Cancer is a complex group of diseases characterized
by the uncontrolled growth and spread of abnormal cells in the
body. With the capacity to afflict any anatomical site and
propagate to adjacent tissues or distant organs, cancer poses a
formidable global health challenge (1,2). The
World Health Organization estimates that cancer accounted for
nearly 10 million deaths worldwide in 2020. Notably, the incidence
of several cancer types continues to increase, imposing an onerous
burden on healthcare systems and affected individuals (3). Research efforts have been instrumental
in advancing our understanding of cancer biology, thereby
facilitating the development of targeted therapeutic modalities and
innovative treatment paradigms. Breakthroughs in precision
medicine, immunotherapy and genetic manipulation have shown
promising results in improving patient outcomes and prolonging
survival rates.
Early detection and screening programs play a
crucial role in cancer management by enabling the timely
identification of cancerous cells through regular screenings, such
as mammograms, colonoscopies and pap smears, ultimately leading to
better treatment outcomes (3).
Given the increasing prevalence of cancer and its
impact on individuals, families and societies, the importance of
current cancer research and treatment cannot be overstated.
Continued investments in scientific advancements and access to
quality care are essential to drive further progress and improve
the lives of cancer patients worldwide. Overall, the importance of
cancer lies in its severe impact on physical, emotional and
socioeconomic aspects of individuals and society, emphasizing the
need for sustained research, prevention efforts and improved access
to quality care (4).
Oncogenes, tumor suppressor gene and DNA repair
genes play pivotal roles in tumorigenesis, which is the process
wherein normal cells undergo malignant transformation (5–7).
Oncogenes, when activated or mutated, harbor the potential to
instigate cancer by fostering uncontrolled cellular proliferation
and aberrant signaling cascades (5). Conversely, tumor suppressor genes act
as guardians of cellular homeostasis, curtailing aberrant growth
and facilitating DNA repair mechanisms (6). DNA repair genes, as the name suggests,
are responsible for repairing DNA damage that occurs naturally or
due to external factors such as exposure to radiation or harmful
chemicals. The intricate interplay among these genes and their
functions is contingent upon the specific cancer type and
implicated genetic aberrations (7).
Understanding these mechanisms is indispensable for identifying
potential therapeutic targets and devising efficacious cancer
interventions (7,8).
The present review centers on elucidating the
pivotal role of LIM domain only 7 (LMO7) in tumorigenesis,
encompassing its molecular attributes, oncogenic functions,
clinical relevance and therapeutic implications. Of note, LMO7, a
multifunctional protein encoded by the LMO7 gene, has garnered
attention for its involvement in diverse physiological processes,
including neuronal development, cardiovascular health (9,10), and
notably, cancer pathogenesis (11,12).
Dysregulation of LMO7 has been implicated in various malignancies,
underscoring its potential as a prognostic marker and therapeutic
target. This review delineates the intricate molecular
characteristics and biological functions of LMO7, elucidates its
implications in cancer development across different organ systems,
and elaborates on the mechanistic underpinnings of its oncogenic
effects. Furthermore, it delineates the challenges inherent in
targeting LMO7 for therapeutic intervention and suggests avenues
for future research aimed at unraveling the complexities of
LMO7-mediated carcinogenesis.
The LMO7 gene encodes a protein called LMO7, which
is found in humans and is located on chromosome 13q14.11. The LMO7
protein is predominantly expressed in cardiac and skeletal muscle
tissues (13). LMO7 protein
encompasses multiple LIM domains, serving as protein-protein
interaction motifs implicated in cell growth, differentiation and
cytoskeletal organization (14–16).
In addition, LMO7 contains PDZ domains, facilitating interaction
modules that typically bind to specific protein sequences, and
which are involved in protein localization, signal transduction and
the assembly of protein complexes within cells (17,18).
LMO7 protein also contains CH domains, an actin-binding motif,
which may exist as a single copy or in tandem repeats, either
functioning autonomously or serving a regulatory role. CH domains
are found in cytoskeletal and signal transduction proteins,
including actin-binding proteins like spectrin, α-actinin,
dystrophin, utrophin and fimbrin, as well as proteins essential for
the regulation of cell shape (cortexillins) and signaling proteins
(Vav), and are crucial for regulating cell shape and signaling
(19,20). It has one CH domain at its
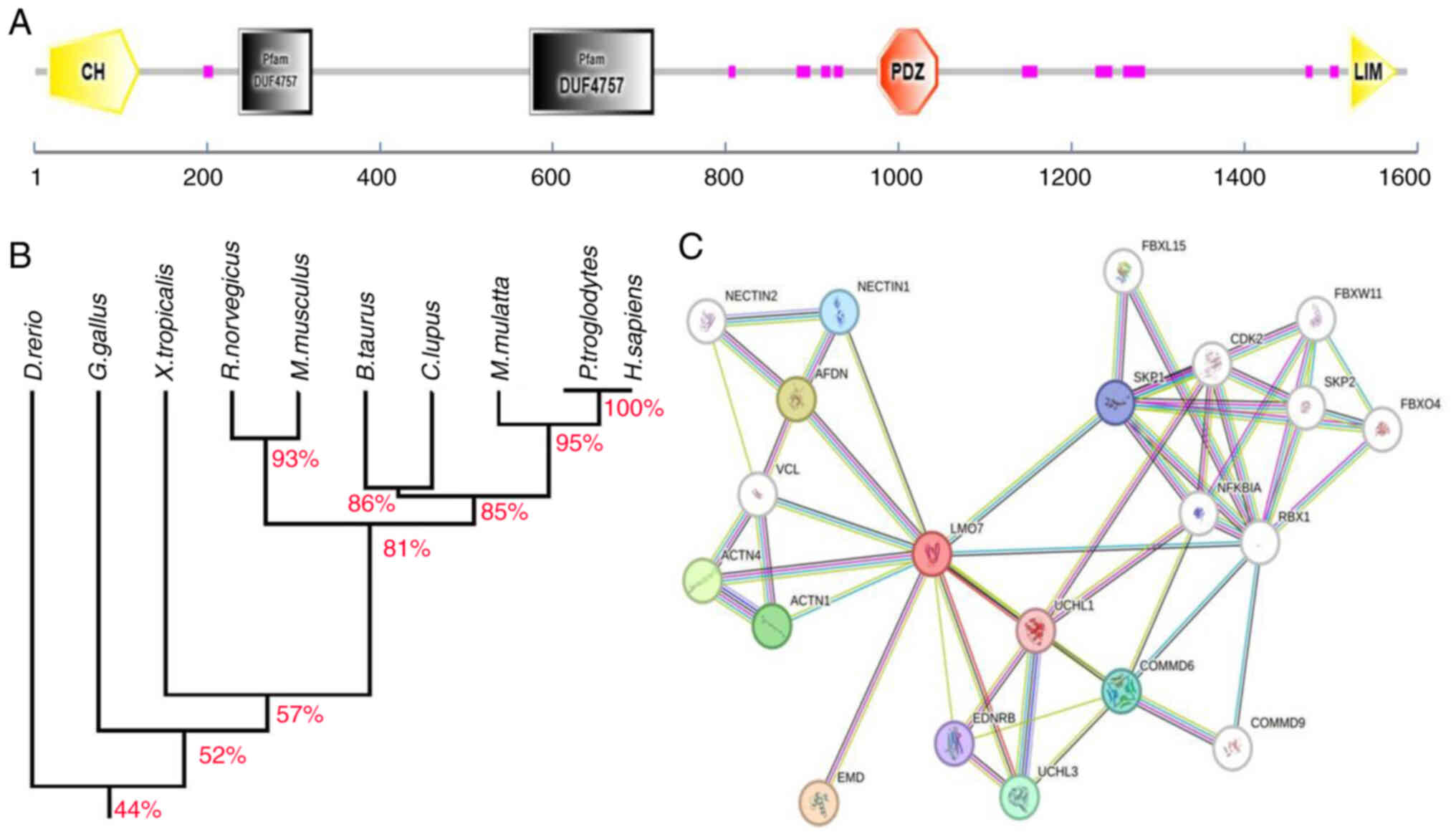
N-terminus, a PDZ domain in the middle region and two DUF4757
domains of unknown function (Fig.
1A). The human LMO7 protein shares a high degree of homology
with LMO7 proteins in other species, particularly in the CH, PDZ
and LIM domains (Fig. 1B).
It has been observed that LMO7 gathers at cell-cell
adhesion sites subsequent to the assembly of nectin-afadin and
E-cadherin-catenin complexes, thereby influencing cell migration,
intercellular communication and tissue organization (21). In Drosophila, the vertebrate
homolog of LMO7, Smash, interacts with Bazooka/Par-3, canoe/afadin,
and the tyrosine kinase Src42A, localizing to the zonula adherens
in a plane-polarized manner. Deletion of smash leads to severe
defects in the morphogenesis of embryonic epithelial tissues and
organs (22). In muscle cells, LMO7
functions as a transcription factor, regulating the expression of
various muscle genes, including paired box gene 3 (Pax3), Pax7,
myogenic differentiation antigen (MyoD) and myogenic factor 5,
thereby controlling myogenesis (11–13).
LMO7-null mice exhibit growth retardation, reduced fiber size, and
impaired skeletal muscle and cardiac function (11). Knockdown of chicken LMO7 diminishes
the number and width of myotubes and the number of MyoD-positive
myoblasts, a phenotype rescued by Wnt/β-catenin activation,
suggesting a role for LMO7 in the initial events of chick skeletal
muscle formation, particularly in myoblast survival (12).
As a critical adapter and scaffolding protein, LMO7
interacts directly or indirectly with ~20 proteins through its
three domains, thereby having vital biological functions (Fig. 1C). Using the Search Tool for the
Retrieval of Interacting Genes and Proteins (STRING) database
(string-db.org) to predict protein interactions of LMO7 revealed
its association with various proteins, such as enamel matrix
derivatives, Afadin (AFDN), actinin α 1/4 and COMM
domain-containing protein 6/9, among others, to exert its functions
in different tumors (Table I). With
these predicted interactions, future investigations could explore
the specific proteins with which LMO7 interacts to regulate
tumorigenesis in various tumor tissues.
LMO7 is a protein that has a role in cell adhesion,
cytoskeletal organization and cellular signaling pathways (23–25).
While extensive research is still needed to fully understand the
precise role of LMO7 in cancer, LMO7 has been implicated as an
oncogene in several types of cancer (11,12).
In breast cancer, elevated LMO7 levels correlate
with aggressive phenotypes characterized by enhanced cell migration
and invasiveness and are positively regulated by CUGBP Elav-like
family member 1 (26,27). Higher levels of LMO7 expression have
been associated with more aggressive breast cancer phenotypes,
including increased cell migration and invasion (26,28).
Serum response factor (SRF) regulates specific functions such as
muscle development and breast cancer metastasis. LMO7 orchestrates
the myocardin-related transcription factors (MRTFs) as coactivators
promoting cytoskeletal rearrangements conducive to cancer cell
motility (26,29). LMO7 is a specific regulator of MRTFs
and plays a vital role in breast cancer cell migration (26,30).
LMO7 activates MRTFs by relieving actin-mediated inhibition in
cooperation with Rho GTPase. Disruption of actin-RPEL interactions
eliminates Rho dependency, allowing strong Rho-independent
activation of LMO7 (30,31). LMO7 reduces the G-actin/F-actin
ratio by colocalizing with F-actin. Knockdown of LMO7 compromises
MRTF activities and impairs breast cancer cell migration (26,31).
LMO7 is upregulated in invasive breast carcinoma and correlates
with increased expression of SRF target genes regulating muscle and
actin cytoskeleton functions (23,32),
suggesting a cell-specific mechanism regulating Rho-MRTF-SRF
signaling and breast cancer cell migration (26).
Transcription factor krüppel-like factor 4 binds to
methylated CpGs at the enhancer regions of LMO7 and activates LMO7
expression via 3D chromatin loop formation with its promoter
regions, influencing cellular functions in human glioblastoma cells
(34).
LMO7 has been suggested to act as a tumor suppressor
for murine lung adenocarcinoma. LMO7-deficient mice develop
irregular and prominent epithelial lesions in terminal and
respiratory bronchioles at a young age, whereas these mice tend to
develop lung adenocarcinoma at an old age (28,35).
Leucine-rich repeats and immunoglobulin-like domains proteins 3
(LRIG3) interacts with LMO7 and LIM and calponin homology domains 1
(LIMCH1), with co-localization and co-immunoprecipitation observed
between LRIG1/3 and LMO7/LIMCH1 (36). LMO7 and LIMCH1 are highly expressed
in normal lung tissue but reduced in malignant tissue. LMO7
immunoreactivity predicts poor prognosis in LRIG1-positive tumors
(36). MicroRNA (miR)-96, as a
serum biomarker for lung cancer, inhibits the expression of LMO7 by
binding to its 3′-UTR, ultimately regulating lung carcinogenesis
through the miR-96-LMO7 axis (37).
LMO7 has been found to be overexpressed in
pancreatic cancer (PC), with recent studies identifying LMO7 as a
potential prognostic marker for PC (38,39).
Its overexpression is associated with tumor progression and poor
patient survival. Studies have shown that knockdown or knockout of
LMO7 in mouse PC cells leads to PC cell cycle arrest and apoptosis,
significantly inhibiting PC cell proliferation, anchorage-free
colony formation, migration in vitro, and slowing down the
growth and metastasis of pancreatic carcinoma in vivo
(28,40).
In inflammatory hepatocellular adenomas (IHCA),
among the five IHCA cases with fyn related Src family tyrosine
kinase (FRK) gene rearrangements, LMO7 was identified to be fused
to the exon 3–8 region of FRK (39). In tumor cells, human genes use
alternative transcription start sites (TSS) to control mRNA levels
and expand transcriptional output, thereby promoting carcinogenesis
(41,42). A study analyzing 108 colorectal
cancer samples using exome arrays identified multiple genes,
including LMO7, relative to normal mucosa, showing
tumor-specific alternative TSS use in both adenoma and cancer
samples (28,43).
LMO7 is upregulated in human malignancies, including
colorectal cancer, with no transcriptional upregulation of the LMO7
observed in adenomas compared with normal mucosa (28,44,45).
However, upregulation of LMO7 transcription was observed in cancer.
LMO7 expression in primary tumors with p53 mutation was
significantly higher than that in tumors without p53 mutation
(45,46).
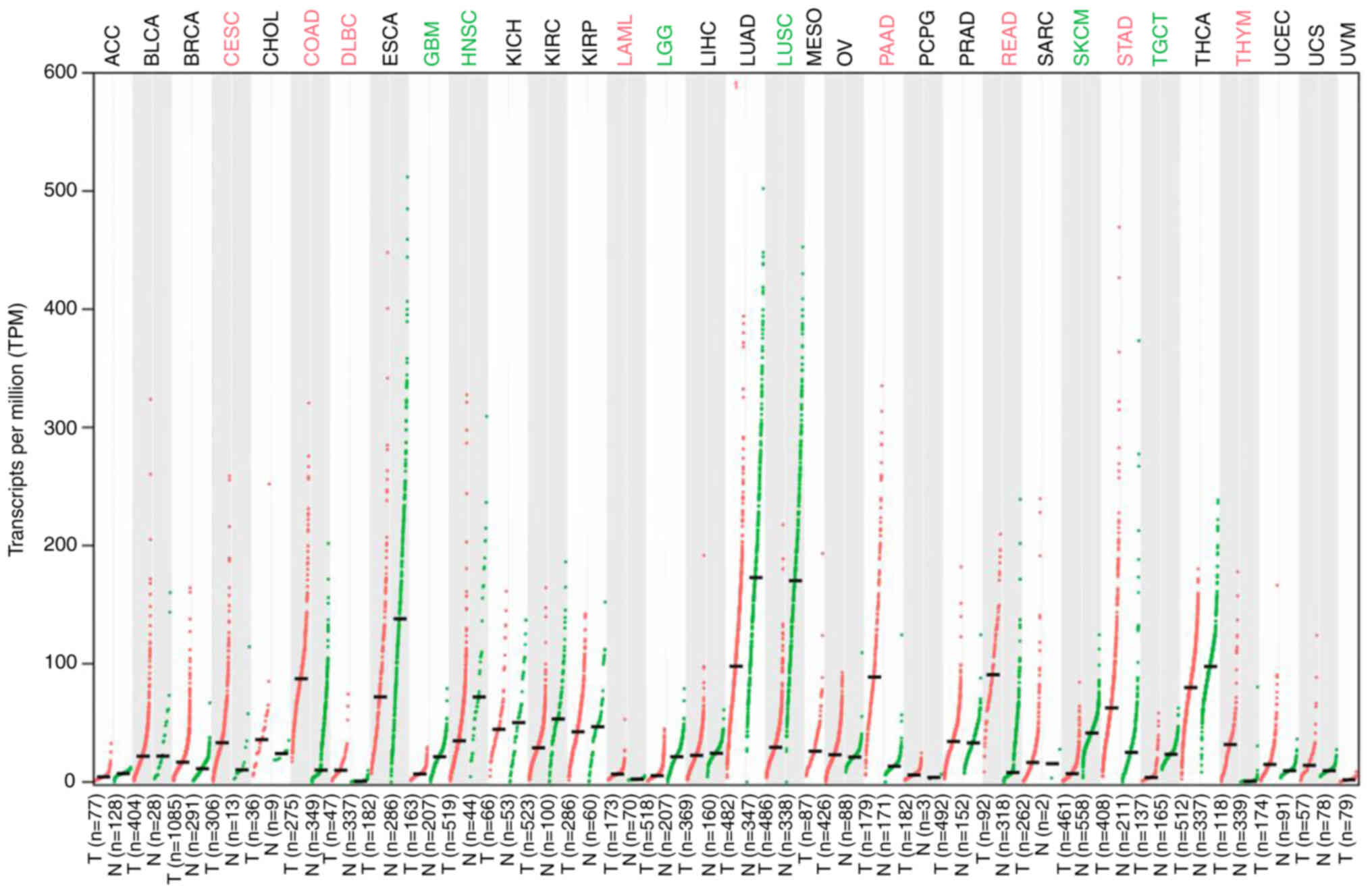
While LMO7 is not commonly discussed as a
well-established oncogene, emerging evidence suggests its potential
oncogenic role in certain cancers; LMO7 was upregulated in most
tumors, but downregulated in certain other tumors (Fig. 2) (28); data were from GEPIA 2 (http://gepia2.cancer-pku.cn/). It is important to note
that more research is needed to fully understand the molecular
mechanisms by which LMO7 functions as an oncogene in cancer. This
will help determine its potential as a therapeutic target and
develop strategies to inhibit its activity for the treatment of
specific cancers.
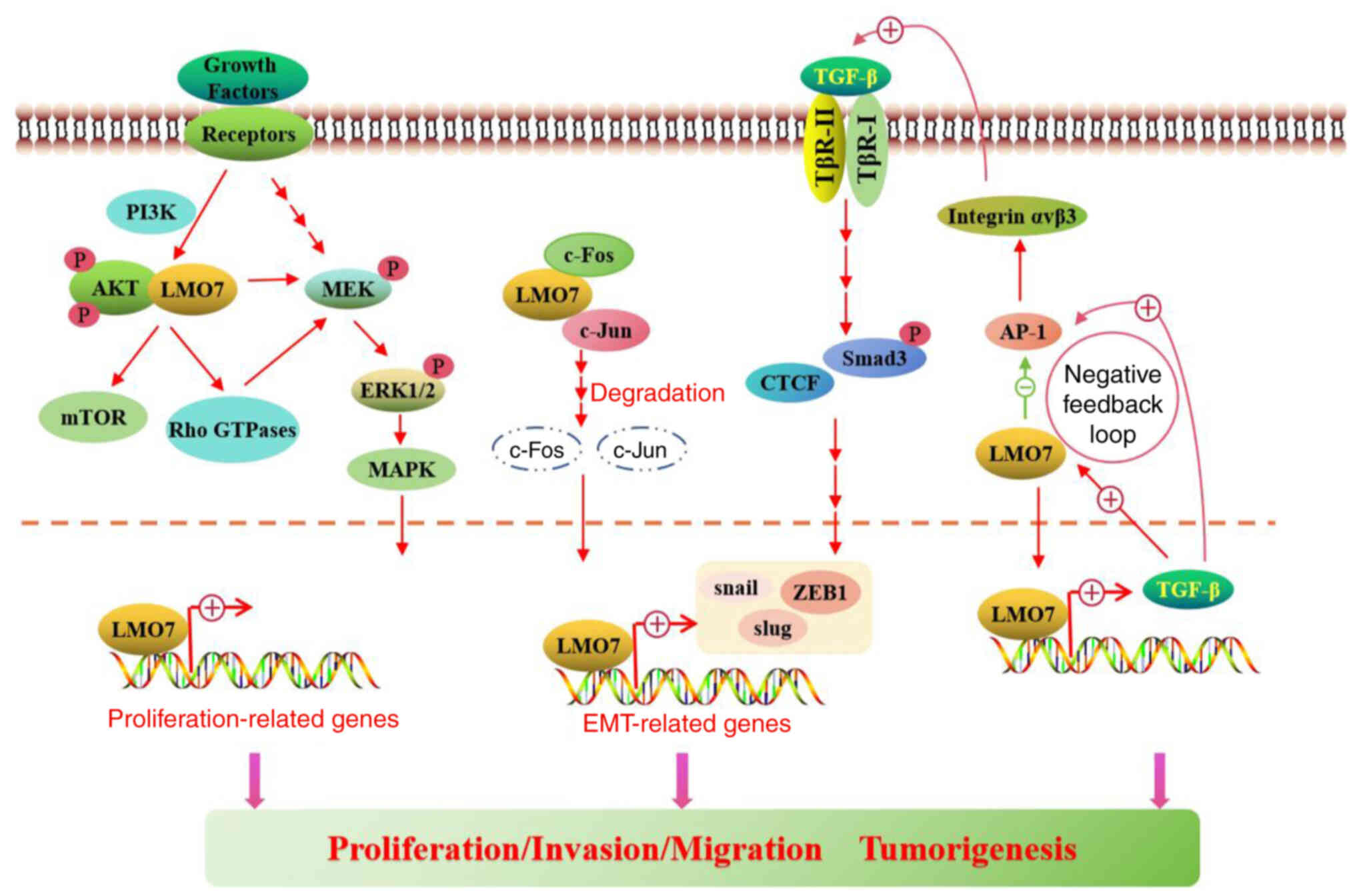
LMO7 interacts with AKT, a downstream effector of
PI3K, potentially contributing to the activation of AKT signaling
(49). This interaction suggests
that LMO7 may promote cell survival and growth by enhancing the
activity of the PI3K/AKT/mTOR pathway (49). Furthermore, LMO7 may modulate
Rho-GTPase activity, impacting cellular processes involved in
cancer progression, such as cell migration and invasion (Fig. 3) (50). In addition, LMO7 has been shown to
bind to and modulate the activity of MEK1, a kinase upstream of
ERK, potentially influencing cell proliferation and survival
through the regulation of the ERK/MAPK pathway (Fig. 3) (49). Further research is necessary to
fully understand the precise mechanisms and consequences of LMO7′s
interactions with these pathways in cancer development and
progression.
Epigenetic modifications play a critical role in
regulating gene expression and can contribute to cancer development
and progression (51). Evidence
suggests that epigenetic alterations may influence LMO7 expression
in different cancer types (29).
For instance, in breast cancer, LMO7 has been found to undergo
hypermethylation, resulting in reduced gene expression (47). Hypermethylation of the LMO7 promoter
region has been associated with tumor progression and poor patient
prognosis (52). Further
investigation into LMO7-associated epigenetic alterations may offer
insight into potential diagnostic, prognostic and therapeutic
strategies for cancer patients.
Furthermore, LMO7 has been identified as a TGF-β1
target gene in hepatoma cells, functioning in vascular physiology
and fibrosis (53). It is triggered
by injury and TGF-β in vascular smooth muscle cells in vitro
(53). Loss of LMO7 enhanced TGF-β
signaling by upregulating TGF-β1 mRNA, TGF-β protein,
integrin-αvβ3, latent TGF-β activation, downstream effectors Smad3
phosphorylation and connective tissue growth factor (53). Mechanistically, LMO7′s LIM domain
interacts with activator protein 1 transcription factors c-Fos and
c-Jun, promoting their degradation and interrupting activator
protein 1-dependent TGF-β autoinduction (53). A study has shown that the increase
in LMO7 mRNA expression synchronizes with TGF-β1-induced invasion,
with higher LMO7 expression in high metastatic cells compared to
low metastatic cells (36). Induced
by TGF-β, LMO7 may limit vascular fibrosis by negative feedback
regulation of the TGF-β pathway (Fig.
3), suggesting implications for intimal hyperplasia, wound
healing and fibrotic diseases and potentially impacting tumor
angiogenesis.
Researchers have found that the curcumin analogue
CA-5f
{(3E,5E)-3-(3,4-dimethoxybenzylidene)-5-[(1H-indol-3-yl)methylene]-1-methylpiperidin-4-one},
as an anticancer therapeutic agent (54), reduced LMO7 protein levels, which
induced the accumulation of microtubule-associated protein 1 light
chain 3β and sequestosome 1, increased mitochondrial reactive
oxygen species levels and then inhibited autophagosome-lysosome
fusion and induced cell death (55). It may thus be suggested that LMO7
may be the target of CA-5f, and related antisense RNA can be
designed from LMO7 to interfere with the expression of LMO7 and
inhibit the growth of tumor cells.
LMO7 also has a role in inflammation. In the mouse
model of autoimmune hepatitis (AIH), injection of bone marrow
mesenchymal stem cells (BMSCs) upregulated the levels of LMO7 and
downregulated the levels of AP-1 and TGF-β, while the expression of
AP-1 and TGF-β was upregulated in the LMO7 interference group.
BMSCs can significantly reduce liver injury in the mouse AIH model
by regulating the LMO7/AP-1/TGF-β signaling pathway to alleviate
liver fibrosis of autoimmune hepatitis (56). The expression of the LMO7 gene was
found to be upregulated in gastric epithelial AGS cells (a gastric
cancer cell line) infected with Helicobacter pylori, which
is involved in the adhesion, invasion and possibly proliferation of
gastric epithelial cells (57). The
expression of LMO7 in H. pylori may be involved in the
carcinogenesis or differentiation of gastric epithelial cells
through direct interactions with other proteins. The proteins that
bind to LMO7 and the functional role of protein interactions in
cell adhesion should be investigated in gastric epithelial cells
stimulated by H. pylori. AFDN, vinculin and other proteins
that were mentioned in Table I play
a role in cell-cell adherens junctions (Table I).
The clinical relevance of LMO7 in cancer extends
beyond its prognostic value to encompass its potential as a
therapeutic target for precision medicine approaches. Of note,
elevated LMO7 expression correlates with adverse clinical outcomes
and poor patient prognosis across various cancer types, thereby
underscoring its utility as a prognostic biomarker (37,47).
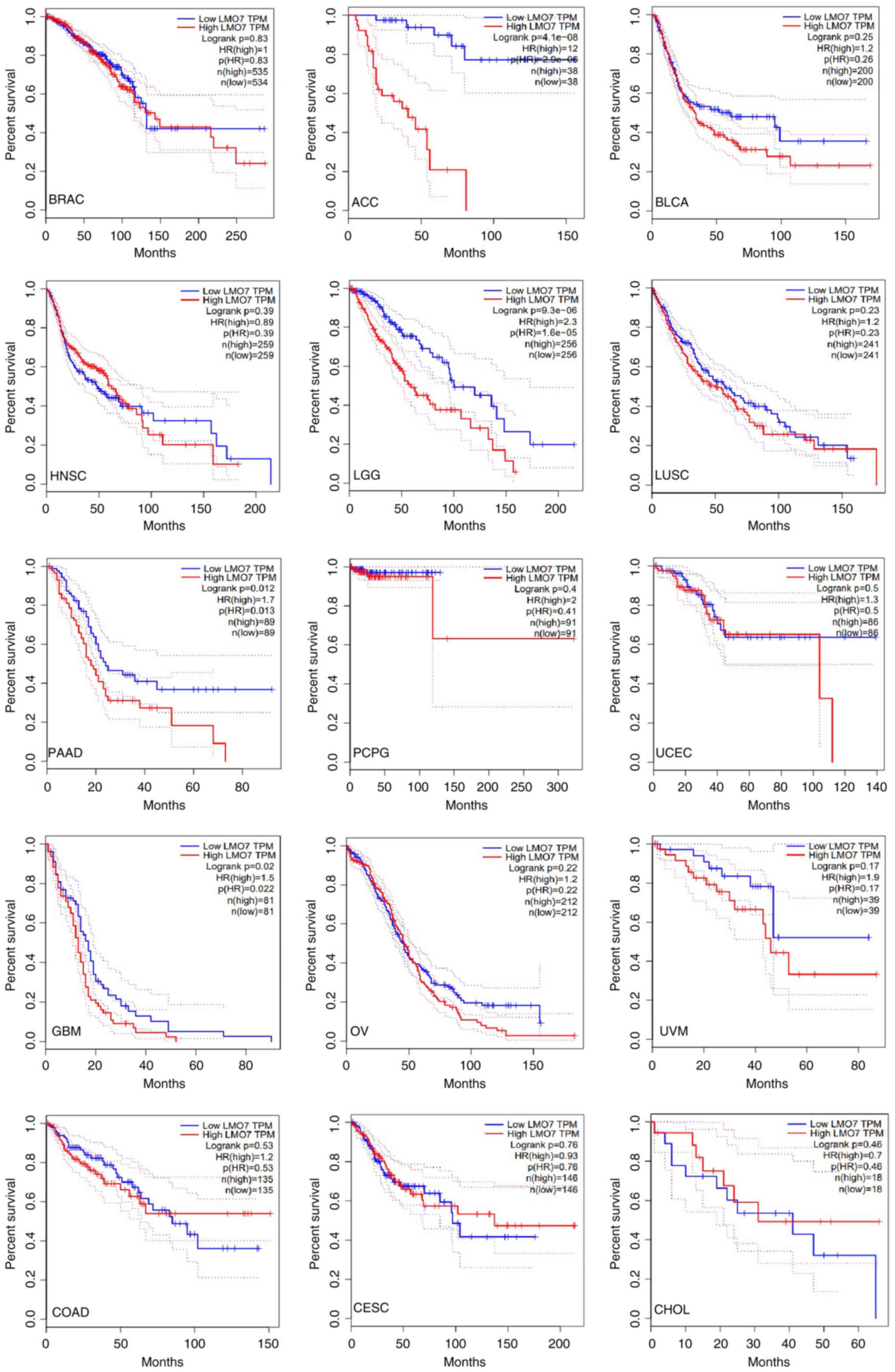
However, it is important to note that the prognostic relevance of
LMO7 may vary across different cancer types and stages (Fig. 4) (28); data were from GEPIA 2.
Compared with human papillomavirus (HPV)-dependent
disease, patients with HPV-independent vulvar squamous cell
carcinoma (VSCC) have a survival deficit and data suggest that
leucine rich repeats and immunoglobulin like domains 2 (LRIG2) and
LMO7 are positive prognostic factors in HPV-independent cases; LMO7
is a positive prognostic factor in the most advanced tumors.
Therefore, these markers may provide tools for individual treatment
strategy selection in patients with VSCC. However, more studies are
needed to further elucidate the functional and prognostic value of
the molecular markers studied in VSCC (58). High expression of LMO7 antisense RNA
1 was associated with poor survival in kidney cancer (59).
LMO7, along with aminopeptidase like 1, von
Willebrand factor, aldehyde dehydrogenase 2 family member, NUAK
family kinase 1 and tumor protein, translationally-controlled 1,
named the castration-resistant PC-derived prognosis signature, is
considered an important molecular signature for predicting
progression-free survival (PFS) of patients with
castration-resistant PC for therapeutic decision-making (60).
In breast cancer, elevated LMO7 expression
correlates with an unfavorable patient prognosis (46,47).
Similarly, in colorectal cancer, LMO7 expression serves as a
potential prognostic marker, with higher levels associated with
adverse patient outcomes, including reduced overall survival and
disease-free survival (46,47,50).
In addition, increased LMO7 expression correlates with advanced
tumor stages, lymph node metastasis and poor tumor differentiation
in colorectal cancer cases (44,45).
In lung cancer, multiple studies consistently link increased LMO7
expression with unfavorable patient outcomes, including diminished
overall survival and disease-free survival rates (Fig. 4) (28,34–36).
However, contradictory evidence suggests that elevated LMO7 levels
may paradoxically be associated with improved overall survival
(Fig. 4) (28). The association between LMO7
expression and prognosis in other cancer types remains to be
elucidated. Further research, considering additional factors, is
essential to establish LMO7 as a reliable and independent
prognostic biomarker in specific cancer contexts.
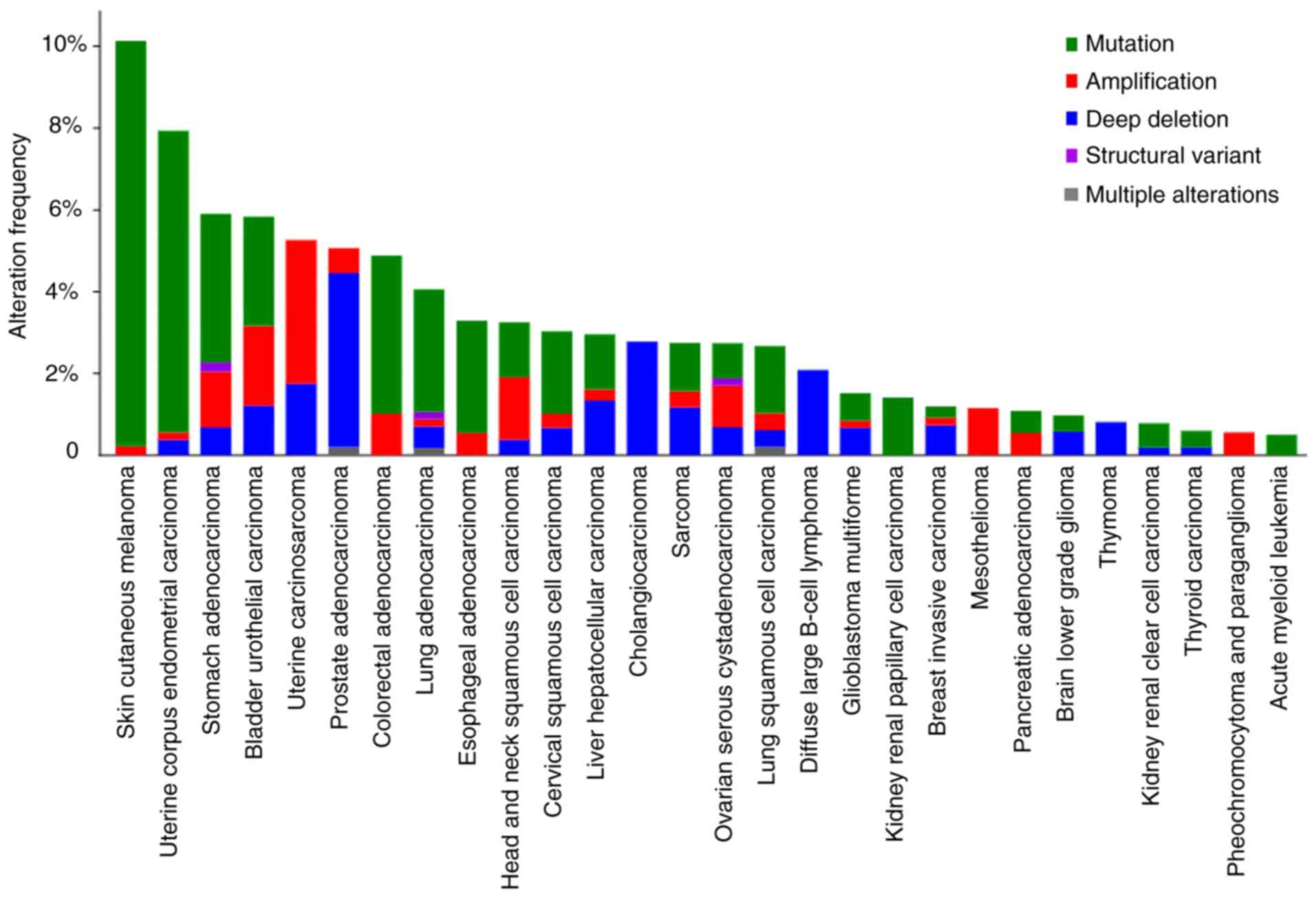
LMO7 mutations have been identified within tumor
tissues, implicating their role in cancer development and
progression. These mutations encompass various types, broadly
categorized as mutation, structural variant, amplification, deep
deletion and multiple alterations (Fig.
5) (61,62); data were from cBioPortal for Cancer
Genomics (http://www.cbioportal.org). An
analysis in cBioPortal revealed the presence of two missense
mutations (P928Q and S1259R) in LMO7 among cancer patients. In
addition, LMO7 demonstrated high-level amplification in lung
adenocarcinoma, leading to LMO7-ITGBL1 fusion and a Q1334* nonsense
mutation (61,62). Deep deletion of LMO7 has been
observed in prostate adenocarcinoma and lung squamous cell
carcinoma genomes, resulting in TPTE2P5-LMO7 fusion and an I774V
missense mutation, respectively (62). It is important to note that the
specific consequences of LMO7 mutations in tumor tissues can vary
based on the mutation and cancer context. Further research is
imperative to comprehensively understand the impact of LMO7
mutations on tumorigenesis and potentially identify novel
therapeutic targets.
Although the expression pattern of LMO7 has been
explored across various cancer types, its precise molecular
functions in tumorigenesis and cancer progression remain largely
elusive. Further research is imperative to identify the specific
mechanisms through which LMO7 promotes cancer development and
metastasis. Numerous studies investigating LMO7 and cancer have
relied on relatively small sample sizes, potentially constraining
the generalizability and statistical power of their findings
(61,62). Robust larger-scale studies are
necessary to corroborate the significance of LMO7 across different
cancer types (63–65). The heterogeneity observed within
cancer samples, including variations in tumor stage, grade and
molecular subtypes, complicates the interpretation of the role of
LMO7. Investigating LMO7 expression and its correlation with
patient outcomes across large, well-characterized cohorts is
necessary to better understand its clinical relevance. Standardized
methodologies for studying LMO7 in cancer research are lacking,
with studies employing diverse experimental approaches like
immunohistochemistry, gene expression profiling and functional
assays, posing challenges for comparison and integration of
findings across studies. The functional characterization of LMO7 in
cancer has mainly relied on in vitro cell culture
experiments and xenograft models (28–30).
The development of more sophisticated preclinical models is crucial
for validating its role in cancer progression, as these models fail
to fully replicate the complex tumor microenvironment and
organismal interactions.
Targeting LMO7 for cancer treatment shows promise
due to its involvement in various cellular processes contributing
to tumorigenesis (26–31,66,67).
However, several challenges need to be addressed before LMO7-based
therapies can be effectively developed. These include the
complexity of LMO7 function, its context-dependent roles in
different cancer types and the absence of specific inhibitors or
modulators targeting LMO7. The diverse functions of LMO7 in cell
adhesion, cytoskeletal organization, signal transduction and gene
regulation make it difficult to selectively target its activity
without interfering with important cellular processes (23–25).
Interfering with LMO7 function may lead to unintended consequences,
undesirable side effects or the limitation of therapeutic efficacy.
The role and expression levels of LMO7 may vary among different
cancer types and even among subtypes of the same cancer, posing
challenges for targeted therapies. Further studies are needed to
understand the precise mechanism by which LMO7 is involved in the
genesis of different cancer types and to identify reliable
biomarkers for patient stratification. Currently, there is a lack
of specific inhibitors or modulators selectively targeting LMO7.
High-throughput screening and rational drug design methods can be
utilized to identify small molecules or peptides interfering with
LMO7 interactions or functional domains.
To overcome the challenges in understanding the role
of LMO7 in various cancer types, continued research is essential.
This includes conducting comprehensive studies to investigate the
molecular mechanisms by which LMO7 promotes tumor progression and
metastasis. Additional experiments using cellular and animal models
can provide valuable insight into its specific contributions to
cancer development.
In PC, prediction of PFS and castration resistance
molecular characteristics is crucial for making treatment
decisions, but current methods lack reliability. Researchers have
applied the Robust Rank Aggregation method to analyze the
transcriptome profile of PC to identify 287 differentially
expressed genes, including LMO7 (68,69).
However, this work has yet to be validated and clinically
applied.
Further research is needed to elucidate the role of
LMO7 as a diagnostic and prognostic biomarker. If LMO7 is confirmed
as an oncogene, strategies for therapeutic targeting can be
explored, such as developing small-molecule inhibitors specific to
LMO7 or gene therapy approaches to suppress its expression.
In-depth molecular characterization of LMO7 and its associated
signaling pathways can help identify potential drug targets and
combination therapies to overcome challenges such as drug
resistance and metastasis. Analyzing LMO7 expression levels in
patient samples through techniques such as immunohistochemistry can
establish correlations between LMO7 expression and clinical
outcomes, including patient survival and response to therapy.
Longitudinal studies can also provide information about dynamic
changes in LMO7 expression during disease progression and its
potential as a predictive biomarker or therapeutic target. In
addition, the development of reliable biomarkers associated with
LMO7 may aid in monitoring treatment response and disease
progression.
The present review delved into the intricate role of
LMO7 in cancer development, elucidating the molecular mechanisms
underlying its oncogenic effects. With its involvement across
various cancer types and potential as a prognostic marker and
therapeutic target, understanding the function of LMO7 offers
valuable insight into cancer biology and paves the way for
precision medicine. However, further research is imperative to
overcome current limitations and fully harness the potential of
targeting LMO7 in cancer therapy. By unraveling the mysteries
surrounding LMO7, we can advance towards personalized and effective
cancer treatments.
Not applicable.
This work was supported by the Natural Science Foundation of
Hunan Province (grant no. 2024JJ5325) and the Excellent Youth
Project of Hunan Provincial Education Department (grant no.
22B0411).
The experimental data and the simulation results
that support the findings of this review are available from the
STRING database (string-db.org), GEPIA 2 (http://gepia2.cancer-pku.cn/) and cBioPortal
(http://www.cbioportal.org).
QZ and TJ: Substantial contributions to conception
and design of the work, acquisition, analysis and interpretation of
data for the study; drafting the manuscript or reviewing it
critically for important intellectual content. JW: Conception and
design, revising the manuscript, final approval of the version to
be published, agreement to be accountable for all aspects of the
work in ensuring that questions related to the accuracy or
integrity of any part of the work are appropriately investigated
and resolved. All authors have read and approved the final version
of the manuscript. Data authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
de Visser KE and Joyce JA: The evolving
tumor microenvironment: From cancer initiation to metastatic
outgrowth. Cancer Cell. 41:374–403. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Feinberg AP and Levchenko A: Epigenetics
as a mediator of plasticity in cancer. Science. 379:eaaw38352023.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang X, Chen G, Zhang Y, Ghareeb WM, Yu Q,
Zhu H, Lu X, Huang Y, Huang S, Hou D and Chi P: The impact of
circumferential tumour location on the clinical outcome of rectal
cancer patients managed with neoadjuvant chemoradiotherapy followed
by total mesorectal excision. Eur J Surg Oncol. 46:1118–1123. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Diori Karidio I and Sanlier SH: Reviewing
cancer's biology: An eclectic approach. J Egypt Natl Canc Inst.
33:322021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zeng Q and Jiang T: The role of FHL1 in
tumors. Gene. 11:1483472024. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dietlein F, Wang AB, Fagre C, Tang A,
Besselink NJM, Cuppen E, Li C, Sunyaev SR, Neal JT and Van Allen
EM: Genome-wide analysis of somatic noncoding mutation patterns in
cancer. Science. 376:eabg56012022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lou Z, Gong YQ, Zhou X and Hu GH: Low
expression of miR-199 in hepatocellular carcinoma contributes to
tumor cell hyper-proliferation by negatively suppressing XBP1.
Oncol Lett. 16:6531–6539. 2018.PubMed/NCBI
|
|
8
|
Li L, Wang S and Zhou W: Balance cell
apoptosis and pyroptosis of caspase-3-activating chemotherapy for
better antitumor therapy. Cancers (Basel). 15:262022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Du TT, Dewey JB, Wagner EL, Cui R, Heo J,
Park JJ, Francis SP, Perez-Reyes E, Guillot SJ, Sherman NE, et al:
LMO7 deficiency reveals the significance of the cuticular plate for
hearing function. Nat Commun. 10:11172019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang W, Xu Q, Su J, Tang L, Hao ZZ, Xu C,
Liu R, Shen Y, Sang X, Xu N, et al: Linking transcriptomes with
morphological and functional phenotypes in mammalian retinal
ganglion cells. Cell Rep. 40:1113222022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mull A, Kim G and Holaska JM: LMO7-null
mice exhibit phenotypes consistent with emery-dreifuss muscular
dystrophy. Muscle Nerve. 51:222–228. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Possidonio AC, Soares CP, Fontenele M,
Morris ER, Mouly V, Costa ML and Mermelstein C: Knockdown of Lmo7
inhibits chick myogenesis. FEBS Lett. 590:317–329. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gomes G, do Amaral MJ, Bagri KM,
Vasconcellos LM, Almeida MDS, Alvares LE and Mermelstein C: New
findings on LMO7 transcripts, proteins and regulatory regions in
human and vertebrate model organisms and the intracellular
distribution in skeletal muscle cells. Int J Mol Sci. 22:128852021.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
She M, Tang M, Jiang T and Zeng Q: The
roles of the LIM domain proteins in Drosophila cardiac and
hematopoietic morphogenesis. Front Cardiovasc Med. 8:6168512021.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
She M, Zhang J, Jiang T, Zhang Y, Liu Y,
Tang M and Zeng Q: The function of Lmpt in Drosophila heart
tissue. Biochem Biophys Res Commun. 612:15–21. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang J, She M, Dai Y, Nie X, Tang M and
Zeng Q: Lmpt regulates the function of Drosophila muscle by
acting as a repressor of Wnt signaling. Gene. 876:1475142023.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sánta A, Czajlik A, Batta G, Péterfia B
and Gáspári Z: Resonance assignment of the Shank1 PDZ domain.
Biomol NMR Assign. 16:121–127. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mieszczanek J, Strutt H, Rutherford TJ,
Strutt D, Bienz M and Gammons MV: Selective function of the PDZ
domain of Dishevelled in noncanonical Wnt signalling. J Cell Sci.
135:jcs2595472022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Palani S, Ghosh S, Ivorra-Molla E, Clarke
S, Suchenko A, Balasubramanian MK and Köster DV: Calponin-homology
domain mediated bending of membrane-associated actin filaments.
Elife. 10:e610782021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mei L, Reynolds MJ, Garbett D, Gong R,
Meyer T and Alushin GM: Structural mechanism for bidirectional
actin cross-linking by T-plastin. Proc Natl Acad Sci USA.
119:e22053701192022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ecke M, Prassler J and Gerisch G:
Expanding ring-shaped cleavage furrows in multinucleate cells. Mol
Biol Cell. 34:ar272023. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cuadrado M and Robles-Valero J: VAV
proteins as double agents in cancer: Oncogenes with tumor
suppressor roles. Biology (Basel). 10:8882021.PubMed/NCBI
|
|
23
|
Guérin A, Roy NH, Kugler EM, Berry L,
Burkhardt JK, Shin JB and Striepen B: Cryptosporidium rhoptry
effector protein ROP1 injected during invasion targets the host
cytoskeletal modulator LMO7. Cell Host Microbe. 29:1407–1420.e5.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li M, An Z, Tang Q, Ma Y, Yan J, Chen S
and Wang Y: Mixed responses to first-line alectinib in non-small
cell lung cancer patients with rare ALK gene fusions: A case series
and literature review. J Cell Mol Med. 25:9476–9481. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hariyanto NI, Yo EC and Wanandi SI:
Regulation and signaling of TGF-β autoinduction. Int J Mol Cell
Med. 10:234–247. 2021.PubMed/NCBI
|
|
26
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xia H, Chen D, Wu Q, Wu G, Zhou Y, Zhang Y
and Zhang L: CELF1 preferentially binds to exon-intron boundary and
regulates alternative splicing in HeLa cells. Biochim Biophys Acta
Gene Regul Mech. 1860:911–921. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45((W1)):
W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Miano JM, Long X and Fujiwara K: Serum
response factor: Master regulator of the actin cytoskeleton and
contractile apparatus. Am J Physiol Cell Physiol. 292:C70–C81.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Medjkane S, Perez-Sanchez C, Gaggioli C,
Sahai E and Treisman R: Myocardin-related transcription factors and
SRF are required for cytoskeletal dynamics and experimental
metastasis. Nat Cell Biol. 11:257–268. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pomiès P, Pashmforoush M, Vegezzi C, Chien
KR, Auffray C and Beckerle MC: The cytoskeleton-associated PDZ-LIM
protein, ALP, acts on serum response factor activity to regulate
muscle differentiation. Mol Biol Cell. 18:1723–1733. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kim D, Jung SH and Chung YJ: Development
of an RNA sequencing panel to detect gene fusions in thyroid
cancer. Genomics Inform. 19:e412021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
He H, Li W, Yan P, Bundschuh R, Killian
JA, Labanowska J, Brock P, Shen R, Heerema NA and de la Chapelle A:
Identification of a recurrent LMO7-BRAF fusion in papillary thyroid
carcinoma. Thyroid. 28:748–754. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Oyinlade O, Wei S, Kammers K, Liu S, Wang
S, Ma D, Huang ZY, Qian J, Zhu H, Wan J and Xia S: Analysis of KLF4
regulated genes in cancer cells reveals a role of DNA methylation
in promoter-enhancer interactions. Epigenetics. 13:751–768. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tanaka-Okamoto M, Hori K, Ishizaki H,
Hosoi A, Itoh Y, Wei M, Wanibuchi H, Mizoguchi A, Nakamura H and
Miyoshi J: Increased susceptibility to spontaneous lung cancer in
mice lacking LIM-domain only 7. Cancer Sci. 100:608–616. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nakamura H, Hori K, Tanaka-Okamoto M,
Higashiyama M, Itoh Y, Inoue M, Morinaka S and Miyoshi J: Decreased
expression of LMO7 and its clinicopathological significance in
human lung adenocarcinoma. Exp Ther Med. 2:1053–1057. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Karlsson T, Kvarnbrink S, Holmlund C,
Botling J, Micke P, Henriksson R, Johansson M and Hedman H: LMO7
and LIMCH1 interact with LRIG proteins in lung cancer, with
prognostic implications for early-stage disease. Lung Cancer.
125:174–184. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wu H, Zhou J, Mei S, Wu D, Mu Z, Chen B,
Xie Y, Ye Y and Liu J: Circulating exosomal microRNA-96 promotes
cell proliferation, migration and drug resistance by targeting
LMO7. J Cell Mol Med. 21:1228–1236. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ren B, Cui M, Yang G, Wang H, Feng M, You
L and Zhao Y: Tumor microenvironment participates in metastasis of
pancreatic cancer. Mol Cancer. 17:1082018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bayard Q, Caruso S, Couchy G, Rebouissou
S, Bioulac Sage P, Balabaud C, Paradis V, Sturm N, de Muret A,
Guettier C, et al: Recurrent chromosomal rearrangements of ROS1,
FRK and IL6 activating JAK/STAT pathway in inflammatory
hepatocellular adenomas. Gut. 69:1667–1676. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu X, Yuan H, Zhou J, Wang Q, Qi X,
Bernal C, Avella D, Kaifi JT, Kimchi ET, Timothy P, et al: LMO7 as
an unrecognized factor promoting pancreatic cancer progression and
metastasis. Front Cell Dev Biol. 9:6473872021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Davuluri RV, Suzuki Y, Sugano S, Plass C
and Huang TH: The functional consequences of alternative promoter
use in mammalian genomes. Trends Genet. 24:167–177. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Inchingolo MA, Diman A, Adamczewski M,
Humphreys T, Jaquier-Gubler P and Curran JA: TP53BP1, a dual-coding
gene, uses promoter switching and translational reinitiation to
express a smORF protein. iScience. 26:1067572023. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Thorsen K, Schepeler T, Øster B, Rasmussen
MH, Vang S, Wang K, Hansen KQ, Lamy P, Pedersen JS, Eller A, et al:
Tumor-specific usage of alternative transcription start sites in
colorectal cancer identified by genome-wide exon array analysis.
BMC Genomics. 12:5052011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Furuya M, Tsuji N, Endoh T, Moriai R,
Kobayashi D, Yagihashi A and Watanabe N: A novel gene containing
PDZ and LIM domains, PCD1, is overexpressed in human colorectal
cancer. Anticancer Res. 22:4183–4186. 2002.PubMed/NCBI
|
|
46
|
Kang S, Xu H, Duan X, Liu JJ, He Z, Yu F,
Zhou S, Meng XQ, Cao M and Kennedy GC: PCD1, a novel gene
containing PDZ and LIM domains, is overexpressed in several human
cancers. Cancer Res. 60:5296–5302. 2000.PubMed/NCBI
|
|
47
|
Jiang ZR, Yang LH, Jin LZ, Yi LM, Bing PP,
Zhou J and Yang JS: Identification of novel cuproptosis-related
lncRNA signatures to predict the prognosis and immune
microenvironment of breast cancer patients. Front Oncol.
12:9886802022. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Bao Z, Zeng W, Zhang D, Wang L, Deng X,
Lai J, Li J, Gong J and Xiang G: SNAIL induces EMT and lung
metastasis of tumours secreting CXCL2 to promote the invasion of
M2-type immunosuppressed macrophages in colorectal cancer. Int J
Biol Sci. 18:2867–2881. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chen J, Chen L, Hua J and Song W:
Long-term dynamic compression enhancement TGF-β3-induced
chondrogenesis in bovine stem cells: A gene expression analysis.
BMC Genom Data. 22:132021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hu Q, Guo C, Li Y, Aronow BJ and Zhang J:
LMO7 mediates cell-specific activation of the Rho-myocardin-related
transcription factor-serum response factor pathway and plays an
important role in breast cancer cell migration. Mol Cell Biol.
31:3223–3240. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sun L, Zhang H and Gao P: Metabolic
reprogramming and epigenetic modifications on the path to cancer.
Protein Cell. 13:877–919. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
He B, Dai C, Lang J, Bing P, Tian G, Wang
B and Yang J: A machine learning framework to trace tumor
tissue-of-origin of 13 types of cancer based on DNA somatic
mutation. Biochim Biophys Acta Mol Basis Dis. 1866:1659162020.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Xie Y, Ostriker AC, Jin Y, Hu H, Sizer AJ,
Peng G, Morris AH, Ryu C, Herzog EL, Kyriakides T, et al: LMO7 is a
negative feedback regulator of transforming growth factor β
signaling and fibrosis. Circulation. 139:679–693. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kim M and Moon A: A curcumin analog CA-5f
inhibits urokinase-type plasminogen activator and invasive
phenotype of triple-negative breast cancer cells. Toxicol Res.
38:19–26. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhang L, Qiang P, Yu J, Miao Y, Chen Z, Qu
J, Zhao Q, Chen Z, Liu Y, Yao X, et al: Identification of compound
CA-5f as a novel late-stage autophagy inhibitor with potent
anti-tumor effect against non-small cell lung cancer. Autophagy.
15:391–406. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Chen ZK, Chen DZ, Cai C, Jin LL, Xu J, Tu
YL, Huang XZ, Xu JL, Chen MZ, Xue FB, et al: BMSCs attenuate
hepatic fibrosis in autoimmune hepatitis through regulation of
LMO7-AP1-TGFβ signaling pathway. Eur Rev Med Pharmacol Sci.
25:1600–1611. 2021.PubMed/NCBI
|
|
57
|
Lim JW, Kim H and Kim KH: Cell
adhesion-related gene expression by Helicobacter pylori in
gastric epithelial AGS cells. Int J Biochem Cell Biol.
35:1284–1296. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhang Y, Liu Q, Cui M, Wang M, Hua S, Gao
J and Liao Q: Comprehensive analysis of expression, prognostic
value, and immune infiltration for ubiquitination-related FBXOs in
pancreatic ductal adenocarcinoma. Front Immunol. 12:7744352022.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Stefansson K, Oda H, Öfverman C, Lundin E,
Hedman H and Lindquist D: LRIG1-2 and LMO7 immunoreactivity in
vulvar squamous cell carcinoma: Association with prognosis in
relation to HPV-DNA and p16INK4a status. Oncol Rep. 42:142–150.
2019.PubMed/NCBI
|
|
60
|
Zheng H, Li BH, Liu C, Jia L and Liu FT:
Comprehensive analysis of lncRNA-mediated ceRNA crosstalk and
identification of prognostic biomarkers in Wilms' tumor. Biomed Res
Int. 2020:49516922020.PubMed/NCBI
|
|
61
|
A J, Zhang B, Zhang Z, Hu H and Dong JT:
Novel gene signatures predictive of patient recurrence-free
survival and castration resistance in prostate cancer. Cancers
(Basel). 13:9172021. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Mao X, Chen Y, Lu X, Jin S, Jiang P, Deng
Z, Zhu X, Cai Q, Wu C and Kang S: Tissue resident memory T cells
are enriched and dysfunctional in effusion of patients with
malignant tumor. J Cancer. 14:1223–1231. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Huang A and Zhou W: Mn-based cGAS-STING
activation for tumor therapy. Chin J Cancer Res. 35:19–43. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Feng S, Song G, Liu L, Liu W, Liang G and
Song Z: Allergen-specific immunotherapy induces monocyte-derived
dendritic cells but attenuates their maturation and cytokine
production in the lesional skin of an atopic dermatitis mouse
model. J Dermatol. 49:1310–1319. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Guo Z, Wang YJ, He BS and Zhou J:
Linc00312 single nucleotide polymorphism as biomarker for
chemoradiotherapy induced hematotoxicity in nasopharyngeal
carcinoma patients. Dis Markers. 2022:67078212022. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wang X, Yang T, Shi S, Xu C, Wang F, Dai
D, Guan G, Zhang Y, Wang S, Wang J, et al: Heterogeneity-induced
NGF-NGFR communication inefficiency promotes mitotic spindle
disorganization in exhausted T cells through PREX1 suppression to
impair the anti-tumor immunotherapy with PD-1 mAb in hepatocellular
carcinoma. Cancer Med. 13:e67362024. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Fu S, Duan L, Zhong Y and Zeng Y:
Comparison of surgical excision followed by adjuvant radiotherapy
and laser combined with steroids for the treatment of keloids: A
systematic review and meta-analysis. Int Wound J. 21:e144492023.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Lee B, Lee S, Lee Y, Park Y and Shim J:
Emerin represses STAT3 signaling through nuclear membrane-based
spatial control. Int J Mol Sci. 22:66692021. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Wu KY, Xie H, Zhang ZL, Li ZX, Shi L, Zhou
W, Zeng J, Tian Z, Zhang Y, Ding YB and Shen WG: Emerin knockdown
induces the migration and invasion of hepatocellular carcinoma
cells by up-regulating the cytoplasmic p21. Neoplasma. 69:59–70.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Awotoye W, Mossey PA, Hetmanski JB, Gowans
LJJ, Eshete MA, Adeyemo WL, Alade A, Zeng E, Adamson O, James O, et
al: Damaging mutations in AFDN contribute to risk of nonsyndromic
cleft lip with or without cleft palate. Cleft Palate Craniofac J.
61:697–705. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Berg HE, Greipp PT, Baughn LB, Falcon CP,
Jackson CC and Peterson JF: Detection of a cryptic KMT2A/AFDN gene
fusion [ins(6;11)(q27;q23q23)] in a pediatric patient with newly
diagnosed acute myeloid leukemia. Lab Med. 53:e95–e99. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Bill M, Mrózek K, Kohlschmidt J, Eisfeld
AK, Walker CJ, Nicolet D, Papaioannou D, Blachly JS, Orwick S,
Carroll AJ, et al: Mutational landscape and clinical outcome of
patients with de novo acute myeloid leukemia and rearrangements
involving 11q23/KMT2A. Proc Natl Acad Sci USA. 117:26340–26346.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Chen Q, Zhou XW, Zhang AJ and He K: ACTN1
supports tumor growth by inhibiting Hippo signaling in
hepatocellular carcinoma. J Exp Clin Cancer Res. 40:232021.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wang R, Gao Y and Zhang H: ACTN1 interacts
with ITGA5 to promote cell proliferation, invasion and
epithelial-mesenchymal transformation in head and neck squamous
cell carcinoma. Iran J Basic Med Sci. 26:200–207. 2023.PubMed/NCBI
|
|
77
|
Chen Q, Wang H, Li Z, Li F, Liang L, Zou
Y, Shen H, Li J, Xia Y, Cheng Z, et al: Circular RNA ACTN4 promotes
intrahepatic cholangiocarcinoma progression by recruiting YBX1 to
initiate FZD7 transcription. J Hepatol. 76:135–147. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Tentler D, Lomert E, Novitskaya K and
Barlev NA: Role of ACTN4 in tumorigenesis, metastasis, and EMT.
Cells. 8:14272019. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Singla A, Chen Q, Suzuki K, Song J,
Fedoseienko A, Wijers M, Lopez A, Billadeau DD, van de Sluis B and
Burstein E: Regulation of murine copper homeostasis by members of
the COMMD protein family. Dis Model Mech. 14:dmm0459632021.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Iwuchukwu I, Nguyen D, Beavers M, Tran V,
Sulaiman W, Fannin E, Lasseigne L, Ramsay E, Wilson J and Bazan NG:
MicroRNA regulatory network as biomarkers of late seizure in
patients with spontaneous intracerebral hemorrhage. Mol Neurobiol.
57:2346–2357. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Neveu B, Richer C, Cassart P, Caron M,
Jimenez-Cortes C, St-Onge P, Fuchs C, Garnier N, Gobeil S and
Sinnett D: Identification of new ETV6 modulators through a
high-throughput functional screening. iScience. 25:1038582022.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
da Silva AN, Ibelli AMG, Savoldi IR,
Cantão ME, Zanella EL, Marques MG, da Silva MVGB, de Peixoto JO,
Ledur MC, Lopes JS, et al: Whole-exome sequencing indicated new
candidate genes associated with unilateral cryptorchidism in pigs.
Sex Dev. 17:56–66. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Barcelo J and Sanz-Moreno V: NECTIN1 is a
melanoma metastasis suppressor gene. Nat Genet. 54:1776–1777. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Ablain J, Al Mahi A, Rothschild H, Prasad
M, Aires S, Yang S, Dokukin ME, Xu S, Dang M, Sokolov I, et al:
Loss of NECTIN1 triggers melanoma dissemination upon local IGF1
depletion. Nat Genet. 54:1839–1852. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Ho DW, Tsui YM, Chan LK, Sze KM, Zhang X,
Cheu JW, Chiu YT, Lee JM, Chan AC, Cheung ET, et al: Single-cell
RNA sequencing shows the immunosuppressive landscape and tumor
heterogeneity of HBV-associated hepatocellular carcinoma. Nat
Commun. 12:36842021. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Zhang S, Jiang C, Su Y, Gui J, Yue Z, Jian
B, He S and Ma X: Nectin2 influences cell apoptosis by regulating
ANXA2 expression in neuroblastoma. Acta Biochim Biophys Sin
(Shanghai). 55:356–366. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Bhave S, Guyer RA, Picard N, Omer M, Hotta
R and Goldstein AM: Ednrb−/− mice with hirschsprung
disease are missing Gad2-expressing enteric neurons in the
ganglionated small intestine. Front Cell Dev Biol. 10:9172432022.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Zheng Z, Gao M, Tang C, Huang L, Gong Y,
Liu Y and Wang J: E. coli JM83 damages the mucosal barrier
in Ednrb knockout mice to promote the development of
Hirschsprung-associated enterocolitis via activation of
TLR4/p-p38/NF-κB signaling. Mol Med Rep. 25:1682022. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Geng B, Wang X, Park KH, Lee KE, Kim J,
Chen P, Zhou X, Tan T, Yang C, Zou X, et al: UCHL1 protects against
ischemic heart injury via activating HIF-1α signal pathway. Redox
Biol. 52:1022952022. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Mondal M, Conole D, Nautiyal J and Tate
EW: UCHL1 as a novel target in breast cancer: Emerging insights
from cell and chemical biology. Br J Cancer. 126:24–33. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Tang J, Yang Q, Mao C, Xiao D, Liu S, Xiao
L, Zhou L, Wu G and Tao Y: The deubiquitinating enzyme UCHL3
promotes anaplastic thyroid cancer progression and metastasis
through Hippo signaling pathway. Cell Death Differ. 30:1247–1259.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
He R, Zhou Y, Liu J, Zhang X, Zhao X, An
L, Li Z and Cheng F: UCHL3 plays an important role in the
occurrence and development of melanoma. Oncol Lett. 22:7562021.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Thompson LL, Rutherford KA, Lepage CC and
McManus KJ: Aberrant SKP1 expression: Diverse mechanisms impacting
genome and chromosome stability. Front Cell Dev Biol.
10:8595822022. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Biryukov M, Dmitrieva A, Vavilova V,
Ustyantsev K, Bazarova E, Sukhikh I, Berezikov E and Blinov A:
Mlig-SKP1 gene is required for spermatogenesis in the flatworm
macrostomum lignano. Int J Mol Sci. 23:151102022. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Engeland K: Cell cycle regulation:
p53-p21-RB signaling. Cell Death Differ. 29:946–960. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Salaroglio IC, Belisario DC, Bironzo P,
Ananthanarayanan P, Ricci L, Digiovanni S, Fontana S, Napoli F,
Sandri A, Facolmatà C, et al: SKP2 drives the sensitivity to
neddylation inhibitors and cisplatin in malignant pleural
mesothelioma. J Exp Clin Cancer Res. 41:752022. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Surka C, Jin L, Mbong N, Lu CC, Jang IS,
Rychak E, Mendy D, Clayton T, Tindall E, Hsu C, et al: CC-90009, a
novel cereblon E3 ligase modulator, targets acute myeloid leukemia
blasts and leukemia stem cells. Blood. 137:661–677. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Jia L and Sun Y: RBX1/ROC1-SCF E3
ubiquitin ligase is required for mouse embryogenesis and cancer
cell survival. Cell Div. 4:162009. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Bays JL and DeMali KA: Vinculin in
cell-cell and cell-matrix adhesions. Cell Mol Life Sci.
74:2999–3009. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Shih YT, Wei SY, Chen JH, Wang WL, Wu HY,
Wang MC, Lin CY, Lee PL, Lin CY, Chiang HC, et al: Vinculin
phosphorylation impairs vascular endothelial junctions promoting
atherosclerosis. Eur Heart J. 44:304–318. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Yang W, Li J, Zhang M, Yu H, Zhuang Y,
Zhao L, Ren L, Gong J, Bi H, Zeng L, et al: Elevated expression of
the rhythm gene NFIL3 promotes the progression of TNBC by
activating NF-κB signaling through suppression of NFKBIA
transcription. J Exp Clin Cancer Res. 41:672022. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Sarkozy C, Hung SS, Chavez EA, Duns G,
Takata K, Chong LC, Aoki T, Jiang A, Miyata-Takata T, Telenius A,
et al: Mutational landscape of gray zone lymphoma. Blood.
137:1765–1776. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Qie S: The E3 ubiquitin ligase fbxo4
functions as a tumor suppressor: Its biological importance and
therapeutic perspectives. Cancers (Basel). 14:21332022. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Mucha B, Qie S, Bajpai S, Tarallo V, Diehl
JN, Tedeschi F, Zhou G, Gao Z, Flashner S, Klein-Szanto AJ, et al:
Tumor suppressor mediated ubiquitylation of hnRNPK is a barrier to
oncogenic translation. Nat Commun. 13:66142022. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Wang L, Piao Y, Zhang D, Feng W, Wang C,
Cui X, Ren Q, Zhu X and Zheng G: Fbxw11 impairs the repopulation
capacity of hematopoietic stem/progenitor cells. Stem Cell Res
Ther. 13:2452022. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Chen C, Zhou H, Zhang X, Liu Z and Ma X:
Association of FBXW11 levels with tumor development and prognosis
in chondrosarcoma. Cancer Biomark. 35:429–437. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Zou C, Chen Y, Smith RM, Snavely C, Li J,
Coon TA, Chen BB, Zhao Y and Mallampalli RK: SCF(Fbxw15) mediates
histone acetyltransferase binding to origin recognition complex
(HBO1) ubiquitin-proteasomal degradation to regulate cell
proliferation. J Biol Chem. 288:6306–6316. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
De La Chesnaye E, Méndez JP, López-Romero
R, De Los Angeles Romero-Tlalolini M, Vergara MD, Salcedo M and
Ojeda SR: FBXW12, a novel F box protein-encoding gene, is deleted
or methylated in some cases of epithelial ovarian cancer. Int J
Clin Exp Pathol. 8:10192–10203. 2015.PubMed/NCBI
|
|
109
|
Zhang J, Gan Y, Li H, Yin J, He X, Lin L,
Xu S, Fang Z, Kim BW, Gao L, et al: Inhibition of the CDK2 and
cyclin A complex leads to autophagic degradation of CDK2 in cancer
cells. Nat Commun. 13:28352022. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Arora M, Moser J, Hoffman TE, Watts LP,
Min M, Musteanu M, Rong Y, Ill CR, Nangia V, Schneider J, et al:
Rapid adaptation to CDK2 inhibition exposes intrinsic cell-cycle
plasticity. Cell. 186:2628–2643.e21. 2023. View Article : Google Scholar : PubMed/NCBI
|