association with abnormal HH signals. In
addition to regulating the known GLI1 target genes in the crosstalk
with STAT3 and other pathways (218), tGLI1 can also regulate the
expression of genes that were not regulated by GLI1, including
vascular endothelial growth factor (VEGF) and heparinase (219). Moreover, tGLI1 displays a strong
correlation with tumor metastasis. The presence of tGLI1 enhances
the expression of MMP-2 and MMP-9 and may target twist and snail
(220). In addition, tGLI1 has
been identified as a brain metastasis-promoting transcription
factor in breast cancer (221).
tGLI1 increases the degree of cancer cell stemness by upregulating
genes such as Nanog and by activating astrocytes to achieve
metastasis (219). In summary, it
is hypothesized that tGLI1 is more likely to be a marker for the
diagnosis and prognosis of cancer metastasis, and thus may become a
new therapeutic target (222).
With the accumulation of research, the unknown role
of ncRNA in EMT progress and MMP expression has been gradually
uncovered (Table II).
The main current view is that the effect of the HH
pathway on angiogenesis ultimately depends on the interaction with
the VEGF pathway. Previously, some scholars proposed the existence
of a HH interacting protein/HH/VEGF/Notch signaling axis, but the
role of GLI1 in this process has gradually been revealed. In a
tissue microarray analysis (223),
GLI1 was linked to the upregulation of VEGF receptor 2
(VEGFR2).
Another view is that tGLI1 is a direct participant
in the VEGF pathway instead of GLI1, and that tGLI1 also inhibits
the expression of soluble VEGFR2, the thrombospondin family and
TIMP metallopeptidase inhibitor 2 (223,224). These molecules are considered to
be potent antagonists of angiogenesis or lymphangiogenesis. In the
presence of VEGF, Rho GTPases are also targets of the SHH
non-classical pathway (225). In
addition, the crosstalk between mTOR and the HH pathway in
angiogenesis is an area of interest (226). It was previously demonstrated that
the SMO-independent activation of GLI1 could be mediated by the
mTOR/S6K1 pathway, blocking the interaction between SUFU and GLI1
(227,228), while also upregulating VEGF
(229). In addition, cysteine-rich
protein 61 is considered to be another angiogenic target of
SHH/GLI1 (211).
Following further research of the basic pathway, it
may be effective to inhibit angiogenesis and reduce the relative
area of tumor blood vessels by using HH inhibitors or by combining
with an mTOR pathway inhibitor after evaluating the expression of
HH, to reduce drug resistance (225,230). In contrast to expectations, it has
been demonstrated that SHH-deficient pancreatic ductal
adenocarcinoma showed higher vascular density and proliferation
activity, and its response to anti-angiogenesis therapy was more
notable (231). A possible
explanation for this is that the inhibition of HH may lead to lower
differentiation. Taken together, the application of HH inhibitors
with anti-angiogenesis therapy requires further attention.
The Warburg effect has been widely discussed in
terms of tumor metabolic changes. This effect allows cells to
replace the mechanism of oxidative phosphorylation with aerobic
glycolysis, thereby obtaining more energy to promote tumor
development. The HH pathway mediates the Warburg effect of tumor
cells and CAFs. Caveolin-1 (Cav-1) is present in the tumor matrix
and participates in the regulation of glycolytic activity (232). The deletion of Cav-1 is associated
with high expression levels of GLI1 (233). It was also revealed that the
activation of SMO promotes glycolysis via GLI upregulation and the
AMPK-mediated activation of hexokinase 2 and pyruvate kinase 2
(234).
In addition, HH activity is involved in the
regulation of metabolism and bioenergy in TAMs. Inhibition of HH
causes metabolically demanding M2 macrophages to shift their
metabolism and bioenergetics from fatty acid oxidation to
glycolysis (148). HH signaling
also acts downstream of metabolic reprogramming to influence
tumorigenesis. HH signaling mediates the hyperglycemia inducing
glycolytic phenotype and promotes EMT via Yes-associated protein 1
(235).
Ornithine decarboxylase (ODC)1 is aberrantly
upregulated in primary (SHH subtype) medulloblastoma, which
increases polyamine metabolism to promote tumors (236). In this instance, AMPK promotes the
stable formation of the SUFU/CCHC type nucleic acid binding protein
(CNBP) complex via phosphorylation of CNBP, and further promotes
the expression of ODC (237). In
addition, a metabolic switch to oxidative phosphorylation was
revealed to be promoted by GLI1 editing (238).
The use of HH pathway inhibitors as anticancer
drugs has gained significance. At present, five HH pathway
inhibitors have been approved for marketing. A large number of new
generation inhibitors and drugs targeting new targets have entered
clinical trials. Among them, SMO inhibitors are classic inhibitors
as inhibition of SMO is a reliable route to blocking activation of
the HH pathway. However, the frequent occurrence of drug-resistant
mutations in SMO means that traditional SMO inhibitors are prone to
failure, a problem that requires urgent attention.
Second-generation SMO inhibitors have been proven to be effective
in preclinical experiments by targeting specific SMO site mutations
or by improving the binding affinity. Directly targeting the
downstream signals is another traditional solution. The HH
inhibitor drugs are summarized in Table III.
HH pathway mutations have been revealed to be
associated with enhanced tumor immunogenicity, and it may be
feasible to further seek immune checkpoint inhibitor treatment for
greater benefits (239). SMO
resistance can also be prevented by altering the epigenetic changes
of histones or transcription factors GLI1 and GLI2, such as by HDAC
(240). Moreover, new technologies
such as CRISPR/Cas9 for pathway site-specific gene editing are also
being developed (241), which may
pave the way for HH therapies. Although drugs for downstream
targets are under study, progress has been slow (242), which may be partly due to
downstream signals, such as GLI1, being activated by other
pathways, such as the TGF-β, Ras and PI3K/AKT pathways. Therefore,
blocking the activity of non-classical pathways is also being
considered as a new alternative strategy (243).
Drugs targeting ncRNA should also be considered as
a future direction for HH pathway-related treatment. However, how
to narrow the scope of the best target ncRNAs and how to improve
recognition of the structure of ncRNA remains to be solved.
As aforementioned, a variety of anti-HH/GLI drugs
have been developed. However, the complex crosstalk between
pathways, compensatory mechanisms, the generation of primary or
secondary drug resistance, as well as toxic side effects, may still
lead to the failure of current drugs. Recently, natural drugs with
multiple targets and a higher safety profile have attracted
attention. The combination of natural drugs and anticancer drugs
has been revealed to be more effective than anticancer drugs alone.
In Table IV, the new progression
of some natural components and their main targets and effects in
research, which may provide support for the future transformation
of natural drugs based on the HH pathway into anticancer drugs, are
summarized.
Through combing the mechanisms of the HH pathway in
different tumor phenotypes, it is not difficult to recognize the
notable role of the HH pathway in tumor formation. The interaction
between HH pathway factors forms several negative feedbacks in cell
function such as promoting autophagy apoptosis, and positive
feedbacks such as the feedback between hypoxia and angiogenesis.
Knowledge of HH pathway signal transduction, the crosstalk
mechanisms and the influence on phenotype may improve the clinical
vigilance of anomalous HH-related test results. Moreover, it may
assist the more accurate use of drugs in tumor treatment and
provide more therapy strategies.
However, the shortcomings in the present study
review of the HH pathway remain undeniable. A common problem is the
lack of accurate methods to clarify the mechanisms of specific
factors in the complex crosstalk of pathways for tumor metastasis.
The exploration of positive and negative regulation of the HH
pathway in different cancer types is also limited. Moreover,
further study is needed to correlate expression of the HH pathway
with tumor characteristics, such as source, metastasis rate,
recurrence rate and other prognosis factors, by referring to HH
gene status. In this way, accurate stratification of tumor subtypes
can be achieved, which is also the mainstream direction of future
oncology development. In terms of drugs, there has been an upsurge
in the field of natural medicines based on HH pathway treatment.
However, very little information is available in the systematic
summary of research progress in this area. Improved understanding
of the HH pathway may assist with improving clinical treatment by
solving the aforementioned problems.
Not applicable.
The present study was supported by the Zhejiang Provincial
Natural Science Foundation of China (grant no. LQ22H270008), the
National Natural Science Foundation of China (grant no. 82204824),
the Scientific Research Fund of Zhejiang Provincial Education
Department (grant no. Y202351288), the Young Elite Scientists
Sponsorship Program by China Association of Chinese Medicine (grant
no. 2021-QNRC2-B13) and the Hangzhou Medical and Health Science and
Technology Project (grant no. A20230054).
Not applicable.
DS and LS conceptualized the study. DS, YX, YF and
WC curated the data. DS, YX, QC, YZ and KG visualized the data. DS,
YX, YF and WC wrote the original draft. LS, QC, YZ and KG wrote,
reviewed and edited the manuscript. DS, KG and LS acquired funding.
Data authentication is not applicable. All authors read and
approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Clara JA, Monge C, Yang Y and Takebe N:
Targeting signalling pathways and the immune microenvironment of
cancer stem cells-a clinical update. Nat Rev Clin Oncol.
17:204–232. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Girardi D, Barrichello A, Fernandes G and
Pereira A: Targeting the hedgehog pathway in cancer: Current
evidence and future perspectives. Cells. 8:1532019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Niyaz M, Khan MS and Mudassar S: Hedgehog
signaling: An Achilles' Heel in cancer. Transl Oncol. 12:1334–1344.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jeng KS, Chang CF and Lin SS: Sonic
hedgehog signaling in organogenesis, tumors, and tumor
microenvironments. Int J Mol Sci. 21:7582020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhou M and Jiang J: Gli phosphorylation
code in hedgehog signal transduction. Front Cell Dev Biol.
10:8469272022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xin M, Ji X, De La Cruz LK, Thareja S and
Wang B: Strategies to target the Hedgehog signaling pathway for
cancer therapy. Med Res Rev. 38:870–913. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Polizio AH, Chinchilla P, Chen X, Kim S,
Manning DR and Riobo NA: Heterotrimeric Gi proteins link Hedgehog
signaling to activation of Rho small GTPases to promote fibroblast
migration. J Biol Chem. 286:19589–19596. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Suchors C and Kim J: Canonical hedgehog
pathway and noncanonical GLI transcription factor activation in
cancer. Cells. 11:25232022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hanahan D: Hallmarks of cancer: New
dimensions. Cancer Discov. 12:31–46. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim BR, Na YJ, Kim JL, Jeong YA, Park SH,
Jo MJ, Jeong S, Kang S, Oh SC and Lee DH: RUNX3 suppresses
metastasis and stemness by inhibiting Hedgehog signaling in
colorectal cancer. Cell Death Differ. 27:676–694. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xia Y, Zhen L, Li H, Wang S, Chen S, Wang
C and Yang X: MIRLET7BHG promotes hepatocellular carcinoma
progression by activating hepatic stellate cells through exosomal
SMO to trigger Hedgehog pathway. Cell Death Dis. 12:3262021.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen L, Li Y, Song Z, Xue S, Liu F, Chang
X, Wu Y, Duan X and Wu H: O-GlcNAcylation promotes cerebellum
development and medulloblastoma oncogenesis via SHH signaling. Proc
Natl Acad Sci USA. 119:e22028211192022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sursal T, Ronecker JS, Dicpinigaitis AJ,
Mohan AL, Tobias ME, Gandhi CD and Jhanwar-Uniyal M: Molecular
stratification of medulloblastoma: Clinical outcomes and
therapeutic interventions. Anticancer Res. 42:2225–2239. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fan Z, Li J, Du J, Zhang H, Shen Y, Wang
CY and Wang S: A missense mutation in PTCH2 underlies dominantly
inherited NBCCS in a Chinese family. J Med Genet. 45:303–308. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Smith MJ and Evans DG: PTCH2 is not a
strong candidate gene for gorlin syndrome predisposition. Fam
Cancer. 21:343–346. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guerrini-Rousseau L, Masliah-Planchon J,
Waszak SM, Alhopuro P, Benusiglio PR, Bourdeaut F, Brecht IB, Del
Baldo G, Dhanda SK, Garrè ML, et al: Cancer risk and tumour
spectrum in 172 patients with a germline SUFU pathogenic variation:
A collaborative study of the SIOPE Host Genome Working Group. J Med
Genet. 59:1123–1132. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bansaccal N, Vieugue P, Sarate R, Song Y,
Minguijon E, Miroshnikova YA, Zeuschner D, Collin A, Allard J,
Engelman D, et al: The extracellular matrix dictates regional
competence for tumour initiation. Nature. 623:828–835. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nicheperovich A and Townsend-Nicholson A:
Towards precision oncology: The role of smoothened and its variants
in cancer. J Pers Med. 12:16482022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pantazi E, Gemenetzidis E, Trigiante G,
Warnes G, Shan L, Mao X, Ikram M, Teh MT, Lu YJ and Philpott MP:
GLI2 induces genomic instability in human keratinocytes by
inhibiting apoptosis. Cell Death Dis. 5:e10282014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kerr DA, Cloutier JM, Margolis M, Mata DA,
Rodrigues Simoes NJ, Faquin WC, Dias-Santagata D, Chopra S,
Charville GW, Wangsiricharoen S, et al: GLI1-altered mesenchymal
tumors with ACTB or PTCH1 fusion: A molecular and clinicopathologic
analysis. Mod Pathol. 37:1003862024. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu B, Chang K, Folpe AL, Kao YC, Wey SL,
Huang HY, Gill AJ, Rooper L, Bishop JA, Dickson BC, et al: Head and
neck mesenchymal neoplasms with GLI1 Gene alterations: A pathologic
entity with distinct histologic features and potential for distant
metastasis. Am J Surg Pathol. 44:729–737. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Agaram NP, Zhang L, Sung YS, Singer S,
Stevens T, Prieto-Granada CN, Bishop JA, Wood BA, Swanson D,
Dickson BC and Antonescu CR: GLI1-amplifications expand the
spectrum of soft tissue neoplasms defined by GLI1 gene fusions. Mod
Pathol. 32:1617–1626. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dawson MA and Kouzarides T: Cancer
epigenetics: From mechanism to therapy. Cell. 150:12–27. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun L, Zhang H and Gao P: Metabolic
reprogramming and epigenetic modifications on the path to cancer.
Protein Cell. 13:877–919. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhu Z, Zhou Q, Sun Y, Lai F, Wang Z, Hao Z
and Li G: MethMarkerDB: A comprehensive cancer DNA methylation
biomarker database. Nucleic Acids Res. 52:D1380–D1392. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Duan ZH, Wang HC, Zhao DM, Ji XX, Song M,
Yang XJ and Cui W: Cooperatively transcriptional and epigenetic
regulation of sonic hedgehog overexpression drives malignant
potential of breast cancer. Cancer Sci. 106:1084–1091. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Garcia N, Al-Hendy A, Baracat EC, Carvalho
KC and Yang Q: Targeting hedgehog pathway and DNA
methyltransferases in uterine leiomyosarcoma cells. Cells.
10:532020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lou H, Li H, Huehn AR, Tarasova NI, Saleh
B, Anderson SK and Dean M: Genetic and epigenetic regulation of the
smoothened gene (SMO) in cancer cells. Cancers (Basel).
12:22192020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang TP, Hsu SH, Feng HC and Huang RF:
Folate deprivation enhances invasiveness of human colon cancer
cells mediated by activation of sonic hedgehog signaling through
promoter hypomethylation and cross action with transcription
nuclear factor-kappa B pathway. Carcinogenesis. 33:1158–1168. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bongiovanni D, Tosello V, Saccomani V,
Dalla Santa S, Amadori A, Zanovello P and Piovan E: Crosstalk
between Hedgehog pathway and the glucocorticoid receptor pathway as
a basis for combination therapy in T-cell acute lymphoblastic
leukemia. Oncogene. 39:6544–6555. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yan Z, Cheng M, Hu G, Wang Y, Zeng S,
Huang A, Xu L, Liu Y, Shi C, Deng L, et al: Positive feedback of
SuFu negating protein 1 on Hedgehog signaling promotes colorectal
tumor growth. Cell Death Dis. 12:1992021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Di Marcotullio L, Ferretti E, Greco A, De
Smaele E, Po A, Sico MA, Alimandi M, Giannini G, Maroder M,
Screpanti I and Gulino A: Numb is a suppressor of Hedgehog
signalling and targets Gli1 for Itch-dependent ubiquitination. Nat
Cell Biol. 8:1415–1423. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liao H, Cai J, Liu C, Shen L, Pu X, Yao Y,
Han B, Yu T, Cheng SY and Yue S: Protein phosphatase 4 promotes
Hedgehog signaling through dephosphorylation of Suppressor of
fused. Cell Death Dis. 11:6862020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hu L, Cao H, Zheng L and Li R: BBOX1-AS1
activates hedgehog signaling pathway to facilitate the
proliferation and stemness of esophageal squamous cell carcinoma
cells via miR-506-5p/EIF5A/PTCH1 axis. Curr Mol Pharmacol.
16:894–904. 2023.PubMed/NCBI
|
|
35
|
Bartl J, Zanini M, Bernardi F, Forget A,
Blümel L, Talbot J, Picard D, Qin N, Cancila G, Gao Q, et al: The
HHIP-AS1 lncRNA promotes tumorigenicity through stabilization of
dynein complex 1 in human SHH-driven tumors. Nat Commun.
13:40612022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hu Z, Liu Y, Tang J, Luo R, Qin J, Mo Z,
Xie J, Jiang X, Wei S and Lin C: LncRNA HHIP-AS1 suppresses lung
squamous cell carcinoma by stabilizing HHIP mRNA. Life Sci.
321:1215782023. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yang Y, Cheng Y, Mou Y, Tang X and Mu X:
Natural antisense long noncoding RNA HHIP-AS1 suppresses
Non-small-cell lung cancer progression by increasing HHIP stability
via interaction with CELF2. Crit Rev Eukaryot Gene Expr. 33:67–77.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li X, Wu Y, Xiao Z, Liu Y, Wang C, Zhou L
and Yang X: Long non-coding RNA HIF1A-AS2 promotes carcinogenesis
by enhancing Gli1-mediated HIF1α expression in clear cell renal
cell carcinoma. Pathol Res Pract. 253:1549842024. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xu F, Li H and Hu C: LIFR-AS1 modulates
Sufu to inhibit cell proliferation and migration by miR-197-3p in
breast cancer. Biosci Rep. 39:BSR201805512019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu J, Zhou F, Lai S, Wang W, Wu T, Liu Y
and Yang L: Propofol inhibits biological function of hepatocellular
carcinoma cells through LINC00475-mediated sonic hedgehog pathway.
Pharmacology. 108:127–137. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Meng D, Zhao S, Wu L, Ma X, Zhao D and Li
Z: LINC00641 impeded the malignant biological behaviors of
papillary thyroid carcinoma cells via interacting with IGF2BP1 to
reduce GLI1 mRNA stability. Hum Exp Toxicol.
42:96032712311808562023. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
He J, Zuo Q, Hu B, Jin H, Wang C, Cheng Z,
Deng X, Yang C, Ruan H, Yu C, et al: A novel, liver-specific long
noncoding RNA LINC01093 suppresses HCC progression by interaction
with IGF2BP1 to facilitate decay of GLI1 mRNA. Cancer Lett.
450:98–109. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Guo K, Gong W, Wang Q, Gu G, Zheng T, Li
Y, Li W, Fang M, Xie H, Yue C, et al: LINC01106 drives colorectal
cancer growth and stemness through a positive feedback loop to
regulate the Gli family factors. Cell Death Dis. 11:8692020.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liu X, Yin Z, Xu L, Liu H, Jiang L, Liu S
and Sun X: Upregulation of LINC01426 promotes the progression and
stemness in lung adenocarcinoma by enhancing the level of SHH
protein to activate the hedgehog pathway. Cell Death Dis.
12:1732021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wei P, Jiang J, Xiao M, Zeng M, Liu X,
Zhao B and Chen F: The transcript ENST00000444125 of lncRNA
LINC01503 promotes cancer stem cell properties of glioblastoma
cells via reducing FBXW1 mediated GLI2 degradation. Exp Cell Res.
412:1130092022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li Q, Wang XJ and Jin JH: SOX2-induced
upregulation of lncRNA LINC01510 promotes papillary thyroid
carcinoma progression by modulating miR-335/SHH and activating
Hedgehog pathway. Biochem Biophys Res Commun. 520:277–283. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Nie H, Liao Z, Wang Y, Zhou J, He X and Ou
C: Exosomal long non-coding RNAs: Emerging players in cancer
metastasis and potential diagnostic biomarkers for personalized
oncology. Genes Dis. 8:769–780. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Guo L, Zhou Y, Chen Y, Sun H, Wang Y and
Qu Y: LncRNA ASAP1-IT1 positively modulates the development of
cholangiocarcinoma via hedgehog signaling pathway. Biomed
Pharmacother. 103:167–173. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wu H, He P, Xie D, Wang J and Wan C:
Long-noncoding RNA ANCR activates the hedgehog signaling pathway to
promote basal cell carcinoma progression by binding to PTCH. Clin
Cosmet Investig Dermatol. 15:955–965. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chen F, Mo J and Zhang L: Long noncoding
RNA BCAR4 promotes osteosarcoma progression through activating
GLI2-dependent gene transcription. Tumour Biol. 37:13403–13412.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yang H, Yan L, Sun K, Sun X, Zhang X, Cai
K and Song T: lncRNA BCAR4 increases viability, invasion, and
migration of Non-small cell lung cancer cells by targeting
glioma-associated oncogene 2 (GLI2). Oncol Res. 27:359–369. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Sun J, Jia J, Yuan W, Liu S, Wang W, Ge L,
Ge L and Liu XJ: LncRNA BLACAT1 accelerates Non-small cell lung
cancer through up-regulating the activation of sonic hedgehog
pathway. Front Oncol. 11:6252532021. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhou H, Xiong Y, Peng L, Wang R, Zhang H
and Fu Z: LncRNA-cCSC1 modulates cancer stem cell properties in
colorectal cancer via activation of the Hedgehog signaling pathway.
J Cell Biochem. 121:2510–2524. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wang Z, Song L, Ye Y and Li W: Long
noncoding RNA DIO3OS hinders cell malignant behaviors of
hepatocellular carcinoma cells through the microRNA-328/Hhip axis.
Cancer Manag Res. 12:3903–3914. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Qiu S, Chen G, Peng J, Liu J, Chen J, Wang
J, Li L and Yang K: LncRNA EGOT decreases breast cancer cell
viability and migration via inactivation of the Hedgehog pathway.
FEBS Open Bio. 10:817–826. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Peng W, Wu J, Fan H, Lu J and Feng J:
LncRNA EGOT promotes tumorigenesis via hedgehog pathway in gastric
cancer. Pathol Oncol Res. 25:883–887. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zheng S, Li M, Miao K and Xu H: lncRNA
GAS5-promoted apoptosis in triple-negative breast cancer by
targeting miR-378a-5p/SUFU signaling. J Cell Biochem.
121:2225–2235. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Li L, Ma TT, Ma YH and Jiang YF: LncRNA
HCG18 contributes to nasopharyngeal carcinoma development by
modulating miR-140/CCND1 and Hedgehog signaling pathway. Eur Rev
Med Pharmacol Sci. 23:10387–10399. 2019.PubMed/NCBI
|
|
59
|
Wu J, Zhu P, Lu T, Du Y, Wang Y, He L, Ye
B, Liu B, Yang L, Wang J, et al: The long non-coding RNA LncHDAC2
drives the self-renewal of liver cancer stem cells via activation
of Hedgehog signaling. J Hepatol. 70:918–929. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Bai JY, Jin B, Ma JB, Liu TJ, Yang C,
Chong Y, Wang X, He D and Guo P: HOTAIR and androgen receptor
synergistically increase GLI2 transcription to promote tumor
angiogenesis and cancer stemness in renal cell carcinoma. Cancer
Lett. 498:70–79. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lu G, Zhong H, Gao J and Zhang Y: Alginate
microspheres encapsulating hox transcript antisense RNA siRNA
regulate the Hedgehog-Gli1 pathway to alleviate epidermal growth
factor receptor tyrosine kinase inhibitors resistance. J Biomater
Appl. 38:877–889. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhou M, Hou Y, Yang G, Zhang H, Tu G, Du
YE, Wen S, Xu L, Tang X, Tang S, et al: LncRNA-Hh strengthen cancer
stem cells generation in Twist-positive breast cancer via
activation of hedgehog signaling pathway. Stem Cells. 34:55–66.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zhang Y, Chen Z, Li MJ, Guo HY and Jing
NC: Long non-coding RNA metastasis-associated lung adenocarcinoma
transcript 1 regulates the expression of Gli2 by miR-202 to
strengthen gastric cancer progression. Biomed Pharmacother.
85:264–271. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Hamada T, Higashi M, Yokoyama S, Akahane
T, Hisaoka M, Noguchi H, Furukawa T and Tanimoto A: MALAT1
functions as a transcriptional promoter of MALAT1::GLI1 fusion for
truncated GLI1 protein expression in cancer. BMC Cancer.
23:4242023. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Chen W, Wang F, Yu X, Qi J, Dong H, Cui B,
Zhang Q, Wu Y, An J, Ni N, et al: LncRNA MIR31HG fosters stemness
malignant features of non-small cell lung cancer via H3K4me1- and
H3K27Ace-mediated GLI2 expression. Oncogene. 43:1328–1340. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Qian CS, Li LJ, Huang HW, Yang HF and Wu
DP: MYC-regulated lncRNA NEAT1 promotes B cell proliferation and
lymphomagenesis via the miR-34b-5p-GLI1 pathway in diffuse large
B-cell lymphoma. Cancer Cell Int. 20:872020. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Dong H, Zeng L, Chen W, Zhang Q, Wang F,
Wu Y, Cui B, Qi J, Zhang X, Liu C, et al: N6-methyladenine-mediated
aberrant activation of the lncRNA SOX2OT-GLI1 loop promotes
non-small-cell lung cancer stemness. Cell Death Discov. 9:1492023.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Herrera-Solorio AM, Peralta-Arrieta I,
Armas López L, Hernández-Cigala N, Mendoza Milla C, Ortiz Quintero
B, Catalán Cárdenas R, Pineda Villegas P, Rodríguez Villanueva E,
Trejo Iriarte CG, et al: LncRNA SOX2-OT regulates AKT/ERK and
SOX2/GLI-1 expression, hinders therapy, and worsens clinical
prognosis in malignant lung diseases. Mol Oncol. 15:1110–1129.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Li J, Zhang Q, Fan X, Mo W, Dai W, Feng J,
Wu L, Liu T, Li S, Xu S, et al: The long noncoding RNA TUG1 acts as
a competing endogenous RNA to regulate the Hedgehog pathway by
targeting miR-132 in hepatocellular carcinoma. Oncotarget.
8:65932–65945. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhang L, Liang H, Zhang J, Yang Y, Ling X
and Jiang H: Long non-coding RNA SNHG16 facilitates esophageal
cancer cell proliferation and Self-renewal through the
microRNA-802/PTCH1 axis. Curr Med Chem. 29:6084–6099. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Ye G, Pan R, Zhu L and Zhou D: Circ_DCAF6
potentiates cell stemness and growth in breast cancer through
GLI1-Hedgehog pathway. Exp Mol Pathol. 116:1044922020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Yang J, Yu L, Yan J, Xiao Y, Li W, Xiao J,
Lei J, Xiang D, Zhang S and Yu X: Circular RNA DGKB promotes the
progression of neuroblastoma by targeting miR-873/GLI1 axis. Front
Oncol. 10:11042020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Zhou Y, Xue X, Luo J, Li P, Xiao Z, Zhang
W, Zhou J, Li P, Zhao J, Ge H, et al: Circular RNA circ-FIRRE
interacts with HNRNPC to promote esophageal squamous cell carcinoma
progression by stabilizing GLI2 mRNA. Cancer Sci. 114:3608–3622.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Liu Y, Song J, Liu Y, Zhou Z and Wang X:
Transcription activation of circ-STAT3 induced by Gli2 promotes the
progression of hepatoblastoma via acting as a sponge for
miR-29a/b/c-3p to upregulate STAT3/Gli2. J Exp Clin Cancer Res.
39:1012020. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Wu B, Xia L, Zhang S, Jin K, Li L, Sun C,
Xia T and Chen G: circRNA-SMO upregulates CEP85 to promote
proliferation and migration of glioblastoma via sponging miR-326.
Histol Histopathol. 38:1307–1319. 2023.PubMed/NCBI
|
|
76
|
Wu X, Xiao S, Zhang M, Yang L, Zhong J, Li
B, Li F, Xia X, Li X, Zhou H, et al: A novel protein encoded by
circular SMO RNA is essential for Hedgehog signaling activation and
glioblastoma tumorigenicity. Genome Biol. 22:332021. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Xiong Z, Zhou C, Wang L, Zhu R, Zhong L,
Wan D and Wang Q: Circular RNA SMO sponges miR-338-3p to promote
the growth of glioma by enhancing the expression of SMO. Aging
(Albany NY). 11:12345–12360. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Wu L, Xia J, Yang J, Shi Y, Xia H, Xiang X
and Yu X: Circ-ZNF609 promotes migration of colorectal cancer by
inhibiting Gli1 expression via microRNA-150. J BUON. 23:1343–1349.
2018.PubMed/NCBI
|
|
79
|
He Y, Huang H, Jin L, Zhang F, Zeng M, Wei
L, Tang S, Chen D and Wang W: CircZNF609 enhances hepatocellular
carcinoma cell proliferation, metastasis, and stemness by
activating the Hedgehog pathway through the regulation of
miR-15a-5p/15b-5p and GLI2 expressions. Cell Death Dis. 11:3582020.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Ye G, He S, Pan R, Zhu L, Zhou D, Cai G
and Chen P: circ_0041732 promotes breast cancer progression. Mol
Cancer Res. 20:1561–1573. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Wang L, Li B, Yi X, Xiao X, Zheng Q and Ma
L: Circ_0036412 affects the proliferation and cell cycle of
hepatocellular carcinoma via hedgehog signaling pathway. J Transl
Med. 20:1542022. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Zhang P, Gao H, Yan R, Yu L, Xia C and
Yang D: has_circ_0070512 promotes prostate cancer progression by
regulating the miR-338-3p/hedgehog signaling pathway. Cancer Sci.
114:1491–1506. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Seidl C, Panzitt K, Bertsch A, Brcic L,
Schein S, Mack M, Leithner K, Prinz F, Olschewski H, Kornmueller K
and Hrzenjak A: MicroRNA-182-5p regulates hedgehog signaling
pathway and chemosensitivity of cisplatin-resistant lung
adenocarcinoma cells via targeting GLI2. Cancer Lett. 469:266–276.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Zhang Z, Wang C, Liu T, Tang Z, Yan R,
Zhang C, Cheng C, Wang J, Wang H, Huang H and Li Y: miRNA-182-5p
promotes human bladder cancer proliferation and migration through
the FOXF2/SHH axis. Neoplasma. 69:321–330. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Li Y, Zhang D, Chen C, Ruan Z, Li Y and
Huang Y: MicroRNA-212 displays tumor-promoting properties in
non-small cell lung cancer cells and targets the hedgehog pathway
receptor PTCH1. Mol Biol Cell. 23:1423–1434. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Chen LT, Xu SD, Xu H, Zhang JF, Ning JF
and Wang SF: MicroRNA-378 is associated with non-small cell lung
cancer brain metastasis by promoting cell migration, invasion and
tumor angiogenesis. Med Oncol. 29:1673–1680. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Xin L, Liu L, Liu C, Zhou LQ, Zhou Q, Yuan
YW, Li SH and Zhang HT: DNA-methylation-mediated silencing of
miR-7-5p promotes gastric cancer stem cell invasion via increasing
Smo and Hes1. J Cell Physiol. 235:2643–2654. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Peng Y, Zhang X, Ma Q, Yan R, Qin Y, Zhao
Y, Cheng Y, Yang M, Wang Q, Feng X, et al: MiRNA-194 activates the
Wnt/β-catenin signaling pathway in gastric cancer by targeting the
negative Wnt regulator, SUFU. Cancer Lett. 385:117–127. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Peng Y, Zhang X, Lin H, Deng S, Qin Y, He
J, Hu F, Zhu X, Feng X, Wang J, et al: Dual activation of Hedgehog
and Wnt/β-catenin signaling pathway caused by downregulation of
SUFU targeted by miRNA-150 in human gastric cancer. Aging (Albany
NY). 13:10749–10769. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Zhang HQ, Sun Y, Li JQ, Huang LM, Tan SS,
Yang FY and Li H: The expression of microRNA-324-3p as a tumor
suppressor in nasopharyngeal carcinoma and its clinical
significance. Onco Targets Ther. 10:4935–4943. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Peng Y, Zhang X, Lin H, Deng S, Qin Y,
Yuan Y, Feng X, Wang J, Chen W, Hu F, et al: SUFU mediates EMT and
Wnt/β-catenin signaling pathway activation promoted by miRNA-324-5p
in human gastric cancer. Cell Cycle. 19:2720–2733. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Xu HS, Zong HL, Shang M, Ming X, Zhao JP,
Ma C and Cao L: MiR-324-5p inhibits proliferation of glioma by
target regulation of GLI1. Eur Rev Med Pharmacol Sci. 18:828–832.
2014.PubMed/NCBI
|
|
93
|
Ysrafil Y and Astuti I: Chitosan
nanoparticle-mediated effect of antimiRNA-324-5p on decreasing the
ovarian cancer cell proliferation by regulation of GLI1 expression.
Bioimpacts. 12:195–202. 2022.PubMed/NCBI
|
|
94
|
Tang B, Xu A, Xu J, Huang H, Chen L, Su Y,
Zhang L, Li J, Fan F, Deng J, et al: MicroRNA-324-5p regulates
stemness, pathogenesis and sensitivity to bortezomib in multiple
myeloma cells by targeting hedgehog signaling. Int J Cancer.
142:109–120. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Rodríguez-García Y, Martínez-Moreno M,
Alonso L, Sánchez-Vencells A, Arranz A, Dagà-Millán R,
Sevilla-Movilla S, Valeri A, Martínez-López J and Teixidó J:
Regulation of miRNA expression by α4β1 integrin-dependent multiple
myeloma cell adhesion. EJHaem. 4:631–638. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Peng Y, Qin Y, Zhang X, Deng S, Yuan Y,
Feng X, Chen W, Hu F, Gao Y, He J, et al: MiRNA-20b/SUFU/Wnt axis
accelerates gastric cancer cell proliferation, migration and EMT.
Heliyon. 7:e066952021. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Yang H, Fu H, Wang B, Zhang X, Mao J, Li
X, Wang M, Sun Z, Qian H and Xu W: Exosomal miR-423-5p targets SUFU
to promote cancer growth and metastasis and serves as a novel
marker for gastric cancer. Mol Carcinog. 57:1223–1236. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Tang CT, Liang Q, Yang L, Lin XL, Wu S,
Chen Y, Zhang XT, Gao YJ and Ge ZZ: RAB31 Targeted by MiR-30c-2-3p
Regulates the GLI1 signaling pathway, affecting gastric cancer cell
proliferation and apoptosis. Front Oncol. 8:5542018. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Cao D, Yu T and Ou X: MiR-873-5P controls
gastric cancer progression by targeting hedgehog-GLI signaling.
Pharmazie. 71:603–606. 2016.PubMed/NCBI
|
|
100
|
Zhou Y, Tang X, Huang Z, Wen J, Xiang Q
and Liu D: KLF5 promotes KIF1A expression through transcriptional
repression of microRNA-338 in the development of pediatric
neuroblastoma. J Pediatr Surg. 57:192–201. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Sun K, Deng HJ, Lei ST, Dong JQ and Li GX:
miRNA-338-3p suppresses cell growth of human colorectal carcinoma
by targeting smoothened. World J Gastroenterol. 19:2197–2207. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Zhao DW, Hou YS, Sun FB, Han B and Li SJ:
Effects of miR-132 on proliferation and apoptosis of pancreatic
cancer cells via Hedgehog signaling pathway. Eur Rev Med Pharmacol
Sci. 23:1978–1985. 2019.PubMed/NCBI
|
|
103
|
Du W, Liu X, Chen L, Dou Z, Lei X, Chang
L, Cai J, Cui Y, Yang D, Sun Y, et al: Targeting the SMO oncogene
by miR-326 inhibits glioma biological behaviors and stemness. Neuro
Oncol. 17:243–253. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Huang JH, Xu Y and Lin FY: The inhibition
of microRNA-326 by SP1/HDAC1 contributes to proliferation and
metastasis of osteosarcoma through promoting SMO expression. J Cell
Mol Med. 24:10876–10888. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Babashah S, Sadeghizadeh M, Hajifathali A,
Tavirani MR, Zomorod MS, Ghadiani M and Soleimani M: Targeting of
the signal transducer Smo links microRNA-326 to the oncogenic
Hedgehog pathway in CD34+ CML stem/progenitor cells. Int J Cancer.
133:579–589. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Sheybani Z, Rahgozar S and Ghodousi ES:
The Hedgehog signal transducer Smoothened and microRNA-326:
Pathogenesis and regulation of drug resistance in pediatric B-cell
acute lymphoblastic leukemia. Cancer Manag Res. 11:7621–7630. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Yin S, Du W, Wang F, Han B, Cui Y, Yang D,
Chen H, Liu D, Liu X, Zhai X and Jiang C: MicroRNA-326 sensitizes
human glioblastoma cells to curcumin via the SHH/GLI1 signaling
pathway. Cancer Biol Ther. 19:260–270. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Miele E, Po A, Begalli F, Antonucci L,
Mastronuzzi A, Marras CE, Carai A, Cucchi D, Abballe L, Besharat
ZM, et al: β-arrestin1-mediated acetylation of Gli1 regulates
Hedgehog/Gli signaling and modulates self-renewal of SHH
medulloblastoma cancer stem cells. BMC Cancer. 17:4882017.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Munoz JL, Rodriguez-Cruz V, Ramkissoon SH,
Ligon KL, Greco SJ and Rameshwar P: Temozolomide resistance in
glioblastoma occurs by miRNA-9-targeted PTCH1, independent of sonic
hedgehog level. Oncotarget. 6:1190–1201. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Wang YF, Yang HY, Shi XQ and Wang Y:
Upregulation of microRNA-129-5p inhibits cell invasion, migration
and tumor angiogenesis by inhibiting ZIC2 via downregulation of the
Hedgehog signaling pathway in cervical cancer. Cancer Biol Ther.
19:1162–1173. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Wang T, Feng J and Zhang A: miR-584
inhibits cell proliferation, migration and invasion in vitro and
enhances the sensitivity to cisplatin in human cervical cancer by
negatively targeting GLI1. Exp Ther Med. 19:2059–2066.
2020.PubMed/NCBI
|
|
112
|
Guan B, Mu L, Zhang L, Wang K, Tian J, Xu
S, Wang X, He D and Du Y: MicroRNA-218 inhibits the migration,
epithelial-mesenchymal transition and cancer stem cell properties
of prostate cancer cells. Oncol Lett. 16:1821–1826. 2018.PubMed/NCBI
|
|
113
|
Zhang J, Li S, Li Y, Liu H, Zhang Y and
Zhang Q: miRNA-218 regulates the proliferation and apoptosis of
cervical cancer cells via targeting Gli3. Exp Ther Med.
16:2433–2441. 2018.PubMed/NCBI
|
|
114
|
Wen SY, Lin Y, Yu YQ, Cao SJ, Zhang R,
Yang XM, Li J, Zhang YL, Wang YH, Ma MZ, et al: miR-506 acts as a
tumor suppressor by directly targeting the hedgehog pathway
transcription factor Gli3 in human cervical cancer. Oncogene.
34:717–725. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Sun Z, Zhang T, Hong H, Liu Q and Zhang H:
miR-202 suppresses proliferation and induces apoptosis of
osteosarcoma cells by downregulating Gli2. Mol Cell Biochem.
397:277–283. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Xu Z, Huang C and Hao D: MicroRNA-1271
inhibits proliferation and promotes apoptosis of multiple myeloma
cells through inhibiting smoothened-mediated Hedgehog signaling
pathway. Oncol Rep. 37:1261–1269. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Li J, Qiu M, An Y, Huang J and Gong C:
miR-7-5p acts as a tumor suppressor in bladder cancer by regulating
the hedgehog pathway factor Gli3. Biochem Biophys Res Commun.
503:2101–2107. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Miao X, Gao H, Liu S, Chen M, Xu W, Ling
X, Deng X and Rao C: Down-regulation of microRNA-224-inhibites
growth and epithelial-to-mesenchymal transition phenotype-via
modulating SUFU expression in bladder cancer cells. Int J Biol
Macromol. 106:234–240. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Zhao D and Cui Z: MicroRNA-361-3p
regulates retinoblastoma cell proliferation and stemness by
targeting hedgehog signaling. Exp Ther Med. 17:1154–1162.
2019.PubMed/NCBI
|
|
120
|
Su YC, Metzen LT, Vélez LM, Bournique E,
Seldin M, Buisson R, Kuo WW, Huang CY and Kaiser P: Induction of
resistance to oxaliplatin in cancer by a microRNA/Fem1B/Gli1
pathway. Am J Cancer Res. 13:6011–6025. 2023.PubMed/NCBI
|
|
121
|
Zheng J, Cheng C, Xu J, Gao P, Wang J and
Chen L: miR-142-3p regulates tumor cell autophagy and promotes
colon cancer progression by targeting TP53INP2. Chemotherapy.
67:57–66. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Wang S, Huang X, Zhang G, Chen Z, Guan H
and Zhou W: Tumor suppressor miR-361-3p inhibits prostate cancer
progression through Gli1 and AKT/mTOR signaling pathway. Cell
Signal. 114:1109982024. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Zhang J, Li Y, Ren Y, Han H and Li J:
Mir-326 potentiates radiosensitivity of cervical squamous cell
carcinoma through downregulating SMO expression in the Hedgehog
signaling pathway. Genes Genomics. 44:981–991. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Rashid S, Rashid S, Das P, Malik N, Dash
NR, Singh N, Pandey RM, Kumar L, Chauhan SS, Chosdol K, et al:
Elucidating the role of miRNA-326 modulating hedgehog signaling in
pancreatic carcinoma. Pancreas. 53:e42–e48. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Xu X, Li Y, Liu G, Li K, Chen P, Gao Y,
Liang W, Xi H, Wang X, Wei B, et al: MiR-378a-3p acts as a tumor
suppressor in gastric cancer via directly targeting RAB31 and
inhibiting the Hedgehog pathway proteins GLI1/2. Cancer Biol Med.
19:1662–1682. 2022.PubMed/NCBI
|
|
126
|
Chang W, Chang Q, Lu H, Li Y and Chen C:
MiR-221-3p facilitates thyroid cancer cell proliferation and
inhibit apoptosis by targeting FOXP2 through hedgehog pathway. Mol
Biotechnol. 64:919–927. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Hong XL, Yu TC, Huang XW, Wang JL, Sun TT,
Yan TT, Zhou CB, Chen HM, Su WY, Du W and Xiong H: Metformin
abrogates Fusobacterium nucleatum-induced chemoresistance in
colorectal cancer by inhibiting miR-361-5p/sonic hedgehog
signaling-regulated stemness. Br J Cancer. 128:363–374. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Huseni MA, Wang L, Klementowicz JE, Yuen
K, Breart B, Orr C, Liu LF, Li Y, Gupta V, Li C, et al:
CD8+ T cell-intrinsic IL-6 signaling promotes resistance
to anti-PD-L1 immunotherapy. Cell Rep Med. 4:1008782023. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Ding L, Sheriff S, Sontz RA and Merchant
JL: Schlafen4+-MDSC in Helicobacter-induced
gastric metaplasia reveals role for GTPases. Front Immunol.
14:11393912023. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Liu Y, Kim HG, Dong E, Dong C, Huang M,
Liu Y, Liangpunsakul S and Dong XC: Sesn3 deficiency promotes
carcinogen-induced hepatocellular carcinoma via regulation of the
hedgehog pathway. Biochim Biophys Acta Mol Basis Dis.
1865:2685–2693. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Bao R, Stapor D and Luke JJ: Molecular
correlates and therapeutic targets in T cell-inflamed versus non-T
cell-inflamed tumors across cancer types. Genome Med. 12:902020.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Wang Y, Jin G, Li Q, Wang Z, Hu W, Li P,
Li S, Wu H, Kong X, Gao J and Li Z: Hedgehog signaling
Non-canonical activated by pro-inflammatory cytokines in pancreatic
ductal adenocarcinoma. J Cancer. 7:2067–2076. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Ding J, Li HY, Zhang L, Zhou Y and Wu J:
Hedgehog signaling, a critical pathway governing the development
and progression of hepatocellular carcinoma. Cells. 10:1232021.
View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Chang WH, Forde D and Lai AG: A novel
signature derived from immunoregulatory and hypoxia genes predicts
prognosis in liver and five other cancers. J Transl Med. 17:142019.
View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Chang WH and Lai AG: Aberrations in
Notch-Hedgehog signalling reveal cancer stem cells harbouring
conserved oncogenic properties associated with hypoxia and
immunoevasion. Br J Cancer. 121:666–678. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Zhang J, Fan J, Zeng X, Nie M, Luan J,
Wang Y, Ju D and Yin K: Hedgehog signaling in gastrointestinal
carcinogenesis and the gastrointestinal tumor microenvironment.
Acta Pharm Sin B. 11:609–620. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Sternberg C, Gruber W, Eberl M, Tesanovic
S, Stadler M, Elmer DP, Schlederer M, Grund S, Roos S, Wolff F, et
al: Synergistic cross-talk of hedgehog and interleukin-6 signaling
drives growth of basal cell carcinoma. Int J Cancer. 143:2943–2954.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Laner-Plamberger S, Wolff F,
Kaser-Eichberger A, Swierczynski S, Hauser-Kronberger C, Frischauf
AM and Eichberger T: Hedgehog/GLI signaling activates suppressor of
cytokine signaling 1 (SOCS1) in epidermal and neural tumor cells.
PLoS One. 8:e753172013. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Lee JJ, Rothenberg ME, Seeley ES, Zimdahl
B, Kawano S, Lu WJ, Shin K, Sakata-Kato T, Chen JK, Diehn M, et al:
Control of inflammation by stromal Hedgehog pathway activation
restrains colitis. Proc Natl Acad Sci USA. 113:E7545–E7553. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Zhou X, Liu Z, Jang F, Xiang C, Li Y and
He Y: Autocrine Sonic hedgehog attenuates inflammation in
cerulein-induced acute pancreatitis in mice via upregulation of
IL-10. PLoS One. 7:e441212012. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Fendrich V, Esni F, Garay MV, Feldmann G,
Habbe N, Jensen JN, Dor Y, Stoffers D, Jensen J, Leach SD and
Maitra A: Hedgehog signaling is required for effective regeneration
of exocrine pancreas. Gastroenterology. 135:621–631. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Nakamura K and Smyth MJ: Targeting
cancer-related inflammation in the era of immunotherapy. Immunol
Cell Biol. 95:325–332. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Wu Y, Guo W, Wang T, Liu Y, Mullor MDMR,
Rodrìguez RA, Zhao S and Wei R: The comprehensive landscape of
prognosis, immunity, and function of the GLI family by pan-cancer
and single-cell analysis. Aging (Albany NY). 16:5123–5148. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Lecoultre M, Dutoit V and Walker PR:
Phagocytic function of tumor-associated macrophages as a key
determinant of tumor progression control: A review. J Immunother
Cancer. 8:e0014082020. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Koh V, Chakrabarti J, Torvund M, Steele N,
Hawkins JA, Ito Y, Wang J, Helmrath MA, Merchant JL, Ahmed SA, et
al: Hedgehog transcriptional effector GLI mediates mTOR-Induced
PD-L1 expression in gastric cancer organoids. Cancer Lett.
518:59–71. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Zhu L, Yang Y, Li H, Xu L, You H, Liu Y,
Liu Z, Liu X, Zheng D, Bie J, et al: Exosomal microRNAs induce
tumor-associated macrophages via PPARγ during tumor progression in
SHH medulloblastoma. Cancer Lett. 535:2156302022. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Grund-Gröschke S, Ortner D, Szenes-Nagy
AB, Zaborsky N, Weiss R, Neureiter D, Wipplinger M, Risch A,
Hammerl P, Greil R, et al: Epidermal activation of Hedgehog
signaling establishes an immunosuppressive microenvironment in
basal cell carcinoma by modulating skin immunity. Mol Oncol.
14:1930–1946. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Hinshaw DC, Hanna A, Lama-Sherpa T, Metge
B, Kammerud SC, Benavides GA, Kumar A, Alsheikh HA, Mota M, Chen D,
et al: Hedgehog signaling regulates metabolism and polarization of
mammary tumor-associated macrophages. Cancer Res. 81:5425–5437.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Petty AJ, Li A, Wang X, Dai R, Heyman B,
Hsu D, Huang X and Yang Y: Hedgehog signaling promotes
tumor-associated macrophage polarization to suppress intratumoral
CD8+ T cell recruitment. J Clin Invest. 129:5151–5162. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Hanna A, Metge BJ, Bailey SK, Chen D,
Chandrashekar DS, Varambally S, Samant RS and Shevde LA: Inhibition
of Hedgehog signaling reprograms the dysfunctional immune
microenvironment in breast cancer. Oncoimmunology. 8:15482412019.
View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Petty AJ, Dai R, Lapalombella R, Baiocchi
RA, Benson DM, Li Z, Huang X and Yang Y: Hedgehog-induced PD-L1 on
tumor-associated macrophages is critical for suppression of
tumor-infiltrating CD8+ T cell function. JCI Insight.
6:e1467072021. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Marcovecchio PM, Thomas G and
Salek-Ardakani S: CXCL9-expressing tumor-associated macrophages:
New players in the fight against cancer. J Immunother Cancer.
9:e0020452021. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Chen Z, Li H, Li Z, Chen S, Huang X, Zheng
Z, Qian X, Zhang L, Long G, Xie J, et al: SHH/GLI2-TGF-β1 feedback
loop between cancer cells and tumor-associated macrophages
maintains epithelial-mesenchymal transition and endoplasmic
reticulum homeostasis in cholangiocarcinoma. Pharmacol Res.
187:1065642023. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Holla S, Stephen-Victor E, Prakhar P,
Sharma M, Saha C, Udupa V, Kaveri SV, Bayry J and Balaji KN:
Mycobacteria-responsive sonic hedgehog signaling mediates
programmed death-ligand 1- and prostaglandin E2-induced regulatory
T cell expansion. Sci Rep. 6:241932016. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Hinshaw DC, Benavides GA, Metge BJ, Swain
CA, Kammerud SC, Alsheikh HA, Elhamamsy A, Chen D, Darley-Usmar V,
Rathmell JC, et al: Hedgehog signaling regulates treg to Th17
conversion through metabolic rewiring in breast cancer. Cancer
Immunol Res. 11:687–702. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Giammona A, Crivaro E and Stecca B:
Emerging roles of hedgehog signaling in cancer immunity. Int J Mol
Sci. 24:13212023. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Ji W, Niu X, Yu Y, Li Z, Gu L and Lu S:
SMO mutation predicts the effect of immune checkpoint inhibitor:
From NSCLC to multiple cancers. Front Immunol. 13:9558002022.
View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Holokai L, Chakrabarti J, Broda T, Chang
J, Hawkins JA, Sundaram N, Wroblewski LE, Peek RM Jr, Wang J,
Helmrath M, et al: Increased programmed Death-ligand 1 is an early
epithelial cell response to Helicobacter pylori Infection.
PLoS Pathog. 15:e10074682019. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Jiang J, Ding Y, Chen Y, Lu J, Chen Y, Wu
G, Xu N, Wang H and Teng L: Pan-cancer analyses reveal that
increased Hedgehog activity correlates with tumor immunosuppression
and resistance to immune checkpoint inhibitors. Cancer Med.
11:847–863. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Mazumdar T, Sandhu R, Qadan M, DeVecchio
J, Magloire V, Agyeman A, Li B and Houghton JA: Hedgehog signaling
regulates telomerase reverse transcriptase in human cancer cells.
PLoS One. 8:e752532013. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Villarinho NJ, Vasconcelos FDC, Mazzoccoli
L, da Silva Robaina MC, Pessoa LS, Siqueira PET, Maia RC, de
Oliveira DM, Leite de Sampaio E, Spohr TC and Lopes GF: Effects of
long-term exposure to MST-312 on lung cancer cells tumorigenesis:
Role of SHH/GLI-1 axis. Cell Biol Int. 46:1468–1479. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Bernabé-García M, Martínez-Balsalobre E,
García-Moreno D, García-Castillo J, Revilla-Nuin B, Blanco-Alcaina
E, Mulero V, Alcaraz-Pérez F and Cayuela ML: Telomerase reverse
transcriptase activates transcription of miR500A to inhibit
Hedgehog signalling and promote cell invasiveness. Mol Oncol.
15:1818–1834. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Wu L, Wang S, Tang B, Tang L, Lei Y, Liu
Y, Yang M, Yang G, Zhang D and Liu E: Human telomerase reverse
transcriptase (hTERT) synergistic with Sp1 upregulate Gli1
expression and increase gastric cancer invasion and metastasis. J
Mol Histol. 52:1165–1175. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Sivakumar S, Qi S, Cheng N, Sathe AA,
Kanchwala M, Kumar A, Evers BM, Xing C and Yu H: TP53 promotes
lineage commitment of human embryonic stem cells through
ciliogenesis and sonic hedgehog signaling. Cell Rep. 38:1103952022.
View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Cain JE and Watkins DN: p53 and RB1
regulate Hedgehog responsiveness via autophagy-mediated
ciliogenesis. Mol Cell Oncol. 7:18050952020. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Zhang L, Zhang Y, Li K and Xue S: Hedgehog
signaling and the glioma-associated oncogene in cancer
radioresistance. Front Cell Dev Biol. 11:12571732023. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Zhou C, Du J, Zhao L, Liu W, Zhao T, Liang
H, Fang P, Zhang K and Zeng H: GLI1 reduces drug sensitivity by
regulating cell cycle through PI3K/AKT/GSK3/CDK pathway in acute
myeloid leukemia. Cell Death Dis. 12:2312021. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Sun Y, Wang Y, Fan C, Gao P, Wang X, Wei G
and Wei J: Estrogen promotes stemness and invasiveness of
ER-positive breast cancer cells through Gli1 activation. Mol
Cancer. 13:1372014. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Wang Y, Wang H, Yan Z, Li G, Hu G, Zhang
H, Huang D, Wang Y, Zhang X, Yan Y, et al: The critical role of
dysregulated Hh-FOXM1-TPX2 signaling in human hepatocellular
carcinoma cell proliferation. Cell Commun Signal. 18:1162020.
View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Zhang Z, Hao C, Zhang R, Pei X, Li J and
Wang L: A Gli inhibitor GANT61 suppresses cell proliferation,
promotes cell apoptosis and induces G1/G0 cycle retardation with a
dose- and time-dependent manner through inhibiting Notch pathway in
multiple myeloma. Cell Cycle. 19:2063–2073. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Qu C, Liu Y, Kunkalla K, Singh RR, Blonska
M, Lin X, Agarwal NK and Vega F: Trimeric G protein-CARMA1 axis
links smoothened, the hedgehog receptor transducer, to NF-κB
activation in diffuse large B-cell lymphoma. Blood. 121:4718–4728.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Kasperczyk H, Baumann B, Debatin KM and
Fulda S: Characterization of sonic hedgehog as a novel NF-kappaB
target gene that promotes NF-kappaB-mediated apoptosis resistance
and tumor growth in vivo. FASEB J. 23:21–33. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Kurita S, Mott JL, Almada LL, Bronk SF,
Werneburg NW, Sun SY, Roberts LR, Fernandez-Zapico ME and Gores GJ:
GLI3-dependent repression of DR4 mediates hedgehog antagonism of
TRAIL-induced apoptosis. Oncogene. 29:4848–4858. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Wu X, Zhang LS, Toombs J, Kuo YC, Piazza
JT, Tuladhar R, Barrett Q, Fan CW, Zhang X, Walensky LD, et al:
Extra-mitochondrial prosurvival BCL-2 proteins regulate gene
transcription by inhibiting the SUFU tumour suppressor. Nat Cell
Biol. 19:1226–1236. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Fingas CD, Mertens JC, Razumilava N, Sydor
S, Bronk SF, Christensen JD, Rizvi SH, Canbay A, Treckmann JW, Paul
A, et al: Polo-like kinase 2 is a mediator of hedgehog survival
signaling in cholangiocarcinoma. Hepatology. 58:1362–1374. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Meister MT, Boedicker C, Linder B, Kögel
D, Klingebiel T and Fulda S: Concomitant targeting of Hedgehog
signaling and MCL-1 synergistically induces cell death in
Hedgehog-driven cancer cells. Cancer Lett. 465:1–11. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Trieu KG, Tsai SY, Eberl M, Ju V, Ford NC,
Doane OJ, Peterson JK, Veniaminova NA, Grachtchouk M, Harms PW, et
al: Basal cell carcinomas acquire secondary mutations to overcome
dormancy and progress from microscopic to macroscopic disease. Cell
Rep. 39:1107792022. View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Koeniger A, Brichkina A, Nee I, Dempwolff
L, Hupfer A, Galperin I, Finkernagel F, Nist A, Stiewe T, Adhikary
T, et al: Activation of Cilia-independent Hedgehog/GLI1 signaling
as a novel concept for neuroblastoma therapy. Cancers (Basel).
13:19082021. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Wang J, Huang S, Tian R, Chen J, Gao H,
Xie C, Shan Y, Zhang Z, Gu S and Xu M: The protective autophagy
activated by GANT-61 in MYCN amplified neuroblastoma cells is
mediated by PERK. Oncotarget. 9:14413–14427. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Azatyan A, Gallo-Oller G, Diao Y,
Selivanova G, Johnsen JI and Zaphiropoulos PG: RITA downregulates
Hedgehog-GLI in medulloblastoma and rhabdomyosarcoma via
JNK-dependent but p53-independent mechanism. Cancer Lett.
442:341–350. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Li S, Wang J, Lu Y, Zhao Y, Prinz RA and
Xu X: Inhibition of the sonic hedgehog pathway activates
TGF-β-activated kinase (TAK1) to induce autophagy and suppress
apoptosis in thyroid tumor cells. Cell Death Dis. 12:4592021.
View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Londero AP, Orsaria M, Viola L, Marzinotto
S, Bertozzi S, Galvano E, Andreetta C and Mariuzzi L: Survivin,
sonic hedgehog, krüppel-like factors, and p53 pathway in serous
ovarian cancer: an immunohistochemical study. Hum Pathol.
127:92–101. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Sun Y, Fang Q, Liu W, Liu Y and Zhang C:
GANT-61 induces cell cycle resting and autophagy by down-regulating
RNAP III signal pathway and tRNA-Gly-CCC synthesis to combate
chondrosarcoma. Cell Death Dis. 14:4612023. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Deng H, Huang L, Liao Z, Liu M, Li Q and
Xu R: Itraconazole inhibits the Hedgehog signaling pathway thereby
inducing autophagy-mediated apoptosis of colon cancer cells. Cell
Death Dis. 11:5392020. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Klingseisen V, Slanovc J, Regouc M and
Hrzenjak A: Bisdemethoxycurcumin sensitizes the response of
cisplatin resistant non-small cell lung carcinoma cell lines by
activating apoptosis and autophagy. J Nutr Biochem. 106:1090032022.
View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Kaushal JB, Bhatia R, Kanchan RK, Raut P,
Mallapragada S, Ly QP, Batra SK and Rachagani S: Repurposing
niclosamide for targeting pancreatic cancer by inhibiting Hh/Gli
Non-canonical axis of Gsk3β. Cancers (Basel). 13:31052021.
View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Chen J, Wen B, Wang Y, Wu S, Zhang X, Gu
Y, Wang Z, Wang J, Zhang W and Yong J: Jervine exhibits anticancer
effects on nasopharyngeal carcinoma through promoting autophagic
apoptosis via the blockage of Hedgehog signaling. Biomed
Pharmacother. 132:1108982020. View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Bissey PA, Mathot P, Guix C, Jasmin M,
Goddard I, Costechareyre C, Gadot N, Delcros JG, Mali SM, Fasan R,
et al: Blocking SHH/patched interaction triggers tumor growth
inhibition through patched-induced apoptosis. Cancer Res.
80:1970–1980. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Delloye-Bourgeois C, Gibert B, Rama N,
Delcros JG, Gadot N, Scoazec JY, Krauss R, Bernet A and Mehlen P:
Sonic Hedgehog promotes tumor cell survival by inhibiting CDON
pro-apoptotic activity. PLoS Biol. 11:e10016232013. View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Kabarriti R and Guha C: Hedgehog signaling
and radiation induced liver injury: A delicate balance. Hepatol
Int. 8:316–320. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Gan GN, Eagles J, Keysar SB, Wang G,
Glogowska MJ, Altunbas C, Anderson RT, Le PN, Morton JJ, Frederick
B, et al: Hedgehog signaling drives radioresistance and
stroma-driven tumor repopulation in head and neck squamous cancers.
Cancer Res. 74:7024–7036. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Wu Z, Huang C, Li R, Li H, Lu H and Lin Z:
PRKCI Mediates radiosensitivity via the Hedgehog/GLI1 pathway in
cervical cancer. Front Oncol. 12:8871392022. View Article : Google Scholar : PubMed/NCBI
|
|
193
|
Sims-Mourtada J, Izzo JG, Apisarnthanarax
S, Wu TT, Malhotra U, Luthra R, Liao Z, Komaki R, van der Kogel A,
Ajani J and Chao KS: Hedgehog: An attribute to tumor regrowth after
chemoradiotherapy and a target to improve radiation response. Clin
Cancer Res. 12:6565–6572. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Metge BJ, Alsheikh HA, Chen D, Elhamamsy
AR, Hinshaw DC, Chen BR, Sleckman BP, Samant RS and Shevde LA:
Ribosome biosynthesis and Hedgehog activity are cooperative
actionable signaling mechanisms in breast cancer following
radiotherapy. NPJ Precis Oncol. 7:612023. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Al-Azab M, Wang B, Elkhider A, Walana W,
Li W, Yuan B, Ye Y, Tang Y, Almoiliqy M, Adlat S, et al: Indian
Hedgehog regulates senescence in bone marrow-derived mesenchymal
stem cell through modulation of ROS/mTOR/4EBP1, p70S6K1/2 pathway.
Aging (Albany NY). 12:5693–5715. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Domen A, Deben C, Verswyvel J, Flieswasser
T, Prenen H, Peeters M, Lardon F and Wouters A: Cellular senescence
in cancer: clinical detection and prognostic implications. J Exp
Clin Cancer Res. 41:3602022. View Article : Google Scholar : PubMed/NCBI
|
|
197
|
Tamayo-Orrego L, Wu CL, Bouchard N,
Khedher A, Swikert SM, Remke M, Skowron P, Taylor MD and Charron F:
Evasion of cell senescence leads to medulloblastoma progression.
Cell Rep. 14:2925–2937. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Liu Z, Tu K, Wang Y, Yao B, Li Q, Wang L,
Dou C, Liu Q and Zheng X: Hypoxia accelerates aggressiveness of
hepatocellular carcinoma cells involving oxidative stress,
epithelial-mesenchymal transition and non-canonical hedgehog
signaling. Cell Physiol Biochem. 44:1856–1868. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Wei M, Ma R, Huang S, Liao Y, Ding Y, Li
Z, Guo Q, Tan R, Zhang L and Zhao L: Oroxylin A increases the
sensitivity of temozolomide on glioma cells by hypoxia-inducible
factor 1α/hedgehog pathway under hypoxia. J Cell Physiol.
234:17392–17404. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Xu QH, Xiao Y, Li XQ, Fan L, Zhou CC,
Cheng L, Jiang ZD and Wang GH: Resveratrol counteracts
Hypoxia-Induced gastric cancer invasion and EMT through hedgehog
pathway suppression. Anticancer Agents Med Chem. 20:1105–1114.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Spivak-Kroizman TR, Hostetter G, Posner R,
Aziz M, Hu C, Demeure MJ, Von Hoff D, Hingorani SR, Palculict TB,
Izzo J, et al: Hypoxia triggers hedgehog-mediated tumor-stromal
interactions in pancreatic cancer. Cancer Res. 73:3235–3247. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Zhou J, Wu K, Gao D, Zhu G, Wu D, Wang X,
Chen Y, Du Y, Song W, Ma Z, et al: Reciprocal regulation of
hypoxia-inducible factor 2α and GLI1 expression associated with the
radioresistance of renal cell carcinoma. Int J Radiat Oncol Biol
Phys. 90:942–951. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
203
|
Xu M, Wang J, Li H, Zhang Z and Cheng Z:
AIM2 inhibits colorectal cancer cell proliferation and migration
through suppression of Gli1. Aging (Albany NY). 13:1017–1031. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Yin L, Cao R, Liu Z, Luo G, Li Y, Zhou X,
Chen X, Wu Y, He J, Zu X and Shen Y: FUNDC2, a mitochondrial outer
membrane protein, mediates triple-negative breast cancer
progression via the AKT/GSK3β/GLI1 pathway. Acta Biochim Biophys
Sin (Shanghai). 55:1770–1783. 2023.PubMed/NCBI
|
|
205
|
Sriramulu S, Sun XF, Malayaperumal S,
Ganesan H, Zhang H, Ramachandran M, Banerjee A and Pathak S:
Emerging role and clinicopathological significance of AEG-1 in
different cancer types: A concise review. Cells. 10:14972021.
View Article : Google Scholar : PubMed/NCBI
|
|
206
|
Inaguma S, Riku M, Ito H, Tsunoda T, Ikeda
H and Kasai K: GLI1 orchestrates CXCR4/CXCR7 signaling to enhance
migration and metastasis of breast cancer cells. Oncotarget.
6:33648–33657. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
207
|
Lu JT, Zhao WD, He W and Wei W: Hedgehog
signaling pathway mediates invasion and metastasis of
hepatocellular carcinoma via ERK pathway. Acta Pharmacol Sin.
33:691–700. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
208
|
Peralta-Arrieta I, Trejo-Villegas OA,
Armas-López L, Ceja-Rangel HA, Ordóñez-Luna MDC, Pineda-Villegas P,
González-López MA, Ortiz-Quintero B, Mendoza-Milla C,
Zatarain-Barrón ZL, et al: Failure to EGFR-TKI-based therapy and
tumoural progression are promoted by MEOX2/GLI1-mediated epigenetic
regulation of EGFR in the human lung cancer. Eur J Cancer.
160:189–205. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Harada K, Ohashi R, Naito K and Kanki K:
Hedgehog signal inhibitor GANT61 inhibits the malignant behavior of
undifferentiated hepatocellular carcinoma cells by targeting
Non-Canonical GLI signaling. Int J Mol Sci. 21:31262020. View Article : Google Scholar : PubMed/NCBI
|
|
210
|
Stecca B, Mas C, Clement V, Zbinden M,
Correa R, Piguet V, Beermann F and Ruiz i Altaba A: Melanomas
require HEDGEHOG-GLI signaling regulated by interactions between
GLI1 and the RAS-MEK/AKT pathways. Proc Natl Acad Sci USA.
104:5895–5900. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
211
|
Chen J, Zhou X, Yang J, Sun Q, Liu Y, Li
N, Zhang Z and Xu H: Circ-GLI1 promotes metastasis in melanoma
through interacting with p70S6K2 to activate Hedgehog/GLI1 and
Wnt/β-catenin pathways and upregulate Cyr61. Cell Death Dis.
11:5962020. View Article : Google Scholar : PubMed/NCBI
|
|
212
|
Wasson CW, Caballero-Ruiz B, Gillespie J,
Derrett-Smith E, Mankouri J, Denton CP, Canettieri G, Riobo-Del
Galdo NA and Del Galdo F: Induction of Pro-Fibrotic CLIC4 in dermal
fibroblasts by TGF-β/Wnt3a is mediated by GLI2 upregulation. Cells.
11:5302022. View Article : Google Scholar : PubMed/NCBI
|
|
213
|
You X, Wu J, Wang Y, Liu Q, Cheng Z, Zhao
X, Liu G, Huang C, Dai J, Zhou Y, et al: Galectin-1 promotes
vasculogenic mimicry in gastric adenocarcinoma via the Hedgehog/GLI
signaling pathway. Aging (Albany NY). 12:21837–21853. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
214
|
Guo E, Liu H and Liu X: Overexpression of
SCUBE2 Inhibits proliferation, migration, and invasion in glioma
cells. Oncol Res. 25:437–444. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
215
|
Gialmanidis IP, Bravou V, Amanetopoulou
SG, Varakis J, Kourea H and Papadaki H: Overexpression of hedgehog
pathway molecules and FOXM1 in non-small cell lung carcinomas. Lung
Cancer. 66:64–74. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
216
|
Xu X, Su B, Xie C, Wei S, Zhou Y, Liu H,
Dai W, Cheng P, Wang F, Xu X and Guo C: Sonic hedgehog-Gli1
signaling pathway regulates the epithelial mesenchymal transition
(EMT) by mediating a new target gene, S100A4, in pancreatic cancer
cells. PLoS One. 9:e964412014. View Article : Google Scholar : PubMed/NCBI
|
|
217
|
Joost S, Almada LL, Rohnalter V, Holz PS,
Vrabel AM, Fernandez-Barrena MG, McWilliams RR, Krause M,
Fernandez-Zapico ME and Lauth M: GLI1 inhibition promotes
epithelial-to-mesenchymal transition in pancreatic cancer cells.
Cancer Res. 72:88–99. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
218
|
Sirkisoon SR, Carpenter RL, Rimkus T,
Anderson A, Harrison A, Lange AM, Jin G, Watabe K and Lo HW:
Interaction between STAT3 and GLI1/tGLI1 oncogenic transcription
factors promotes the aggressiveness of triple-negative breast
cancers and HER2-enriched breast cancer. Oncogene. 37:2502–2514.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
219
|
Sirkisoon SR, Carpenter RL, Rimkus T,
Doheny D, Zhu D, Aguayo NR, Xing F, Chan M, Ruiz J, Metheny-Barlow
LJ, et al: TGLI1 transcription factor mediates breast cancer brain
metastasis via activating metastasis-initiating cancer stem cells
and astrocytes in the tumor microenvironment. Oncogene. 39:64–78.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
220
|
Fan YH, Ding J, Nguyen S, Liu XJ, Xu G,
Zhou HY, Duan NN, Yang SM, Zern MA and Wu J: Aberrant hedgehog
signaling is responsible for the highly invasive behavior of a
subpopulation of hepatoma cells. Oncogene. 35:116–124. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
221
|
Sirkisoon SR, Wong GL, Aguayo NR, Doheny
DL, Zhu D, Regua AT, Arrigo A, Manore SG, Wagner C, Thomas A, et
al: Breast cancer extracellular vesicles-derived miR-1290 activates
astrocytes in the brain metastatic microenvironment via the
FOXA2→CNTF axis to promote progression of brain metastases. Cancer
Lett. 540:2157262022. View Article : Google Scholar : PubMed/NCBI
|
|
222
|
Doheny D, Manore S, Sirkisoon SR, Zhu D,
Aguayo NR, Harrison A, Najjar M, Anguelov M, Cox AO, Furdui CM, et
al: An FDA-Approved antifungal, ketoconazole, and its novel
derivative suppress tGLI1-Mediated breast cancer brain metastasis
by inhibiting the DNA-Binding activity of brain
metastasis-promoting transcription factor tGLI1. Cancers (Basel).
14:42562022. View Article : Google Scholar : PubMed/NCBI
|
|
223
|
Cao X, Geradts J, Dewhirst MW and Lo HW:
Upregulation of VEGF-A and CD24 gene expression by the tGLI1
transcription factor contributes to the aggressive behavior of
breast cancer cells. Oncogene. 31:104–115. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
224
|
Di Mauro C, Rosa R, D'Amato V, Ciciola P,
Servetto A, Marciano R, Orsini RC, Formisano L, De Falco S,
Cicatiello V, et al: Hedgehog signalling pathway orchestrates
angiogenesis in triple-negative breast cancers. Br J Cancer.
116:1425–1435. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
225
|
Bausch D, Fritz S, Bolm L, Wellner UF,
Fernandez-Del-Castillo C, Warshaw AL, Thayer SP and Liss AS:
Hedgehog signaling promotes angiogenesis directly and indirectly in
pancreatic cancer. Angiogenesis. 23:479–492. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
226
|
Maiti S, Mondal S, Satyavarapu EM and
Mandal C: mTORC2 regulates hedgehog pathway activity by promoting
stability to Gli2 protein and its nuclear translocation. Cell Death
Dis. 8:e29262017. View Article : Google Scholar : PubMed/NCBI
|
|
227
|
Wang Y, Ding Q, Yen CJ, Xia W, Izzo JG,
Lang JY, Li CW, Hsu JL, Miller SA, Wang X, et al: The crosstalk of
mTOR/S6K1 and hedgehog pathways. Cancer Cell. 21:374–387. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
228
|
Wang W, Yan T, Guo W, Niu J, Zhao Z, Sun
K, Zhang H, Yu Y and Ren T: Constitutive GLI1 expression in
chondrosarcoma is regulated by major vault protein via mTOR/S6K1
signaling cascade. Cell Death Differ. 28:2221–2237. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
229
|
Youssef M, Moussa N, W Helmy M and Haroun
M: Unraveling the therapeutic potential of GANT61/Dactolisib
combination as a novel prostate cancer modality. Med Oncol.
39:1432022. View Article : Google Scholar : PubMed/NCBI
|
|
230
|
Griffiths SC, Schwab RA, El Omari K,
Bishop B, Iverson EJ, Malinauskas T, Dubey R, Qian M, Covey DF,
Gilbert RJC, et al: Hedgehog-Interacting protein is a multimodal
antagonist of hedgehog signalling. Nat Commun. 12:71712021.
View Article : Google Scholar : PubMed/NCBI
|
|
231
|
Rhim AD, Oberstein PE, Thomas DH, Mirek
ET, Palermo CF, Sastra SA, Dekleva EN, Saunders T, Becerra CP,
Tattersall IW, et al: Stromal elements act to restrain, rather than
support, pancreatic ductal adenocarcinoma. Cancer Cell. 25:735–747.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
232
|
Johar D, Elmehrath AO, Khalil RM, Elberry
MH, Zaky S, Shalabi SA and Bernstein LH: Protein networks linking
Warburg and reverse Warburg effects to cancer cell metabolism.
Biofactors. 47:713–728. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
233
|
Wu J, Di D, Zhao C, Pan Q, Liu Y, Zhang X,
Zhao X and Chen H: Clinical significance of Gli-1 And Caveolin-1
expression in the human small cell lung cancer. Asian Pac J Cancer
Prev. 19:401–406. 2018.PubMed/NCBI
|
|
234
|
Di Magno L, Manzi D, D'Amico D, Coni S,
Macone A, Infante P, Di Marcotullio L, De Smaele E, Ferretti E,
Screpanti I, et al: Druggable glycolytic requirement for
Hedgehog-dependent neuronal and medulloblastoma growth. Cell Cycle.
13:3404–3413. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
235
|
Liu Z, Hayashi H, Matsumura K, Ogata Y,
Sato H, Shiraishi Y, Uemura N, Miyata T, Higashi T, Nakagawa S, et
al: Hyperglycaemia induces metabolic reprogramming into a
glycolytic phenotype and promotes epithelial-mesenchymal
transitions via YAP/TAZ-Hedgehog signalling axis in pancreatic
cancer. Br J Cancer. 128:844–856. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
236
|
Su J, Xie Q and Xie L: Identification and
validation of a metabolism-related gene signature for predicting
the prognosis of paediatric medulloblastoma. Sci Rep. 14:75402024.
View Article : Google Scholar : PubMed/NCBI
|
|
237
|
D'Amico D, Antonucci L, Di Magno L, Coni
S, Sdruscia G, Macone A, Miele E, Infante P, Di Marcotullio L, De
Smaele E, et al: Non-canonical Hedgehog/AMPK-Mediated control of
polyamine metabolism supports neuronal and medulloblastoma cell
growth. Dev Cell. 35:21–35. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
238
|
Luo J, Gong L, Yang Y, Zhang Y, Liu Q, Bai
L, Fang X, Zhang B, Huang J, Liu M, et al: Enhanced mitophagy
driven by ADAR1-GLI1 editing supports the self-renewal of cancer
stem cells in HCC. Hepatology. 79:61–78. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
239
|
Deng S, Gu H, Chen Z, Liu Y, Zhang Q, Chen
D and Yi S: PTCH1 mutation as a potential predictive biomarker for
immune checkpoint inhibitors in gastrointestinal cancer.
Carcinogenesis. 45:351–357. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
240
|
Pak E, MacKenzie EL, Zhao X, Pazyra-Murphy
MF, Park PMC, Wu L, Shaw DL, Addleson EC, Cayer SS, Lopez BG, et
al: A large-scale drug screen identifies selective inhibitors of
class I HDACs as a potential therapeutic option for SHH
medulloblastoma. Neuro Oncol. 21:1150–1163. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
241
|
Pandey S, Lee M, Lim J, Park S, Choung YH,
Kim JE, Garg P and Chung JH: SMO-CRISPR-mediated apoptosis in
CD133-targeted cancer stem cells and tumor growth inhibition. J
Control Release. 357:94–108. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
242
|
Thazhackavayal Baby B, Kulkarni AM, Gayam
PKR, Harikumar KB and Aranjani JM: Beyond cyclopamine: Targeting
Hedgehog signaling for cancer intervention. Arch Biochem Biophys.
754:1099522024. View Article : Google Scholar : PubMed/NCBI
|
|
243
|
Whitson RJ, Lee A, Urman NM, Mirza A, Yao
CY, Brown AS, Li JR, Shankar G, Fry MA, Atwood SX, et al:
Noncanonical hedgehog pathway activation through SRF-MKL1 promotes
drug resistance in basal cell carcinomas. Nat Med. 24:271–281.
2018. View Article : Google Scholar : PubMed/NCBI
|