Introduction
The salt-induced kinase (SIK) family is one of the
14 members of the AMPK superfamily (1,2). The
SIK subfamily comprises three kinases, namely SIK1, SIK2 and SIK3
(3), which are routinely expressed
in humans. SIK1 is the founding member of this subfamily (4), and the induction of SIK1 expression
was first demonstrated in the adrenal glands of rats fed on a
high-salt diet (5). Although the
members of the SIK subfamily possess a common domain in terms of
their structure, the expression of SIK1 is regulated by external
factors, unlike the other members of the subfamily, which are
constitutively expressed (4). SIK1
features an extended C-terminal region, a sucrose non-fermenting
protein (SNF-1) homologous (SNH) domain, and several
phosphorylation sites (6).
Furthermore, SIK1 is abundant in nerve tissue, fat and the adrenal
cortex (7–9). Based on its special structure and high
expression, SIK1 has been shown to regulate numerous physiological
processes involved in energy production, gluconeogenesis (10–12)
and lipid metabolism (10,13,14).
SIK1 has also been found to be involved in apoptosis (15) and the regulation of circadian
rhythms and sleep (16,17). Furthermore, SIK1 has been implicated
in the development of diseases of the nervous system, including
hypertension (18,19) and epilepsy (20–22).
Cytokines involved in these processes create an extensive signaling
network, featuring SIK1 as a central hub. Consequently, SIK1 exerts
a crucial role in regulating both physiological and pathological
processes, and, as a result, inhibitors targeting SIK1 have been
developed, ranging from broad-spectrum SIK inhibitors to those
specifically targeting SIK1. The efficacy of these inhibitors has
progressively improved both in terms of their sensitivity and their
safety (23).
Moreover, accumulating evidence has suggested that
SIK1 is involved in regulating the onset and progression of various
types of tumors through cancer-associated signaling pathways. The
expression of SIK1 was found to be downregulated in samples from
patients with hepatocellular carcinoma, gastric cancer and
colorectal cancer (24–29). This downregulated expression of SIK1
was found to be correlated with poor prognosis of the disease
(27). Moreover, previous studies
have shown that SIK1 inhibits the proliferation, invasion and
migration of cancer cells in the majority of tumors (1,25,26,29);
however, in certain tumors, the regulation of SIK1 has been
revealed to have the opposite effect (30,31).
This contradiction necessitates a further clarification of the
association between SIK1 and cancer.
In the present review, the experimental studies that
have been published on SIK1 over the course of the last 20 years
are summarized, and the function of SIK1 is explained based on the
experimental results and data that have been obtained. At the time
of writing and preparing this review, the current state of
knowledge on tumor signaling pathways in which SIK1 has been
identified to participate is summarized, and this theme is
developed with a discussion of the SIK1 regulatory factors in the
second part of the review. Through analyses of the molecules,
pathways and results of SIK1 research in different types of tumors,
the present review highlights the independent effects of SIK1 that
differentiate it from the other members of the SIK family, which
should be of interest for researchers to refer to prior to
designing further experiments, and which will aid scientists in
terms of providing new ideas for research. Moreover, the present
review provides robust evidence for the clinical development of
drugs targeting SIK1, a novel class of molecules with great
potential for future tumor therapeutic interventions.
Structure of SIK1 and its associated
regulatory factors
Structure of SIK1 and its
phosphorylation site
SIK1 is a member of the SIK subfamily of the
AMPK-activated serine/threonine family of protein kinases (32). AMPK family members are activated by
LKB1 phosphorylation (33). AMPK
suppresses lung cancer through indirectly inhibiting the activation
of mammalian target of rapamycin complex 1 oncogenic signaling via
phosphorylating the downstream protein, tuberin complex subunit 2
(TSC2), to inhibit tumor growth pathways, including the fatty acid
synthesis pathway that is required for cell proliferation. However,
AMPK deficiency has been revealed to be detrimental to cancer cell
survival in certain leukemia models (1). SIKs are a subfamily within the AMPK
family. They share two key structural domains with other AMPK
family members, one of which is the N-terminal serine/threonine
kinase domain (KD), the other being the ubiquitin-associated (UBA)
structural domain, which is responsible for the enzymatic activity
of these proteins. The UBA domain fulfills a role in
protein-protein interactions (32,34);
however, unlike AMPK, which forms a complex with an ABC tetramer,
SIKs are markedly simpler molecules (35). Each SIK consists of one monomer.
Furthermore, the SIK family also has structural differences; for
example, SIK2 and SIK3 share 70 and 37% sequence similarity in the
SNH structural domain with SIK1, respectively (36). In addition to the common structural
domain of the AMPK family, the SIK members also have a long
C-terminal structure containing multiple cyclic AMP-dependent
protein kinase A (PKA) activation sites (6). SIK1 was the first SIK to be identified
in the adrenal cortical tissue of rats fed on a high-salt diet,
thus becoming the founding member of this subfamily (5). The gene encoding SIK1 is located
differently in different species; for example, it is located on
chromosome 17 in mice, and on chromosome 21 in humans (17). SIK1 is a protein encoding 776 amino
acids (32) (Fig. 1). The KD of SIK1 is located at
residues 27–278 (2), and an
activation loop (T-loop) has been identified in the KD (33) (Fig.
1). SIK activity is influenced by phosphorylation;
specifically, phosphorylation at residue Thr-182 within the T-loop
region is crucial for the enzymatic activity of SIKs (17). This phosphorylation may be triggered
by LKB1, another kinase. The importance of LKB1 to SIK activation
has been clearly demonstrated in cells lacking LKB1 activity or
expression, such as HeLa cells or cells generated from
LKB1-knockout mice (33,37,38).
These cells exhibit significantly reduced SIK activity.
Furthermore, SIK1 itself may have an additional autophosphorylation
site within the T-loop, which is also essential for its activity
(39). Glycogen synthase kinase 3
also phosphorylates SIK1 at the Thr-182 site, thereby forming a
positive feedback regulation of SIK1 activation (36). Moreover, SIK1 contains an SNH
structural domain at residues 301–354 (32). The Thr-322 residue located within
this structural domain can be phosphorylated by
Ca2+/calmodulin-dependent protein kinase (CaMK) I
(Fig. 1) (34,40).
In addition, there is an UBA domain located in the SNH structural
domains (41). The UBA domain
participates in the activation of SIK1 mediated via LKB1 (42). Mutations in this structural domain
have been shown to markedly reduce the SIK1 activity (41), thereby promoting SIK nuclear
translocation through preventing SIK from interacting with the
14-3-3 adaptor protein (41,43). A
structural domain containing a PKA-dependent phosphorylation site
at residues 567–613 (32) is
located at the C-terminus, which contains multiple PKA
phosphorylation sites (32). PKA
phosphorylation promotes the association of SIK1 and protein
14-3-3; moreover, SIK1 phosphorylation at Thr-473 and Ser-575
promotes both the relocalization of SIK1 to the cytoplasm, and its
binding to protein 14-3-3 (17,44).
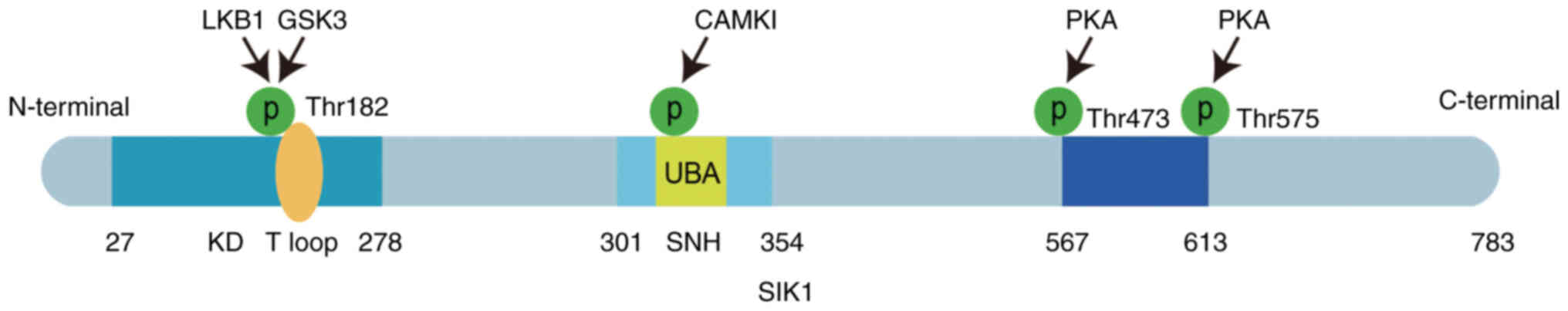 | Figure 1.SIK1 structure and phosphorylation
sites: SIK1 can be divided into three structural domains, namely:
The KD structural domain, the SNH structural domain, and the
C-terminal structural domain. From N-terminal to C-terminal, 27–278
amino acid residues have a KD structural domain, which contains an
activation loop (T-loop) with phosphorylation sites. Amino acid
residues 301–354 have an SNH structural domain, which contains a
UBA structural domain with CAMP phosphorylation sites. Residues
567–613 have a structural domain containing multiple PKA
phosphorylation sites. SIK1, salt-induced kinase 1; KD,
silk-threonine kinase structural domain; SNH, sucrose
nonfermentable homologous structural domain; UBA,
ubiquitin-associated structural domain; LKB1, liver kinase B1;
CaMKI, Ca2+/calmodulin-dependent protein kinase I; PKA,
protein kinase A; GSK3, glycogen synthase kinase 3. |
Moderating factors
Upstream regulators
LKB1 activates AMPK in vivo, subsequently
activating SIK1. LKB1 phosphorylates the Thr-182 site of SIK1,
which induces the kinase activity of SIK1 (Fig. 2) (10). LKB1 encodes a serine/threonine
kinase mediating the phosphorylation of downstream AMPK, and is
considered a potent tumor suppressor gene (45,46).
Hollstein et al (1) showed
that LKB1-activated SIK1 and SIK3 are key targets for lung cancer.
In addition, LKB1 is able to mediate SIK1 apoptosis and to regulate
E-calmodulin (15), E-calmodulin
expression and intercellular junction stability (47). Ca2+/CaMK is also a major
upstream regulator, although it operates in a stand-alone manner.
Phospholipase C promotes the movement of Ca2+ ions from
the endoplasmic reticulum to the cytoplasm via the inositol
trisphosphate receptor, especially in the central nervous system
and during the regulation of T cells (48). This process activates CaMK, leading
to phosphorylation of SIK1 within the UBA structural domains,
thereby increasing SIK1 activity (49), followed by SIK1 activation of
Na-K-ATPase (Fig. 2). SIK family
members are induced by cAMP signaling (4). An increase in the concentration of
cAMP increase is mediated by adenylate cyclase (AC), after which
cAMP activates PKA (Fig. 2). PKA is
another major upstream regulator of SIK1 that phosphorylates SIK1
at the Thr-475 site. SIK1 interacts with 14-3-3 following
phosphorylation (50). SIK1
contains two PKA/14-3-3 sites, and its sensitivity to cAMP depends
on these two PKA sites that mediate the interaction with 14-3-3
protein. Deletion of either of the 14-3-3 protein binding sites in
SIK1 results in 14-3-3 protein binding failure, rendering it
insensitive to cAMP (51). The
absence of a single 14-3-3 binding site in SIK1 makes it
insensitive to cAMP. There are several other upstream regulators,
including the transcriptional repressor, inducible cAMP early
repressor (ICER). ICER is induced by gastrin, which binds to the
CRE promoter element and negatively regulates gene expression
(52,53). The SIK1 promoter includes CRE
binding sites (54), and SIK1 is a
potential target gene for ICER. Selvik et al (55) demonstrated that ICER is a
gastrin-induced factor that negatively regulates SIK1
expression.
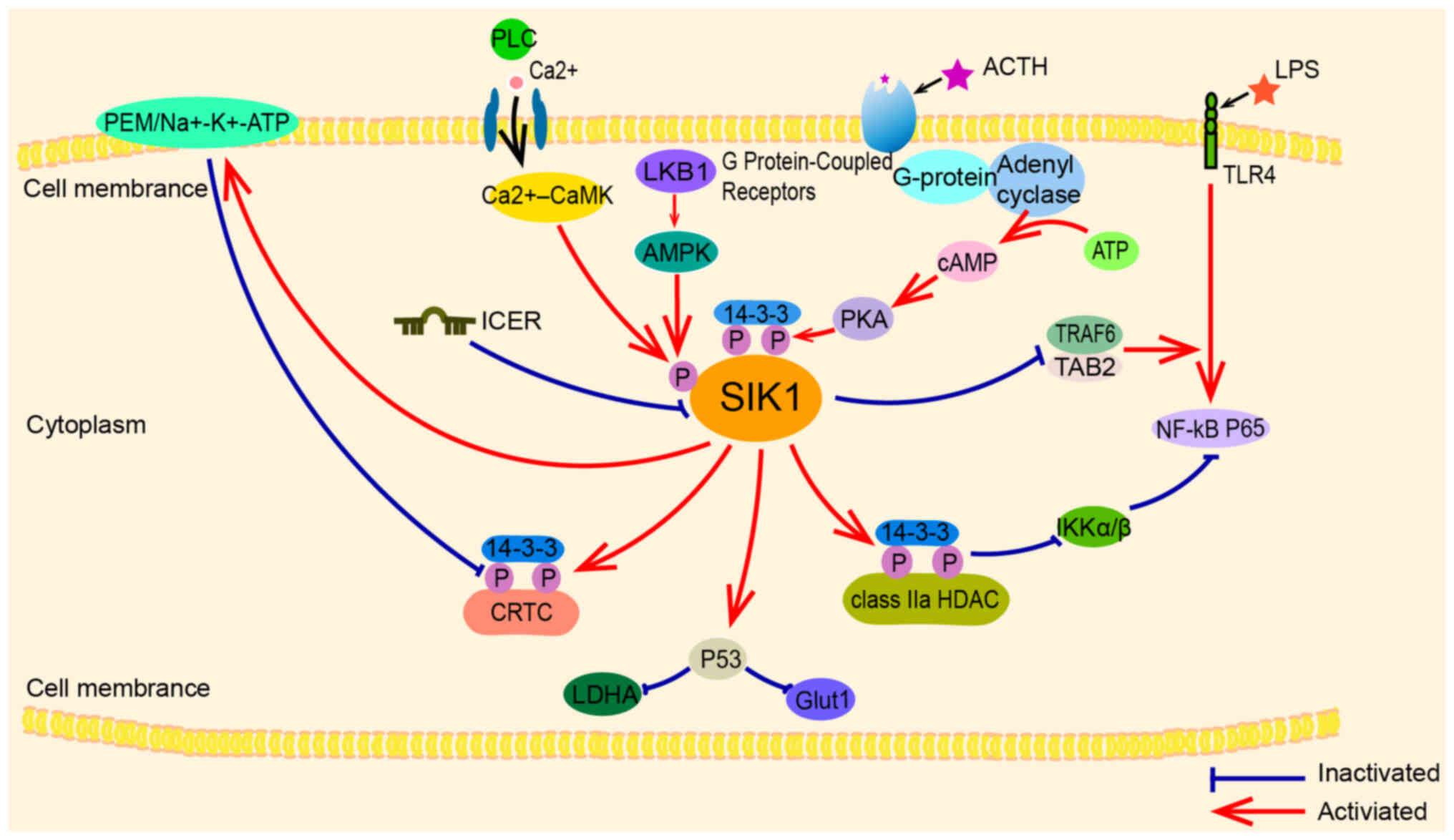 | Figure 2.SIK1 upstream and downstream
regulators: Upstream mainly includes LKB1, Ca2+ -CaMK,
cAMP-PKA and some other regulators such as ICER; Downstream mainly
identifies substrates such as CRTC and class IIa HDACs, and there
are also some other signaling pathways such as
PME-1/Na+, K+-ATP, NF-kB, P53 and other
signaling pathways that play a regulatory role in tumor and other
events. SIK1, salt-induced kinase 1; PLC, phospholipase C; CaMK,
Ca2+/calmodulin-dependent protein kinase; LKB1, liver
kinase B1; PKA, protein kinase A; ICER, inducible cAMP early
repressor; CRTC, cAMP-responsive element-binding protein
(CREB)-regulated transcription coactivators; HDAC, histone
deacetylase; LDHA, lactate dehydrogenase A; ACTH,
adrenocorticotropic hormone; LPS, lipopolysaccharide. |
Downstream regulators
SIK1 has two key groups of common substrates:
cAMP-responsive element-binding protein (CREB)-regulated
transcription coactivators (CRTCs) and class IIa histone
deacetylases (HDACs). The CRTCs are regulated by CREB (17). SIKs proteins affect gene expression
through phosphorylation. SIKs function as gene repressors in the
nucleus by targeting another protein group: The class IIa HDACs,
which act as gene repressors through binding to a protein called
myoblast enhancer (51,54,56,57).
When SIKs phosphorylate class IIa HDACs, this influences their
ability to bind to DNA and to repress gene expression. As far as
CRTCs are concerned, the mechanism via which these proteins operate
is opposite to that of HDACs. They bind to CREB, and can also
enhance the activity of bZIP transcription factors (4). Class IIa HDACs and CRTCs remain in the
cytoplasm following phosphorylation due to their association with
14-3-3 protein in the cytoplasm (Fig.
2) (4,58). The dephosphorylated SIK substrates
are transferred to the nucleus to participate in the regulation of
gene expression (4). SIK1 controls
the shuttling of these substrates between the cytoplasm and the
nucleus through binding to 14-3-3. In a previous study, deletion of
CRTC2 was shown to result in the complete inhibition of IL-6
production and SIK1 deletion-induced soft agar growth proliferation
in non-small cell lung cancer (NSCLC), suggesting that CRTC2 is a
major downstream target for LKB1 and SIK1/3-mediated inhibition of
tumor formation (1).
Phosphatase methyl esterase-1(PME-1)/Na+
and K+-ATPases provide another set of downstream
substrates of SIK1 (Fig. 2).
PME-1/Na+ and K+-ATPases are predominantly
found in cell membranes, and are able to regulate intracellular
electrophysiological homeostasis by transporting Na+ and
K+ ions. PME1 alone demethylates and inactivates protein
phosphatase 2A (PP2A) (59), and
when SIK1 is activated by CaMK, it phosphorylates PME-1. This
phosphorylation event causes PME-1 to dissociate from the
PP2A-Na+, K+/ATPase complex, leading to
activation of PP2A, which ultimately influences how the
Na+/K+-ATPase pump functions in the cell
(40,60,61).
SIK1 also regulates important signaling pathway factors, such as
NF-kB and p53. Under inflammatory conditions, an increased
expression of SIK1 interferes with protein interactions in the
NF-κB pathway that are triggered by immune cells by preventing the
phosphorylation of IKKα/β (62).
Through activating the tumor suppressor protein p53, SIK1 reduces
the expression of the glucose transporter Glut1 and lactate
dehydrogenase A, which both participate in aerobic glycolysis, a
process that is important for cancer cell proliferation.
Additionally, SIK1 can inhibit the proliferation of breast cancer
cells under conditions when the supply of nutrients is limited
(24).
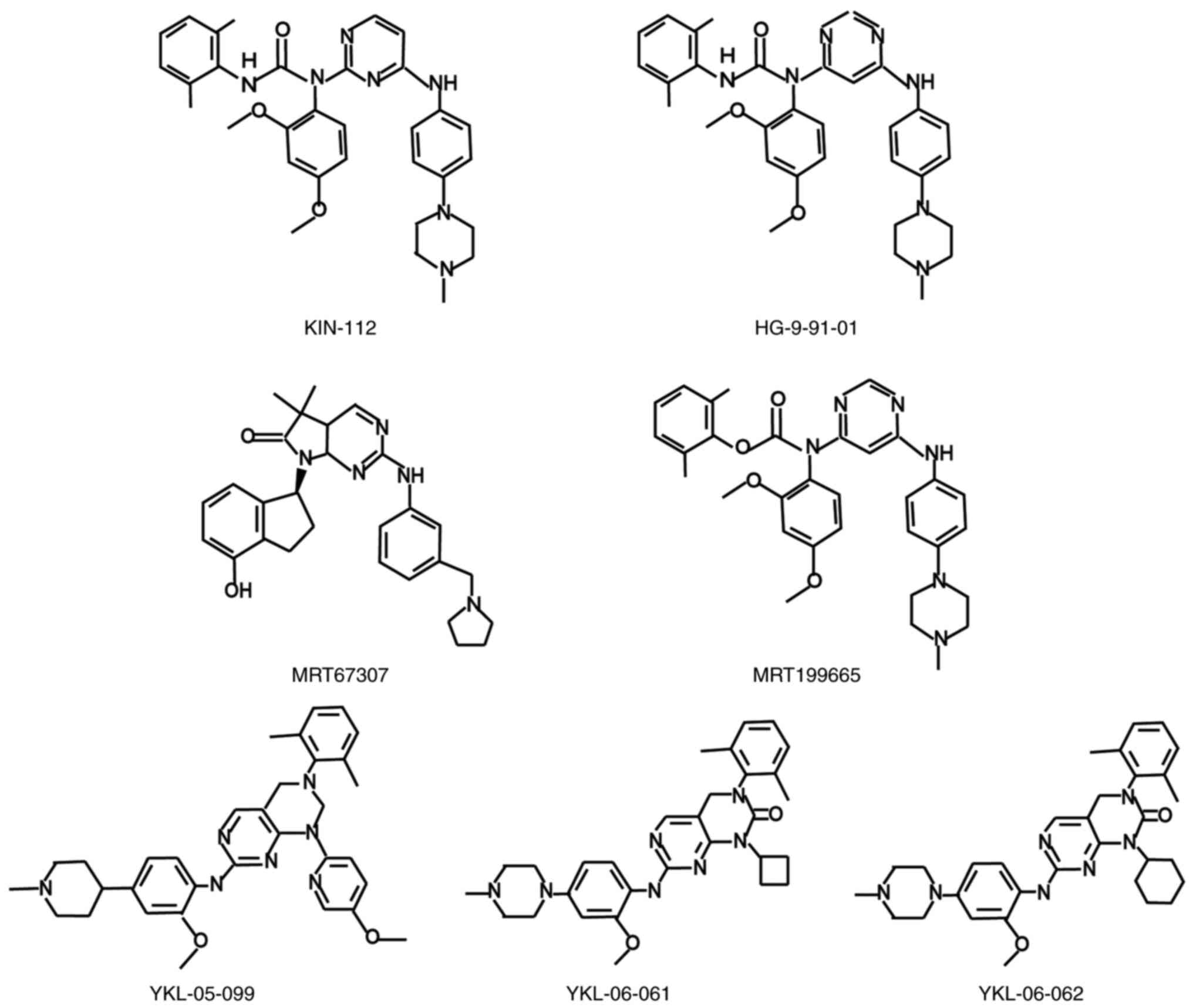
SIK1 inhibitors
SIK1 inhibitors also regulate SIK1. Given that
current pan-SIK inhibitors target SIK isozymes, these are valuable
for understanding the role of SIK1. The inhibitor HG-9-91-01, a
4,6-diaminopyrimidine derivative, has become an important tool for
studying the specific role of SIK1 (63), and this highly selective inhibitor
specifically targets the SIK family without inhibiting other AMPK
family members (22,23,64). A
pathophysiological model that exploited the selectivity of
HG-9-91-01 revealed that SIK inhibition induces the production of
IL-10 through macrophage-like cells, while downregulating IL-6,
IL-12 and TNF-α (63). In another
study, upon converting the 2,4-diaminopyrimidine derivative KIN-112
into HG-9-91-01, the activity of SIK1 was found to be significantly
enhanced (~10-fold) without significantly changing its selectivity
(Fig. 3) (63). Given that HG-9-91-01 showed
promising effects in vitro, but poor outcomes in
vivo, the HG-9-91-01 analogues YKL-05-099, YKL-06-061 and
YKL-06-062 (Fig. 3) were
subsequently developed for in vivo studies (17). Studies on two other inhibitors,
MRT-67307 and MRT-199665 (63,65)
(Fig. 3), primarily explored their
regulatory effects on IL-10 and its functions, as well as the
kinase selectivity of both these compounds and HG-9-91-01. The
microtubule affinity-regulating kinases (MARKs)1-4, and NUAK family
SNF1-like kinases (NUAKs)-1 and −2, are other members of the AMPK
family. In contrast to HG-9-91-01, the MRT-67307 and MRT-199665
inhibitors target several other AMPK-associated kinases. Although
these inhibitors can inhibit all the kinases MARK1-4 and NUAK1 and
NUAK2, MRT-67307 has been shown to have greater potency against
MARK3, whereas MRT-199665 is more effective against MARK1, MARK2
and MARK4 (63). In a separate
study, Peng et al (22)
investigated the limitations of pan-SIK inhibitors through
developing selective inhibitors targeting only SIK1 and SIK2. These
inhibitors were designed to be safer than HG-9-91-01. Notably, key
differences were identified in the amino acid sequences of these
new inhibitors compared with other similar kinases, as the focus
was on specific residues such as threonine ‘gatekeepers’ and the
nearby glutamate residue (Glu-103). Using this information and
structure-based drug-design techniques, JRD-SIK 1/2 i-3 and JRD-SIK
1/2 i-4, selective inhibitors for SIK1 and SIK2, were developed
(23). These SIK1/2-specific
inhibitors were found to hold promise for treating inflammatory
bowel disease through promoting the production of the
anti-inflammatory factor IL-10, while simultaneously inhibiting
pro-inflammatory factors by interfering with SIK1/2-dependent
signaling pathways. In addition, endogenous proteins in the body
are able to significantly inhibit SIK; for example, 14-3-3 proteins
modulate SIK1 inhibition through the cAMP-mediated regulation of
gene expression in vertebrate cells. Camp-mediated phosphorylation
of PKA inhibits SIK family dephosphorylation, thereby promoting
14-3-3 protein release and causing nuclear translocation of the
CRTCs (51).
SIK1 regulates tumor progression
Role of SIK1 in inflammation
Inflammation is closely associated with cancer
(66). The majority of the
experimental and population-based studies that have been published
have confirmed that chronic inflammatory mediators promote tumor
initiation, development and progression, thereby mediating
tumorigenesis (67). The causes of
chronic inflammation, such as infectious agents, immune-mediated
diseases and allergies, have been shown to create a
cancer-triggering microenvironment (66).
SIK1 is able to act as a key molecular switch in the
occurrence of inflammation. The dephosphorylation of CRTC3 enables
the transformation of classically activated macrophages (M1
macrophages) into regulatory macrophages (M2 macrophages) through
the inhibition of SIK1, which leads to the production of higher
levels of the anti-inflammatory molecule IL-10 and lower levels of
the pro-inflammatory factor IL-12, thereby preventing and
alleviating chronic inflammation and autoimmune diseases (38). Alcohol consumption is known to
increase the likelihood of developing neuroinflammatory diseases.
Microglia are significantly associated with alcohol-induced
neuroinflammation and apoptosis. An increased expression of SIK1
has been demonstrated to occur in the primary microglia of
alcohol-consuming mice (62). SIK 1
inhibits microglial inflammation through the NF-κB signaling
pathway (62). SIK1 participates in
the process of epithelial-mesenchymal transition (EMT) as well as
inflammation; in addition, SIK1 is a marker of the transition from
acute kidney injury (AKI) to chronic kidney disease (CKD). SIK1
overexpression inhibits Wnt/β-catenin signaling and its downstream
transcription factor, Twist1, leading to an attenuation of the
inflammatory response and slowing the transition from AKI to CKD
(68). NLR family pyrin
domain-containing 3 (NLRP3) inflammasomes are protein complexes
that promote inflammation through producing cytokines and
triggering cellular pyroptosis. A recent study suggested that SIK1,
when phosphorylated, has a role in regulating NLRP3 (69). This regulation appears to involve
reducing the mRNA expression levels of the protein absent in
melanoma 2, which prevents the development of autoimmune uveitis in
the eye. Similarly, Pirie et al (70) observed that reducing SIK1 expression
in healthy C57B16/J mouse primary splenocytes from mice enhanced
their acute inflammatory responses, with acute inflammation being a
healthy physiological response associated with tissue recovery or
reconstruction (67). However,
elucidating the precise details of the association between chronic
inflammation and tumorigenesis requires further studies. SIK1
inhibitors can also exert anti-inflammatory effects. Lombardi et
al (71) demonstrated that SIK
inhibition (through the use of the inhibitors HG-9-91-01 and
ARN-3236) significantly reduced the production of pro-inflammatory
factors (such as TNF α and IL6) in human bone marrow cells, and
increased the secretion of anti-inflammatory factor IL-10 thereby
preventing and mitigating the development of inflammation (71). Timely blockade of SIK is able to
effectively prevent disease progression to tumors from occurring.
Although HG-9-91-01 has a high selectivity for SIK1, it has poor
pharmacokinetic and pharmacodynamic characteristics, including a
fast clearance rate, a slow exposure rate in vivo, and a
high binding rate with plasma proteins. Cai et al (72) designed and synthesized several
pyrimidine-5-carboxamide derivatives, where compound 8h was shown
to have favorable activity and selectivity for SIK1, avoiding the
aforementioned shortcomings of HG-9-91-01. This compound had the
capability of increasing the anti-inflammatory effects of IL-10,
while significantly reducing the pro-inflammatory effects of IL-12;
in addition, compound 8h was shown to exert favorable effects on
colitis models.
Expression of SIK1 in clinical
samples
Several studies have been performed to explore the
influence of SIK1 in tumorigenesis and development. The
downregulation of SIK1 was shown to promote tumor enlargement and
distant metastasis in liver cancer cells (P<0.001) (25). In addition, SIK1 downregulation is
associated with shorter overall survival (OS) rates and worsened
disease-free survival rates. Moreover, SIK1 is an independent
predictor of OS. SIK1 expression was found to be significantly
decreased in all the studied liver cancer cell lines compared with
normal liver cell lines (25).
Another study has shown that SIK1 levels are reduced in liver
cancer tissues (73); the level of
SIK1 was higher in normal liver tissues compared with that in liver
cancer tissues (73) (5.15±0.41 vs.
3.12±0.29; P<0.006). In addition, survival analysis identified
that low SIK1 expression was positively associated with a poor
prognosis (73). Similarly,
survival analysis of the prognosis of patients with breast cancer
showed that the high expression of SIKs was positively associated
with OS and recurrence-free survival (RFS) (26). Moreover, the Klein score of SIK1 in
patients with breast cancer was found to be higher in ER-positive
patients compared with ER-negative patients (26). Ponnusamy and Manoharan (24) revealed that low expression of SIK1
is associated with breast carcinogenesis. Based on an analysis of
the Oncomine data, SIK1 expression was found to be lower in breast
cancer tissues compared with normal tissues, and the most prominent
subtype of breast cancer present was the luminal subtype (24). Compared with paraneoplastic
non-tumor tissues, SIK1 was shown to be downregulated in 36 pairs
of gastric cancer tissues (29).
SIK1 is also downregulated in colorectal cancer cell lines; the
downregulation of SIK1 was found to be an independent risk factor
of patients with colorectal cancer (27). Notably, SIK1 is also downregulated
in patients with pancreatic cancer, suggesting that SIK1 may be a
tumor suppressor in pancreatic cancer (28).
Functions of SIK1 in tumors
The roles of SIK1 in tumor growth may be divided
into two broad categories (Fig. 4):
One is the inhibition of cancer (Table
I), and the other is promotion of cancer (Table I). These two opposite effects have
been verified in a large number of experiments.
 | Table I.Model types and functional
characteristics of SIK1 used in different tumors. |
Table I.
Model types and functional
characteristics of SIK1 used in different tumors.
| First author,
year | Tumors | Model types | Functional
characteristics | (Refs.) |
|---|
| Hollstein et
al, 2019 | Lung cancer | KP
(KrasLSLG12D/+; p53fl/fl; R26LSL;
luc/luc) mice induced by sgSIK1 | SIK1 inhibits lung
cancer growth. | (1) |
| Murray et
al, 2019 |
|
KrasLSL-G12D,
p53flox, Lkb1flox, H11LSL-Cas9 and
Rosa26LSL-tdTomato mice | SIK1 is a
downstream gene of the oncogene LKB1, which is regulated by LKB1 to
inhibit lung cancer growth. | (74) |
| Qu et al,
2016 | Liver cancer | Nude mice
orthotopic liver cancer cell model overexpressing SIK1 and HepG2
model silencing SIK expression. | SIK1 inhibits
invasion and lung metastasis of liver cancer cells. | 25 |
| Fu et al,
2017 |
| Nude mice injected
with siSIK1 Hep3B and siSIK1 Huh-7 | SIK can inhibit the
proliferation and apoptosis of liver cancer cells. | (76) |
| Qu et al,
2017 |
| MHCC97H cells
treated with siRNF2, shRNF2 and shSIK1 respectively | SIK1 is the target
of RNF2 E3 ligase, which can reverse RNF2′s promotion of liver
cancer growth. | (73) |
| Ponnusamy and
Manoharan, 2021 | Breast cancer | MCF-7, ZR-75-1 and
MCF-10A transfected with siSIK1 | SIK significantly
inhibits the proliferation of breast cancer, promotes oxidative
phosphorylation and inhibits aerobic glycolysis. | (24) |
| Xin et al,
2021 |
| MDA-MB-231 and
MCF-7 treated with SIK1 si/shRNA | SIK1 inhibits
chemotherapy resistance of breast cancer more obviously than other
subtypes. | (26) |
| Zang et al,
2022 | Gastric cancer | Human gastric
cancer cell lines (AGS, HGC-27, MKN-45 and SGC-7901) and siSIK1
transduced AGS and HGC-27 cell lines | SIK1 is an
intermediate molecule that circEIF4G3 regulates gastric cancer
cells and inhibits gastric cancer growth when it exists alone. | (29) |
| Selvik et
al, 2014 |
| AGS-GR cells
transfected with siSIK1 | SIK1 participates
in the migration of gastric adenocarcinoma cells induced by
gastrin. | (55) |
| Gao et al,
2023 | Colorectal
cancer | RKO and SW480 cell
lines of OE-SIK1; HCT116 cell line of KD-SIK1; Female BALB/c nude
mice (6–8 weeks old) injected with HCT116shSIK1. | SIK1 inhibits
metastasis and chemotherapy resistance of colorectal cancer. | (77) |
| Jin et al,
2022 | Ovarian cancer | HEY and ES-2 cell
lines of pc-DNA over-expression vector | SIK1 inhibits the
migration and invasion of ovarian cancer cells and promotes
apoptosis. | (78) |
| Ren et al,
2016 | Pancreatic
cancer |
Gemcitabine-resistant cells (PANC-1,
Hs766T, ASPC-1, MiaPaCa-2 and MPanc96 cells) and
gemcitabine-sensitive cells (HPAC, L3.6pl, CFPAC, BxPC-3 and
SU86.86 cells) | SIK1 inhibits
gemcitabine resistance and inhibits DNA synthesis in pancreatic
cancer cells. | (28) |
| Kou et al,
2022 | Thyroid cancer | LKB1 overexpressed
TPC-1 and BCPAP | SIK1 as the main
downstream regulator of LKB1 has an inhibitory effect on thyroid
cancer. | (79) |
| Wang et al,
2022 | Osteosar-coma | Transfected 143B
and U-2OS cells with different siRNA (siRNA#1 and siRNA#2) | SIK1 inhibits the
proliferation and migration of osteo-sarcoma cells. | (82) |
| Huang et al,
2020 |
Medullo-blastoma | Specific siRNA
knocks down Daoy cells; 6-week-old nude mice implanted with Daoy
cells stably expressing sh-SIK 1 | SIK1 is an
intermediate molecule connecting miR-130b-3p and p53, which
promotes medulloblastoma growth when it exists alone. | (30) |
| Hartono et
al, 2022 | Desmo- plastic
small round cell tumor | Dox-inducible
shSIK1 stable JN-DSRCT-1 (JN) and BER-DSRCT (BER) cells; Stable
injection of JN- and BER-shSIK1 and sh-Renilla Mice with
immune-deficient NOD.SCID/IL2Rγ-null (NSG) mice | SIK1 may mediate
the regulation of desmoplastic small round cell tumor by EWSR1-WT1
and participate in DNA replication of tumor cells. | (31) |
Anticancer effects of SIK1
Lung cancer
Human A549 lung adenocarcinoma cells with LKB1
deleted have been used for colony formation on soft agar plates to
explore the role of SIK family members in KRAS-dependent lung
tumors. The results obtained showed that only SIK family members
were able to restore the soft agar colonies after the inhibition of
recombinant colonies by LKB1 (1).
Lung tumor size was found to be significantly higher in
SIK1-disrupted mice compared with the control mice. Furthermore,
H&E staining of lung tissue from mice transfected with sgSIK1
revealed that the lung tumor size of virus-treated mice carrying
sgRNA targeting SIK1 was significantly increased, consistent with
floating Sik1 allele analysis (1).
Another experiment investigating the effect of the LKB1-SIK axis
pair in lung cancer showed that the tumor load was increased in
mice transduced with vectors carrying sgSik1 or sgSik1+3. In
another study, genes downstream of SIK were found to be highly
enriched when LKB1 was re-expressed, thereby confirming that SIK1
is a downstream gene of LKB1, and is regulated by LKB1 to inhibit
tumor growth (74).
Liver cancer
Liver cancer cells are able to stably express SIK1.
SIK1 significantly reduces growth efficiency; however, SIK1
deletion led to an increased efficiency of lesion formation, a
larger number of colonies as shown by soft-agar colony formation
assay, and an increased liver cancer growth rate (25). In nude mouse orthotopic MHCC97H
liver cancer cells, SIK1-overexpressing cells were found to have a
smaller tumor volume compared with control cells. By contrast, SIK1
silencing led to a marked promotion of the proliferation of liver
cancer cells in vivo (25).
These experiments also investigated the presence of metastatic
cancers (25), such as bone
metastases, which are common in lung cancer (75); however, in liver cancer, lung
metastasis is frequently detected. Fewer lung metastases and
smaller metastatic foci of liver cancer were found in nude mice
compared with the control mice following hydrostatic injection of
MHCC97H-SIK1. These experiments confirmed that SIK1 inhibits the
invasion and metastasis of liver cancer (25). MicroRNA (miR)-25 inhibitors have
also been shown to attenuate tumor metastasis and proliferation in
mice, and to promote apoptosis in liver cancer cell lines; however,
SIK1 silencing is able to reverse these effects (76). The E3 ubiquitin ligase RNF2
regulates downstream SIK1 activity (73). Downregulation of RNF2 can induce
restoration of the levels of SIK1, which thereby leads to the
inhibition of liver cancer cell proliferation and promotion of
apoptosis of liver cancer cells, both changes of which were found
to be significant (73). Moreover,
the presence of SIK1 following RNF2 knockdown reduced the ability
of cells to migrate (P<0.01); however, knocking down SIK1 led to
a reversal of the aforementioned effects (73).
Breast cancer
Ponnusamy and Manoharan (24) transduced SIK1-specific siRNA into
the malignant breast cancer cell lines MCF7, ZR-75-1 and MCF10A,
revealing that SIK1 knockdown (SIK1-KD) is able to promote cancer
cell proliferation based on MTT assays. SIK1-KD also led to
significant increases in energy expenditure, inhibition of ATP
production, and promotion of the proliferation of the luminal
subtypes of breast cancer cells. Another experiment also showed
that high SIK1 expression is positively correlated with RFS
(P=0.0026). Furthermore, an in vitro stromal cell invasion
assay was performed to reveal that SIK1-KD can increase the
invasive potential of breast cancer cells. Further studies found
that SIK1 knockdown can increase the resistance of breast cancer
cells to chemotherapeutic drugs paclitaxel (IC50:
SIK1-KD, 18.62 µM vs. 5.279 µM). Notably, the resistance of the
breast cancer cell lines to chemotherapy was more pronounced when
the three SIK family members were simultaneously knocked down
(26). Furthermore, The Cancer
Genome Atlas database showed that the patients who responded better
to chemotherapy with paclitaxel had a higher expression level of
SIK1 (P=0.019).
Gastric cancer
Overexpression of the circular RNA (circRNA)
circEIF4G3 inhibits the proliferation of malignant cells in gastric
cancer. However, SIK1-KD was found to attenuate this inhibition.
Moreover, cells of the circEIF4G3-transfected HGC-27 gastric cancer
cell line exhibited inhibited tumor growth with an increasing level
of SIK1. These findings suggested that circEIF4G3 can inhibit the
proliferation of gastric cancer cells via regulating SIK1 (29). In another study, SIK1 was
demonstrated to affect gastrin-induced migration of gastric
adenocarcinoma cells; notably, SIK1-KD gastric cancer cell line AGS
exhibits stronger cell migration (55).
Colorectal cancer
A recent study showed that SIK1 is downregulated in
colorectal cancer tissues (P<0.001), with a reduced migratory
capability (P<0.001) and a reduced capacity for trauma healing
(P<0.01) in two cell lines RKO and SW480 with OE-SIK1. However,
the opposite trend was observed in SIK1-KD strain HCT116 (both
migratory capability and capacity for trauma healing, P<0.01)
(77). Lentiviral transfection of
HCT116 cells with downregulated SIK1 led to a significant increase
in the chemotherapy resistance of nude mice, thereby decreasing the
efficacy of the therapy. Furthermore, H&E staining and Ki67
immunohistochemistry experiments showed that the SIK1 + oxaliplatin
(OXA) chemotherapy group had an increased rate of proliferation, as
indicated by higher Ki67 values compared with the short hairpin
(sh) NC + OXA chemotherapy group. Taken together, these findings
suggested that SIK1 may be useful as an in vivo colorectal
cancer chemotherapeutic drug-binding target (77).
Ovarian cancer
In an investigation of the regulatory mechanism of
ovarian cancer, upon transfection of circ_0078607 into the HEY and
ES-2 cell lines, overexpression of circ_0078607 led to a
significant promotion of the expression of SIK1 in HEY and ES-2
cells (P<0.001), although the expression of miR-32-5p was
inhibited. Furthermore, an overexpression vector, pc-DNA SIK1, was
constructed. Overexpressed SIK1 was revealed to significantly
inhibit the migration (P<0.01) and invasion (P<0.01) of HEY
and ES-2 cells, and to promote cell apoptosis (P<0.001). A
series of experiments were performed, including reverse
transcription-quantitative PCR experiments, which confirmed that
overexpressed circ_0078607 inhibited the progression of ovarian
cancer via targeting miR-32-5p to upregulate SIK1 (78).
Pancreatic cancer
In a study which explored the resistance of SIK1 to
gemcitabine, the 10 pancreatic cancer cell lines employed in the
study were divided into two categories: The PANC-1, Hs766T, ASPC-1,
MiaPaCa-2 and MPanc96 cell lines, which were resistant to
gemcitabine, and the HPAC, L3.6pl, CFPAC, BxPC-3 and SU86.86 cell
lines, which were sensitive to gemcitabine. Western blot analysis
identified that the expression of SIK1 in gemcitabine-resistant
cells was lower compared with that in gemcitabine-sensitive cells.
Through quantitative image analysis again, it was shown that SIK1
was downregulated in gemcitabine-resistant pancreatic cancer.
Finally, according to a bromodeoxyuridine incorporation test, SIK1
was able to significantly inhibit DNA synthesis in pancreatic
cancer cells (28).
Thyroid cancer
High expression of LKB1 inhibits the growth of
thyroid cancer (79). Using OE-LKB1
cell lines TPC-1 and BCPAP, it was found that the upregulation of
LK1 increased the levels of p-SIK1 and SIK1 proteins (79). Moreover, SIK1 inhibitors can
eliminate the inhibition of proliferation caused by LKB1
overexpression (P<0.05) and reverse the elevation of E-Cadherin
(79). These findings demonstrated
SIK1′s role as a key downstream inhibitor of LKB1 in thyroid
cancer. Western blot analysis of nude mice injected with TPC-1 or
OE-LKB1 cells revealed significant reductions in thyroid tumor mass
and volume in the LKB1 overexpression group. Furthermore, levels of
p-SIK1, SIK1 and E-Cadherin proteins were elevated (79).
Osteosarcoma
The proto-oncogene B lymphoma Mo-MLV insertion
region 1 (BMI 1) is a transcriptional repressor that is known to
modulate tumorigenesis (80,81).
In osteosarcoma, BMI1 was found to promote osteosarcoma cell
proliferation, migration and invasion in vitro.
Bioinformatics analysis of osteosarcoma lines with BMI using
chromatin immunoprecipitation sequencing data revealed the presence
of the target gene SIK1 in BMI. The levels of SIK1 mRNA were
increased in osteosarcoma cells treated with PTC-209 (BMI1-specific
inhibitor). SIK1-KD in osteosarcoma cell lines 143B and U-2OS
enhances the proliferation (P<0.001) and migration (P<0.001)
of tumor cells (82). Taken
together, these studies have shown that various means of inhibiting
SIK1 can lead to an enhancement of the proliferation and migration
of osteosarcoma cells.
Pro-cancer effects of SIK1
Medulloblastoma
Transfection with siSIK1 led to an inhibition of the
migration (P<0.05) and invasion (P<0.05) of medulloblastoma
cell Daoy (30). In addition, six
downstream candidate target genes (SIK 1, SIK 3, ESR 1, SMAD 4, MAP
2 and TSC1) were also detected in medulloblastoma cells transfected
with miR 130b 3p. Among these six genes, the SIK1 gene demonstrated
the most pronounced level of downregulation (P<0.01). Further
studies reported that miR-130b-3p upregulation and SIK1
downregulation in medulloblastoma cells led to an increase in the
activity of the p53 oncogenic pathway (30). Injection of medulloblastoma cells
with a SIK1-KD vector caused a significant decrease in the average
tumor volume and tumor weight of mice inoculated with shSIK1 cells
at week 8 compared with sh-GFP mice (P<0.05), demonstrating that
SIK1 was able to promote medulloblastoma formation (30).
Desmoplastic small round cell
tumor
Desmoplastic small round cell tumor is a rare and
aggressive type of cancer that is caused by a specific genetic
abnormality (31). This abnormality
involves a chromosomal translocation, where two portions of
chromosomes 11 and 22 swap positions (31). The resulting fusion gene creates a
protein called EWSR1-WT1, which acts as an oncogenic transcription
factor, causing an abnormal promotion of cell proliferation,
ultimately leading to cancer (31).
Researchers have found that SIK1 is expressed at markedly higher
levels in desmoplastic small round cell tumor compared with other
types of sarcomas (cancers that arise in connective tissues). This
suggests that SIK1 may be a regulatory factor which acts downstream
of EWSR1-WT1, potentially fulfilling a role in mediating the
effects of EWSR1-WT1 on desmoplastic small round cell tumor
(31). Later, under the induction
of DOX, the levels of SIK1 protein in JN and BER of shSIK1 stable
desmoplastic small round cell tumor cells decreased, thus reducing
or inhibiting DNA replication in cancer cells (P<0.0001).
Injection of desmoplastic small round cell tumor cells stably
expressing shSIK1 in immunodeficient mice revealed that SIK1
deficiency could inhibit the growth of xenograft tumors
(P<0.001). Intraperitoneal injection of EdU into mice before
euthanasia revealed that the deletion of SIK1 gene could reduce the
number of EdU cells in xenograft tumors (P<0.0001), which
indicated that SIK1 was necessary for DNA replication of tumor
cells in vivo (31).
SIK1 regulates tumor progression through
different signals
SIK1 is expressed in diverse types of tumors,
either promoting or inhibiting tumor growth by participating in
tumor signaling pathways and influencing upstream or downstream
molecules (Table II). Therefore,
SIK1 is a potential target in the treatment of solid tumors, and
SKI participates in different signaling pathways that are
associated with each other to jointly regulate tumorigenesis
(Fig. 5).
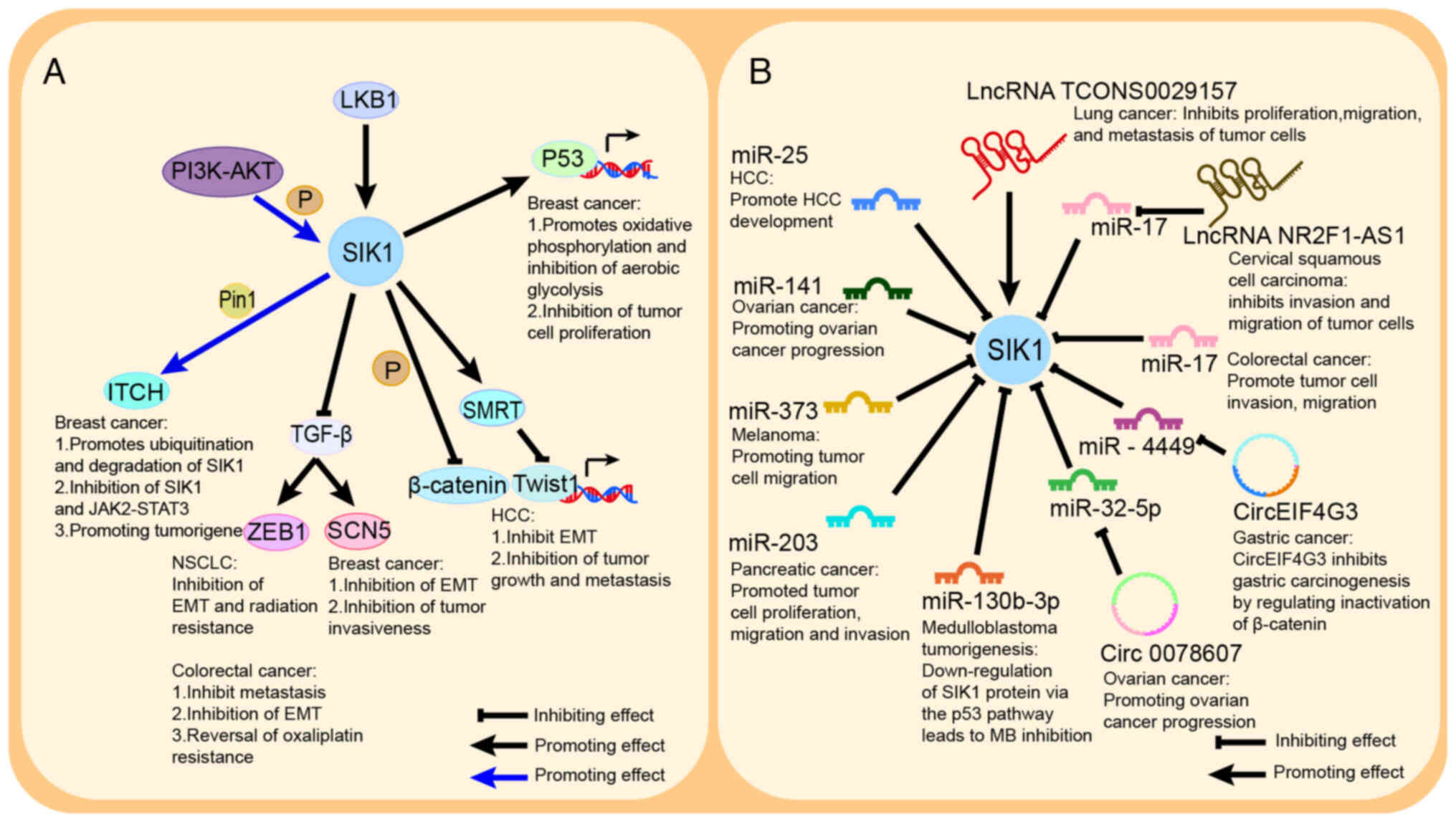 | Figure 5.SIK1 regulates tumor development
through different signaling pathways: (A) PI3K-AKT phosphorylates
SIK1, which interacts with ITCH under the action of Pin1 to promote
the occurrence and development of breast cancer; LKB1 activates
SIK1 to target and regulate different downstream pathways; SIK1
inhibits the TGF-β pathway and participate in NSCLC and colorectal
cancers; SIK1 inhibits β-catenin pathway and inhibits Twist
transcription factor by phosphorylating SMRT in HCC; and SIK1
promotes p53 transcription and plays an inhibitory role in breast
cancer. (B) Non-coding RNAs are involved in regulating SIK1
expression. miR-130b-3p, miR-141, miR-25, miR-17, miR-373 and
miR-203 directly inhibit SIK1 to promote tumor development; lncRNA
NR2F1-AS1, Circ0078607 and CircEIF4G3 inhibit SIK1 to promote tumor
development by regulating downstream miR to inhibit SIK1 to promote
tumor development; lncRNA TCONS0029157 activates SIK1 to inhibit
tumor development. SIK 1, salt-induced kinase 1; LKB1, liver kinase
B1; NSCLC, non-small cell lung cancer; SMRT, silencing mediator of
retinoic acid and thyroid hormone receptor; HCC, hepatocellular
carcinoma; miR, microRNA; lncRNA, long non-coding RNA; circ-,
circular; EMT, epithelial-mesenchymal transition. |
 | Table II.The signaling pathways and regulatory
mechanisms of SIK1 in tumors. |
Table II.
The signaling pathways and regulatory
mechanisms of SIK1 in tumors.
| First author,
year | Signaling
pathways | Tumors | Regulatory
mechanisms | (Refs.) |
|---|
| Yao et al,
2016 | LKB1-SIK1 | NSCLC | Suppression of EMT
and radiation resistance in NSCLC. | (84) |
| Murray et
al, 2019 |
| Lung cancer | Inhibits the growth
of lung cancer cells. | (74) |
| Gao et al,
2023 | TGF-β | Colorectal
cancer | SIK1 inhibits TGF-β
pathway and metastasis of colorectal cancer, inhibits EMT, and
reverses drug resistance. | (77) |
| Gradek et
al, 2019 |
| Brest cancer | Downregulated SIK1
upregulates Nav1.5 to increase EMT and cancer cell
invasiveness. | (89) |
| Qu et al,
2016 | Wnt/β-catenin | Hepatocellular
carcinoma | SIK1 inhibits
hepatocellular carcinoma cell invasion, migration, and EMT through
inhibition of β-catenin and Twist1. | (25) |
| Huang et al,
2020 | p53 |
Medulloblastoma | miR-130b-3p
inhibits medulloblastoma by downregulating SIK1 factor through the
p53 pathway. | (30) |
| Ponnusamy and
Manoharan, 2021 |
| Brest cancer | SIK1 activates p53
to inhibit glycolysis and suppress breast cancer cell
proliferation. | (24) |
| Sun et al,
2023 | PI3K-AKT | Brest cancer | Inhibits SIK1
binding to JAK2-STAT3 after activation of AKT signaling to promote
breast carcinogenesis. | (95) |
LKB1-SIK1 signaling pathway
LKB1 kinase is an important tumor suppressor in
human cancers (1,83). LKaB1 can target and regulate SIK1
and its downstream factors (Fig.
5A), thereby exerting inhibitory effects in numerous tumors
(1,74,79,83).
Through constructing the radiation-resistant cell lines A549R and
H1299R, Yao et al (84)
showed that an attenuation of LKB1-SIK1 signaling led to an
upregulation of the expression of the EMT driver ZEB1, which
promotes EMT and radiation-resistance in NSCLC. Murray et al
(74) also demonstrated that tumor
growth inhibition occurs through the LKB1-SIK axis in lung cancer
growth and differentiation.
Role of the transforming growth
factor-β (TGF-β) signaling pathway
TGF-β induces multiple signaling pathways, and is
integrated into different signaling regulatory pathways (85). TGF-β is a downstream substrate of
LKB1-SIK1 (86), which stimulates
the downstream genes ZEB1 and α-subunit of the channel protein
called Nav1.5 (SCN5) to control tumor progression (87–89).
SIK1 can also inhibit Smad7 (90)
and other targets of the TGF-β pathway in colorectal cancer,
thereby inhibiting colorectal cancer metastasis (77). Furthermore, SIK1 inhibits EMT by
regulating the level of ZEB1 and reversing oxaliplatin resistance
in colorectal cancer (Fig. 5A)
(77). An elevated level of ZEB1
expression was shown to promote EMT development and tumor
metastasis in both ovarian cancer and NSCLC (84,91).
In metastatic breast cancer, lower levels of SIK1 protein are
associated with an increase in the level of Nav1.5. This channel
protein, encoded by the SCN5A gene, is a voltage-gated
Na+ channel that fulfils a crucial role in nerve and
muscle cells. In cancer cells, Nav1.5 also has a role in cell
adhesion and movement. When Nav1.5 levels are high due to a
reduction in the level of SIK1, cancer cells become more
aggressive. This happens as increased Nav1.5 activity triggers
increases in the levels of two other proteins, SNAI1 and ZEB1,
which are known to be involved in EMT. EMT allows cancer cells to
lose their original form and become more motile, contributing to
their spread (89,92). Nav1.5 overexpression has also been
shown to promote Na+-mediated invasiveness, since the
tumor cells are sensitive to Na+ (93).
Role of the Wnt/β-catenin signaling
pathway
SIK1 suppresses EMT by inhibiting the
transcriptional activity of β-catenin, thereby inhibiting tumor
growth and metastasis in liver cancer (25). Twist1 is a protein that functions as
a brake on gene expression. When Twist1 levels are high, this
protein is able to promote cancer cell invasion, migration and
resistance to cell death signals (94). In liver cancer, SIK1 and β-catenin
have been revealed to work together to regulate Twist1. They
phosphorylate the co-repressor, silencing mediator of retinoic acid
and thyroid hormone receptor (SMRT), which then inhibits Twist1
activity (Fig. 5A). This pathway
inhibits the growth and spread of liver cancer. Interestingly,
researchers have also found that Twist1 can target the SIK1 gene
directly in vivo. Through binding to the SIK1 promoter,
Twist1 can suppress SIK1 production: This creates a negative
feedback loop, where high Twist1 levels can lead to lower SIK1
levels, which partially counteracts the initial inhibitory effect
of SIK1 on Twist1. A low SIK1 expression in liver cancer has been
shown to be suggestive of a poor prognosis, indicating that SIK1
may be a valuable biomarker for liver cancer (25).
Role of the p53 signaling pathway
p53 is also a signaling molecule that lies
downstream of the LKB1-SIK1 axis with its own associated regulatory
pathway. Studies have shown that SIK1 inhibits breast cancer cell
proliferation through activating p53 transcriptional activity,
which promotes oxidative phosphorylation and inhibits aerobic
glycolysis (Fig. 5A) (24). SIK1 also connects LKB1 with
p53-dependent anoikis, thereby suppressing breast cancer metastasis
(15). Furthermore, p53
participates in the regulation of medulloblastoma tumorigenesis
mediated by SIK1. miR-130b-3p was identified to inhibit
medulloblastoma tumor progression by downregulating the expression
of SIK1 factor via p53 targeting (Fig.
5B) (30).
Role of the PI3K-AKT signaling
pathway
Overactive ATK signaling has been found to
contribute towards breast cancer. In breast cancer, SIK1 functions
as a downstream target of PI3K-AKT, acting as the bridge between
AKT and STAT3. SIK1 binds AKT and is phosphorylated at Ser-435.
Following phosphorylation, SIK1 interacts with 14-3-3 proteins, and
is translocated to the cytoplasm. The presence of the prolyl
isomerase Pin1 promotes the interaction between SIK1 and the E3
ubiquitin-protein ligase ITCH, which enhances SIK1 ubiquitination
and degradation (Fig. 5A).
Considered overall, these factors have been shown to ameliorate the
inhibitory effect of SIK1 on STAT3, thereby promoting breast cancer
development (95).
Regulation of SIK1 via other signaling
mechanisms
In addition to the aforementioned molecular
pathways, several other molecules have also been reported to
regulate SIK1, among which miRs are the most extensively
investigated (Fig. 5B). Through the
previous research articles on SIK1, the inhibitory effects of eight
miRs on SIK1 and tumor growth have been summarized (Table III). miR-141 was shown to
downregulate SIK1 protein, which led to the promotion of cancer
cell proliferation (96). Increased
miR-17 levels were also identified to enhance the aggression and
mobilization of colorectal cancer cells. Dual-luciferase reporter
gene assay demonstrated that SIK1 directly targets miR-17.
Consequently, the upregulation of miR-17 may exert oncogenic
effects via targeting SIK1. The SIK1 protein presents itself as a
potential therapeutic target for colorectal cancer (27). miR-25 targets SIK1 to downregulate
the expression of SIK1 protein, which consequently increases the
activity of the Wnt/β-catenin signaling pathway, promoting the
progression of liver cancer and potentially providing a novel
therapeutic strategy for the targeted therapy of liver cancer
(76). miR-373 has been found to
inhibit SIK1 expression to enhance melanoma cell migration
(97). Moreover, inhibition of SIK1
by miR-203 promoted the proliferation, migration and invasion of
pancreatic cancer cells (28).
Other types of non-coding RNAs, such as long non-coding RNAs
(lncRNAs) and circRNAs, have been shown to mediate the regulatory
effects of SIK1 on tumor growth (Fig.
5B). Among them, circEIF4G3, circ0078607 and the lncRNA
NR2F1-AS1 indirectly inhibit SIK1 expression through regulating
miRs (29,78,98);
the lncRNA TCONS 0029157 directly targeted SIK1 to modulate tumor
development (99). circEIF4G3 was
found to regulate the miR-4449-SIK1 signaling axis, thereby
inhibiting the β-catenin signaling pathway and tumor growth
(29). Eventually, circEIF4G3 was
also shown to stimulate the β-catenin signaling pathway, thereby
inhibiting gastric carcinogenesis (29). A recent study on ovarian cancer
showed that circ0078607 may act as an miR-32-5p ‘sponge’ to
modulate the expression of the SIK1 protein, thereby suppressing
ovarian cancer progression (78).
NR2F1-AS1 inhibits the migration and invasion of cervical squamous
cell carcinoma through modulating the miR-17-SIK1 axis (98). Finally, activation of SIK1 by TCONS
0029157 was shown to inhibit the proliferation, migration and
metastasis of lung cancer (99).
 | Table III.Summary of eight miRs regulated by
SIK1. |
Table III.
Summary of eight miRs regulated by
SIK1.
| First author,
year | Name | Targeted
cancer | Action on SIK1 | Functions | (Refs.) |
|---|
| Chen et al,
2016 | miR-141 | Ovarian cancer | Inhibitory
effect | MiR-141 can promote
the proliferation of ovarian cancer cells by inhibiting the
expression of SIK1 protein. | (96) |
| Huang et al,
2019 | miR-17 | Colorectal
cancer | Inhibitory
effect | MiR-17 can inhibit
the invasion and migration of colorectal cancer cells by targeting
the inhibition of SIK1. | (27) |
| Peng et al,
2020 |
| Cervical squamous
cell carcinoma | Inhibitory
effect | NR2F1-AS1 inhibits
the invasion and migration of cervical squamous cell carcinoma by
regulating the miR-17/SIK1 axis. | (98) |
| Fu et al,
2017 | miR-25 | Liver cancer | Inhibitory
effect | MiR-25 targets SIK1
and promotes liver cancer development by decreasing the expression
of SIK1. | (76) |
| Bai et al,
2018 | miR-373 | Melanoma | Inhibitory
effect | MiR-373 inhibits
the expression of SIK1 and promotes the migration of melanoma
cells. | (97) |
| Ren et al,
2016 | miR-203 | Pancreatic
cancer | Inhibitory
effect | The inhibition of
SIK1 by miR-203 promotes the proliferation, migration and invasion
of pancreatic cancer cells. | (28) |
| Huang et al,
2020 | miR-130b-3p |
Medulloblastoma | Inhibitory
effect | SIK1 promotes the
development of medulloblastoma, and miR130b-3p downregulates SIK1
to inhibit the migration and invasion of medulloblastoma
cells. | (30) |
| Zang et al,
2022 | miR-4449 | Gastric cancer | Inhibitory
effect | CircEIF4G3 can
inhibit the development of gastric cancer by regulating the
miR-4449/SIK1 axis to inactivate β-catenin signaling. | (29) |
| Jin and Wang,
2022 | miR-32-5p | Ovarian cancer | Inhibitory
effect | Circ0078607 acts as
a molecular sponge of miR-32-5p to regulate the miR-32-5p/SIK1 axis
and thus inhibit the progression of ovarian cancer. | (78) |
Discussion
SIK1 phosphorylates the Thr-1391 site of SMRT,
thereby promoting nuclear localization, inhibiting the
Wnt/β-catenin/Twist1 signaling pathway, and silencing the progress
of liver cancer (25). Under
identical conditions, SIK2 and SIK3 do not exhibit these effects.
The structural differences among the three SIK proteins,
particularly in their C-terminal domains, contribute to their
varied roles in tumor development. The potential for further
distinguishing and deepening the understanding of these
differences, especially associated with the C-terminal long carbon
chain and UBA domain in the SIK family, may be explored through
X-ray crystallization analysis of the SIK structures. The three
members of the SIK family have different functions in tumor
regulation. The overexpression of SIK2/3 leads to progression of
the cell cycle (promoting the G1/S phase), which provides tumor
cells with the capability of rapid reproduction (100–104). By contrast, SIK1 exerts an
inhibitory role in the majority of tumor types. An enhanced
understanding of the structure of the SIK family members would be
helpful in terms of understanding the reasons why SIK exerts
different roles in tumor regulation. Additionally, SIK1 has been
found to promote cell proliferation in mouse neuro-endothelial
cells (105) under conditions of
oxygen and glucose deprivation, as well as in hypoglycemic mouse
neuro-endothelial cells (30). This
situation is similar to the roles that have been identified for
SIK1 in medulloblastoma and desmoplastic small round cell tumor
cells. These results suggest that the functions of SIK1 may be
cell-specific, and further research is required to delineate fully
the mechanisms of SIK1 in tumorigenesis. SIK1 has been shown to
influence the progression of breast cancer through the PI3K/AKT and
P53 signal-transduction pathways (24,95).
Via the same P53 signal pathway, SIK1 is able to inhibit the
development of breast cancer and promote medulloblastoma
proliferation (24,30). In addition, the SIK1 gene may be
utilized as a downstream target of various regulatory RNA species,
fulfilling regulatory roles in the occurrence of different types of
cancer; for example, targeting SIK1 with miR-17 can regulate both
colorectal cancer and cervical squamous cell carcinoma (27,98).
In the future, all types of RNA that are targeted to regulate the
SIK1 gene should be investigated; at the same time, larger studies
enrolling more patients with cancer are required to evaluate more
comprehensively the potential biological functions of RNAs in terms
of their regulation of SIK1.
Conclusion and future prospects
SIK1 is a member of AMPK kinase family, which
contains multiple domains and phosphorylation sites. These special
structures ensure that SIK1 can be easily phosphorylated and
activated, and it can be used as an intermediate component for
connecting various cytokines, thus playing a connecting role in
signal molecular networks. In the present review, it is considered
that SIK1 has inhibitory effects on lung, liver, breast, gastric,
colorectal, ovarian, pancreatic and thyroid cancers, as well as
osteosarcoma. By contrast, it can promote medulloblastoma and
desmoplastic small round cell tumor with adhesion and
proliferation, although it is not known whether the inhibitory
function comes from the difference of genes themselves or from the
difference of tumor cells, which needs further experimental
verification. SIK1 is involved in most classical tumor signaling
pathways, including TGFβ, Wnt/β catenin and PI3K/AKT pathways. In
addition, different types of RNA, such as miRs, lncRNAs and
circRNAs, also indirectly participate in the regulation of SIK1 in
tumor signaling pathway, or they directly affect the expression of
SIK1. Moreover, most of the recent studies on SIK1 are related to
RNA, and the combination of SIK1 with various RNA molecules is
expected to make a further breakthrough in targeted regulation. In
conclusion, the present review provides novel insights into the
intricate regulatory mechanisms of SIK1 and its diverse
implications in cancer progression. By elucidating the multifaceted
functions of SIK1, the way is paved for the development of
precision medicinal approaches that leverage SIK1 as a strategic
target for cancer intervention. The current findings emphasize the
importance of understanding the complex roles of SIK1 in cancer,
which may lead to the discovery of more effective therapies and
improved patient outcomes.
Acknowledgements
Not applicable.
Funding
The present study was supported by the project exploring the
mechanism of SIK1 resistance to FGFR-positive non-small cell lung
cancer (grant no. 2022BCE038) and the project research on the
mechanism of CX3CR1 regulating the PI3K/AKT/mTOR signaling pathway
in non-small cell lung cancer through the G-protein-coupled
receptor (grant no. 2022HC44).
Availability of data and materials
Not applicable.
Authors' contributions
JC received funding, designed and directed the
study. YS, DXA and YZ reviewed the references. XRZ and CYZ wrote
the manuscript. JC, QQY, XCP and JYX revised the manuscript. ZXR
and JL contributed to the figures and diagrams. All authors read
and approved the final version of the manuscript. Data
authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hollstein PE, Eichner LJ, Brun SN,
Kamireddy A, Svensson RU, Vera LI, Ross DS, Rymoff TJ, Hutchins A,
Galvez HM, et al: The AMPK-related kinases SIK1 and SIK3 mediate
key tumor-suppressive effects of LKB1 in NSCLC. Cancer Discov.
9:1606–1627. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bright NJ, Thornton C and Carling D: The
regulation and function of mammalian AMPK-related kinases. Acta
Physiol (Oxf). 196:15–26. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sun Z, Jiang Q, Li J and Guo J: The potent
roles of Salt-inducible kinases (SIKs) in metabolic homeostasis and
tumorigenesis. Signal Transduct Target Ther. 5:1502020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wein MN, Foretz M, Fisher DE, Xavier RJ
and Kronenberg HM: Salt-inducible kinases: Physiology, regulation
by cAMP, and therapeutic potential. Trends Endocrinol Metab.
29:723–735. 2028. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang Z, Takemori H, Halder SK, Nonaka Y
and Okamoto M: Cloning of a novel kinase (SIK) of the SNF1/AMPK
family from high salt diet-treated rat adrenal. FEBS Lett.
453:135–139. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Okamoto M, Takemori H and Katoh Y:
Salt-inducible kinase in steroidogenesis and adipogenesis. Trends
Endocrinol Metab. 15:21–26. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Feldman JD, Vician L, Crispino M, Hoe W,
Baudry M and Herschman HR: The salt-inducible kinase, SIK, is
induced by depolarization in brain. J Neurochem. 74:2227–2238.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Horike N, Takemori H, Katoh Y, Doi J, Min
L, Asano T, Sun XJ, Yamamoto H, Kasayama S, Muraoka M, et al:
Adipose-specific expression, phosphorylation of Ser794 in insulin
receptor substrate-1, and activation in diabetic animals of
salt-inducible kinase-2. J Biol Chem. 278:18440–18447. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen F, Chen L, Qin Q and Sun X:
Salt-Inducible Kinase 2: An oncogenic signal transmitter and
potential target for cancer therapy. Front Oncol. 9:182019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Song D, Yin L, Wang C and Wen X:
Adenovirus-mediated expression of SIK1 improves hepatic glucose and
lipid metabolism in type 2 diabetes mellitus rats. PLoS One.
14:e02109302019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang C, Song D, Fu J and Wen X: SIK1
Regulates CRTC2-Mediated gluconeogenesis signaling pathway in human
and mouse liver cells. Front Endocrinol (Lausanne). 11:5802020.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gao WW, Tang HV, Cheng Y, Chan CP, Chan CP
and Jin DY: Suppression of gluconeogenic gene transcription by
SIK1-induced ubiquitination and degradation of CRTC1. Biochim
Biophys Acta Gene Regul Mech. 1861:211–223. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Song D, Yin L, Wang C and Wen X: Zhenqing
recipe attenuates Non-alcoholic fatty liver disease by regulating
the SIK1/CRTC2 signaling in experimental diabetic rats. BMC
Complement Med Ther. 20:272020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang Y, Takemori H, Wang C, Fu J, Xu M,
Xiong L, Li N and Wen X: Role of salt inducible kinase 1 in high
glucose-induced lipid accumulation in HepG2 cells and metformin
intervention. Life Sci. 173:107–115. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cheng H, Liu P, Wang ZC, Zou L, Santiago
S, Garbitt V, Gjoerup OV, Iglehart JD, Miron A, Richardson AL, et
al: SIK1 couples LKB1 to p53-dependent anoikis and suppresses
metastasis. Sci Signal. 2:ra352009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Park M, Miyoshi C, Fujiyama T, Kakizaki M,
Ikkyu A, Honda T, Choi J, Asano F, Mizuno S, Takahashi S, et al:
Loss of the conserved PKA sites of SIK1 and SIK2 increases sleep
need. Sci Rep. 10:86762020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Darling NJ and Cohen P: Nuts and bolts of
the salt-inducible kinases (SIKs). Biochem J. 478:1377–1397. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pires NM, Igreja B and Soares-da-Silva P:
Antagonistic modulation of SIK1 and SIK2 isoforms in high blood
pressure and cardiac hypertrophy triggered by high-salt intake.
Clin Exp Hypertens. 43:428–435. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jaitovich A and Bertorello AM:
Intracellular sodium sensing: SIK1 network, hormone action and high
blood pressure. Biochim Biophys Acta. 1802:1140–1149. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hansen J, Snow C, Tuttle E, Ghoneim DH,
Yang CS, Spencer A, Gunter SA, Smyser CD, Gurnett CA, Shinawi M, et
al: De novo mutations in SIK1 cause a spectrum of developmental
epilepsies. Am J Hum Genet. 96:682–690. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pröschel C, Hansen JN, Ali A, Tuttle E,
Lacagnina M, Buscaglia G, Halterman MW and Paciorkowski AR:
Epilepsy-causing sequence variations in SIK1 disrupt synaptic
activity response gene expression and affect neuronal morphology.
Eur J Hum Genet. 25:216–221. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Peng L, Li C, Tang X, Xiang Y, Xu Y, Cao
W, Zhou H and Li S: Blocking salt-inducible kinases with YKL-06-061
prevents PTZ-induced seizures in mice. Brain Behav. 13:e33052023.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Babbe H, Sundberg TB, Tichenor M,
Seierstad M, Bacani G, Berstler J, Chai W, Chang L, Chung M, Coe K,
et al: Identification of highly selective SIK1/2 inhibitors that
modulate innate immune activation and suppress intestinal
inflammation. Proc Natl Acad Sci USA. 121:e23070861202024.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ponnusamy L and Manoharan R: Distinctive
role of SIK1 and SIK3 isoforms in aerobic glycolysis and cell
growth of breast cancer through the regulation of p53 and mTOR
signaling pathways. Biochim Biophys Acta Mol Cell Res.
1868:1189752021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Qu C, He D, Lu X, Dong L, Zhu Y, Zhao Q,
Jiang X, Chang P, Jiang X, Wang L, et al: Salt-inducible Kinase
(SIK1) regulates HCC progression and WNT/β-catenin activation. J
Hepatol. 64:1076–1089. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xin L, Liu C, Liu Y, Mansel RE, Ruge F,
Davies E, Jiang WG and Martin TA: SIKs suppress tumor function and
regulate drug resistance in breast cancer. Am J Cancer Res.
11:3537–3557. 2021.PubMed/NCBI
|
|
27
|
Huang C, Liu J, Xu L, Hu W, Wang J, Wang M
and Yao X: MicroRNA-17 promotes cell proliferation and migration in
human colorectal cancer by downregulating SIK1. Cancer Manag Res.
11:3521–3534. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ren ZG, Dong SX, Han P and Qi J: miR-203
promotes proliferation, migration and invasion by degrading SIK1 in
pancreatic cancer. Oncol Rep. 35:1365–1374. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zang X, Jiang J, Gu J, Chen Y, Wang M,
Zhang Y, Fu M, Shi H, Cai H, Qian H, et al: Circular RNA EIF4G3
suppresses gastric cancer progression through inhi bition of
β-catenin by promoting δ-catenin ubiquitin degradation and u
pregulating SIK1. Mol Cancer. 21:1412022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang S, Xue P, Han X, Zhang C, Yang L,
Liu L, Wang X, Li H, Fu J and Zhou Y: Exosomal miR-130b-3p targets
SIK1 to inhibit medulloblastoma tumorigenesis. Cell Death Dis.
11:4082020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hartono AB, Kang HJ, Shi L, Phipps W,
Ungerleider N, Giardina A, Chen W, Spraggon L, Somwar R, Moroz K,
et al: Salt-inducible Kinase 1 is a potential therapeutic target in
desmoplastic small round cell tumor. Oncogenesis. 11:182022.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Katoh Y, Takemori H, Horike N, Doi J,
Muraoka M, Min L and Okamoto M: Salt-inducible kinase (SIK)
isoforms: Their involvement in steroidogenesis and adipogenesis.
Mol Cell Endocrinol. 217:109–112. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lizcano JM, Göransson O, Toth R, Deak M,
Morrice NA, Boudeau J, Hawley SA, Udd L, Mäkelä TP, Hardie DG and
Alessi DR: LKB1 is a master kinase that activates 13 kinases of the
AMPK subfamily, including MARK/PAR-1. EMBO J. 23:833–843. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sakamoto K, Bultot L and Göransson O: The
Salt-inducible kinases: Emerging metabolic regulators. Trends
Endocrinol Metab. 29:827–840. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Taub M, Springate JE and Cutuli F:
Targeting of renal proximal tubule Na,K-ATPase by Salt-inducible
kinase. Biochem Biophys Res Commun. 393:339–344. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Feng S, Wei F, Shi H, Chen S, Wang B,
Huang D and Luo L: Roles of Salt-inducible kinases in cancer
(Review). Int J Oncol. 63:1182023. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Katoh Y, Takemori H, Lin XZ, Tamura M,
Muraoka M, Satoh T, Tsuchiya Y, Min L, Doi J, Miyauchi A, et al:
Silencing the constitutive active transcription factor CREB by the
LKB1-SIK signaling cascade. FEBS J. 273:2730–2748. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Clark K, MacKenzie KF, Petkevicius K,
Kristariyanto Y, Zhang J, Choi HG, Peggie M, Plater L, Pedrioli PG,
McIver E, et al: Phosphorylation of CRTC3 by the salt-inducible
kinases controls the interconversion of classically activated and
regulatory macrophages. Proc Natl Acad Sci USA. 109:16986–16991.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hashimoto YK, Satoh T, Okamoto M and
Takemori H: Importance of autophosphorylation at Ser186 in the
A-loop of salt inducible kinase 1 for its sustained kinase
activity. J Cell Biochem. 104:1724–1739. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bertorello AM and Zhu JK: SIK1/SOS2
networks: Decoding sodium signals via calcium-responsive protein
kinase pathways. Pflugers Arch. 458:613–619. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jaleel M, Villa F, Deak M, Toth R,
Prescott AR, Van Aalten DM and Alessi DR: The Ubiquitin-associated
domain of AMPK-related kinases regulates conformation and
LKB1-mediated phosphorylation and activation. Biochem J.
394:545–555. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jaleel M, McBride A, Lizcano JM, Deak M,
Toth R, Morrice NA and Alessi DR: Identification of the sucrose
non-fermenting related kinase SNRK, as a novel LKB1 substrate. FEBS
Lett. 579:1417–1423. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Al-Hakim AK, Göransson O, Deak M, Toth R,
Campbell DG, Morrice NA, Prescott AR and Alessi DR: 14-3-3
cooperates with LKB1 to regulate the activity and localization of
QSK and SIK. J Cell Sci. 118:5661–5673. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Berggreen C, Henriksson E, Jones HA,
Morrice N and Göransson O: cAMP-elevation mediated by β-adrenergic
stimulation inhibits salt-inducible kinase (SIK) 3 activity in
adipocytes. Cell Signal. 24:1863–1871. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Matsumoto S, Iwakawa R, Takahashi K, Kohno
T, Nakanishi Y, Matsuno Y, Suzuki K, Nakamoto M, Shimizu E, Minna
JD and Yokota J: Prevalence and specificity of LKB1 genetic
alterations in lung cancers. Oncogene. 26:5911–5918. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shackelford DB: Unravelling the connection
between metabolism and tumorigenesis through studies of the liver
kinase B1 tumour suppressor. J Carcinog. 12:162013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Eneling K, Brion L, Pinto V, Pinho MJ,
Sznajder JI, Mochizuki N, Emoto K, Soares-da-Silva P and Bertorello
AM: Salt-inducible kinase 1 regulates E-cadherin expression and
intercellular junction stability. FASEB J. 26:3230–3239. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Grahame Hardie D: AMP-activated protein
kinase: A key regulator of energy balance with many roles in human
disease. J Intern Med. 276:543–559. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Taub M: Salt inducible kinase signaling
networks: Implications for acute kidney injury and therapeutic
potential. Int J Mol Sci. 20:32192019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yang Z, Wang C, Xue Y, Liu X, Chen S, Song
C, Yang Y and Guo Y: Calcium-activated 14-3-3 proteins as a
molecular switch in salt stress tolerance. Nat Commun. 10:11992019.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sonntag T, Vaughan JM and Montminy M:
14-3-3 proteins mediate inhibitory effects of cAMP on
salt-inducible kinases (SIKs). FEBS J. 285:467–480. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Thommesen L, Nørsett K, Sandvik AK, Hofsli
E and Laegreid A: Regulation of inducible cAMP early repressor
expression by gastrin and cholecystokinin in the pancreatic cell
line AR42J. J Biol Chem. 275:4244–4250. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Thommesen L, Hofsli E, Paulssen RH,
Anthonsen MW and Laegreid A: Molecular mechanisms involved in
gastrin-mediated regulation of cAMP-responsive promoter elements.
Am J Physiol Endocrinol Metab. 281:E1316–E1325. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Berdeaux R, Goebel N, Banaszynski L,
Takemori H, Wandless T, Shelton GD and Montminy M: SIK1 is a class
II HDAC kinase that promotes survival of skeletal myocytes. Nat
Med. 13:597–603. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
55
|
Selvik LK, Rao S, Steigedal TS, Haltbakk
I, Misund K, Bruland T, Prestvik WS, Lægreid A and Thommesen L:
Salt-inducible kinase 1 (SIK1) is induced by gastrin and inhibits
migration of gastric adenocarcinoma cells. PLoS One. 9:e1124852014.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
van der Linden AM, Nolan KM and Sengupta
P: KIN-29 SIK regulates chemoreceptor gene expression via an MEF2
transcription factor and a class II HDAC. EMBO J. 26:358–370. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chan JK, Sun L, Yang XJ, Zhu G and Wu Z:
Functional characterization of an amino-terminal region of HDAC4
that possesses MEF2 binding and transcriptional repressive
activity. J Biol Chem. 278:23515–23521. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Haberland M, Montgomery RL and Olson EN:
The many roles of histone deacetylases in development and
physiology: Implications for disease and therapy. Nat Rev Genet.
10:32–42. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Longin S, Jordens J, Martens E, Stevens I,
Janssens V, Rondelez E, De Baere I, Derua R, Waelkens E, Goris J,
et al: An inactive protein phosphatase 2A population is associated
with methylesterase and can be re-activated by the phosphotyrosyl
phosphatase activator. Biochem J. 380:111–119. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Pagel P, Zatti A, Kimura T, Duffield A,
Chauvet V, Rajendran V and Caplan MJ: Ion pump-interacting
proteins: Promising new partners. Ann N Y Acad Sci. 986:360–368.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Sjöström M, Stenström K, Eneling K,
Zwiller J, Katz AI, Takemori H and Bertorello AM: SIK1 is part of a
cell sodium-sensing network that regulates active sodium transport
through a calcium-dependent process. Proc Natl Acad Sci USA.
104:16922–16927. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhang Y, Gao W, Yang K, Tao H and Yang H:
Salt-Inducible Kinase 1 (SIK1) is induced by alcohol and suppresses
microglia inflammation via NF-κB signaling. Cell Physiol Biochem.
47:1411–1421. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Norman P: The use of salt-inducible kinase
inhibitors to treat autoimmune and inflammatory diseases:
Evaluation of WO2013136070. Expert Opin Ther Pat. 24:943–946. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Sundberg TB, Liang Y, Wu H, Choi HG, Kim
ND, Sim T, Johannessen L, Petrone A, Khor B, Graham DB, et al:
Development of chemical probes for investigation of Salt-inducible
kinase function in vivo. ACS Chem Biol. 11:2105–2111. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
McIver Edward G, Bryans Justin S,
Smiljanic ELA, Lewis Stephen J, Hough J and Drake T: Pyrimidine
derivatives capable of inhibiting one or more kinases. 2009.
|
|
66
|
Raposo TP, Beirão BC, Pang LY, Queiroga FL
and Argyle DJ: Inflammation and cancer: Till death tears them
apart. Vet J. 205:161–174. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Chai EZ, Siveen KS, Shanmugam MK, Arfuso F
and Sethi G: Analysis of the intricate relationship between chronic
inflammation and cancer. Biochem J. 468:1–15. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Hu J, Qiao J, Yu Q, Liu B, Zhen J, Liu Y,
Ma Q, Li Y, Wang Q, Wang C, et al: Role of SIK1 in the transition
of acute kidney injury into chronic kidney disease. J Transl Med.
19:692021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Meng J, Li N, Liu X, Qiao S, Zhou Q, Tan
J, Zhang T, Dong Z, Qi X, Kijlstra A, et al: NLRP3 attenuates
intraocular inflammation by inhibiting AIM2-mediated pyroptosis
through the phosphorylated salt-inducible kinase 1/Sterol
regulatory element binding transcription factor 1 pathway.
Arthritis Rheumatol. 75:842–855. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Pirie E, Cauntay P, Fu W, Ray S, Pan C,
Lusis AJ, Hsiao J, Burel SA, Narayanan P, Crooke RM, et al: Hybrid
mouse diversity panel identifies genetic architecture associated
with the acute antisense oligonucleotide-mediated inflammatory
response to a 2'-O-Methoxyethyl antisense oligonucleotide. Nucleic
Acid Ther. 29:266–277. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Lombardi MS, Gilliéron C, Dietrich D and
Gabay C: SIK inhibition in human myeloid cells modulates TLR and
IL-1R signaling and induces an anti-inflammatory phenotype. J
Leukoc Biol. 99:711–721. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Cai X, Wang L, Yi Y, Deng D, Shi M, Tang
M, Li N, Wei H, Zhang R, Su K, et al: Discovery of
pyrimidine-5-carboxamide derivatives as novel salt-inducible
kinases (SIKs) inhibitors for inflammatory bowel disease (IBD)
treatment. Eur J Med Chem. 256:1154692023
|
|
73
|
Qu C and Qu Y: Down-regulation of
salt-inducible kinase 1 (SIK1) is mediated by RNF2 in
hepatocarcinogenesis. Oncotarget. 8:3144–3155. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Murray CW, Brady JJ, Tsai MK, Li C,
Winters IP, Tang R, Andrejka L, Ma RK, Kunder CA, Chu P and Winslow
MM: An LKB1-SIK axis suppresses lung tumor growth and controls
differentiation. Cancer Discov. 9:1590–1605. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Cheng D, Wang J, Wang Y, Xue Y, Yang Q,
Yang Q, Zhao H, Huang J and Peng X: Chemokines: Function and
therapeutic potential in bone metastasis of lung cancer. Cytokine.
172:1564032023. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Fu X, Tang Y, Wu W, Ouyang Y, Tan D and
Huang Y: Exosomal microRNA-25 released from cancer cells targets
SIK1 to promote hepatocellular carcinoma tumorigenesis. Dig Liver
Dis. 54:954–963. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Gao Y, Li H, Wang P, Wang J and Yao X:
SIK1 suppresses colorectal cancer metastasis and chemoresistance
via the TGF-β signaling pathway. J Cancer. 14:2455–2467. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Jin Y and Wang H: Circ_0078607 inhibits
the progression of ovarian cancer via regulating the miR-32-5p/SIK1
network. J Ovarian Res. 15:32022. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Kou B, Wang XD, Sun XP, Qi Q, Yang M, Yun
YN, Zhou JS and Liu W: LKB1 inhibits proliferation, metastasis and
angiogenesis of thyroid cancer by upregulating SIK1. J Cancer.
13:2872–2883. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Li B, Chen Y, Wang F, Guo J, Fu W, Li M,
Zheng Q, Liu Y, Fan L, Li L and Xu C: Bmi1 drives
hepatocarcinogenesis by repressing the TGFβ2/SMAD signalling axis.
Oncogene. 39:1063–1079. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
De Faveri LE, Hurst CD, Roulson JA, Wood
H, Sanchez-Carbayo M, Knowles MA and Chapman EJ: Polycomb repressor
Complex 1 member, BMI1 contributes to urothelial tumorigenesis
through p16-independent mechanisms. Transl Oncol. 8:387–399. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wang Q, Wu Y, Lin M, Wang G, Liu J, Xie M,
Zheng B, Shen C and Shen J: BMI1 promotes osteosarcoma
proliferation and metastasis by repressing the transcription of
SIK1. Cancer Cell Int. 22:1362022. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Nguyen K, Hebert K, McConnell E, Cullen N,
Cheng T, Awoyode S, Martin E, Chen W, Wu T, Alahari SK, et al: LKB1
signaling and patient survival outcomes in hepatocellular
carcinoma. Pharmacol Res. 192:1067572023. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Yao YH, Cui Y, Qiu XN, Zhang LZ, Zhang W,
Li H and Yu JM: Attenuated LKB1-SIK1 signaling promotes
epithelial-mesenchymal transition and radioresistance of non-small
cell lung cancer cells. Chin J Cancer. 35:502016. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Seoane J and Gomis RR: TGF-β Family
signaling in tumor suppression and cancer progression. Cold Spring
Harb Perspect Biol. 9:a0222772017. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Kowanetz M, Lönn P, Vanlandewijck M,
Kowanetz K, Heldin CH and Moustakas A: TGFbeta induces SIK to
negatively regulate type I receptor kinase signaling. J Cell Biol.
182:655–662. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Ang HL, Mohan CD, Shanmugam MK, Leong HC,
Makvandi P, Rangappa KS, Bishayee A, Kumar AP and Sethi G:
Mechanism of epithelial-mesenchymal transition in cancer and its
regulation by natural compounds. Med Res Rev. 43:1141–1200. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Saitoh M: Transcriptional regulation of
EMT transcription factors in cancer. Semin Cancer Biol. 97:21–29.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Gradek F, Lopez-Charcas O, Chadet S,
Poisson L, Ouldamer L, Goupille C, Jourdan ML, Chevalier S,
Moussata D, Besson P, et al: Sodium Channel Na(v)1.5 controls
Epithelial-to-mesenchymal transition and invasiveness in breast
cancer cells through its regulation by the salt-inducible Kinase-1.
Sci Rep. 9:186522019. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Lönn P, Vanlandewijck M, Raja E, Kowanetz
M, Watanabe Y, Kowanetz K, Vasilaki E, Heldin CH and Moustakas A:
Transcriptional induction of salt-inducible kinase 1 by
transforming growth factor β leads to negative regulation of type I
receptor signaling in cooperation with the Smurf2 ubiquitin ligase.
J Biol Chem. 287:12867–12878. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Sánchez-Tilló E, Liu Y, de Barrios O,
Siles L, Fanlo L, Cuatrecasas M, Darling DS, Dean DC, Castells A
and Postigo A: EMT-activating transcription factors in cancer:
Beyond EMT and tumor invasiveness. Cell Mol Life Sci. 69:3429–3456.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Nelson M, Yang M, Millican-Slater R and
Brackenbury WJ: Nav1.5 regulates breast tumor growth and metastatic
dissemination in vivo. Oncotarget. 6:32914–32929. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Yang M, Kozminski DJ, Wold LA, Modak R,
Calhoun JD, Isom LL and Brackenbury WJ: Therapeutic potential for
phenytoin: Targeting Na(v)1.5 sodium channels to reduce migration
and invasion in metastatic breast cancer. Breast Cancer Res Treat.
134:603–615. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Zhu QQ, Ma C, Wang Q, Song Y and Lv T: The
role of TWIST1 in epithelial-mesenchymal transition and cancers.
Tumour Biol. 37:185–197. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Sun Z, Jiang Q, Gao B, Zhang X, Bu L, Wang
L, Lin Y, Xie W, Li J and Guo J: AKT blocks SIK1-mediated
repression of STAT3 to promote breast tumorigenesis. Cancer Res.
83:1264–1279. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Chen JL, Chen F, Zhang TT and Liu NF:
Suppression of SIK1 by miR-141 in human ovarian cancer cell lines
and tissues. Int J Mol Med. 37:1601–1610. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Bai X, Yang M and Xu Y: MicroRNA-373
promotes cell migration via targeting salt-inducible kinase 1
expression in melanoma. Exp Ther Med. 16:4759–4764. 2018.PubMed/NCBI
|
|
98
|
Peng J, Hou F, Zhu W, Li J and Teng Z:
lncRNA NR2F1-AS1 regulates miR-17/SIK1 axis to suppress the
invasion and migration of cervical squamous cell carcinoma cells.
Reprod Sci. 27:1534–1539. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Bawa P, Zackaria S, Verma M, Gupta S,
Srivatsan R, Chaudhary B and Srinivasan S: Integrative analysis of
normal long intergenic non-coding RNAs in prostate cancer. PLoS
One. 10:e01221432015. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Bon H, Wadhwa K, Schreiner A, Osborne M,
Carroll T, Ramos-Montoya A, Ross-Adams H, Visser M, Hoffmann R,
Ahmed AA, et al: Salt-inducible kinase 2 regulates mitotic
progression and transcription in prostate cancer. Mol Cancer Res.
13:620–635. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Gao T, Zhang X, Zhao J, Zhou F, Wang Y,
Zhao Z, Xing J, Chen B, Li J and Liu S: SIK2 promotes reprogramming
of glucose metabolism through PI3K/AKT/HIF-1α pathway and
Drp1-mediated mitochondrial fission in ovarian cancer. Cancer Lett.
469:89–101. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Zhou J, Alfraidi A, Zhang S,
Santiago-O'Farrill JM, Yerramreddy Reddy VK, Alsaadi A, Ahmed AA,
Yang H, Liu J, Mao W, et al: A novel compound ARN-3236 inhibits
Salt-inducible kinase 2 and sensitizes ovarian cancer cell lines
and xenografts to paclitaxel. Clin Cancer Res. 23:1945–1954. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Du WQ, Zheng JN and Pei DS: The diverse
oncogenic and tumor suppressor roles of salt-inducible kinase (SIK)
in cancer. Expert Opin Ther Targets. 20:477–485. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Charoenfuprasert S, Yang YY, Lee YC, Chao
KC, Chu PY, Lai CR, Hsu KF, Chang KC, Chen YC, Chen LT, et al:
Identification of salt-inducible kinase 3 as a novel tumor antigen
associated with tumorigenesis of ovarian cancer. Oncogene.
30:3570–3584. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Zhou X, Xu B, Gu Y, Ji N, Meng P and Dong
L: Long noncoding RNA SNHG1 protects brain microvascular
endothelial cells against oxygen-glucose
deprivation/reoxygenation-induced injury by sponging miR-298 and
upregulating SIK1 expression. Biotechnol Lett. 43:1163–1174. 2021.
View Article : Google Scholar : PubMed/NCBI
|