Introduction
Lung cancer has high morbidity (~12.4% of new cases
of cancer) and mortality (~18.7% of cancer deaths) rates, making it
the leading cause of cancer deaths globally (1–3). Lung
cancer is categorized into small cell lung cancer and non-small
cell lung cancer (NSCLC) including lung squamous carcinoma, lung
adenocarcinoma (LUAD) and lung large cell carcinoma, of which NSCLC
is the predominant histologic type (3). Although there have been considerable
advancements in the early detection strategies of NSCLC, such as
early screening and minimally invasive diagnostics, and in the
treatment of NSCLC with radiation therapy, targeted therapies
against disease-associated oncogenic driver molecules and
immunotherapies related to immune checkpoint inhibitors, the
identification of emerging biomarkers is important for guiding the
prognosis of NSCLC (4,5). Understanding the molecular
carcinogenesis and studying oncogenic drivers can greatly aid in
the development of targeted therapeutics for NSCLC and provide
additional treatment options, thereby increasing patient
survival.
Homeobox (HOX) genes are a highly conserved family
of genes encoding a type of DNA-binding transcription factor, which
are involved in cell differentiation, metastasis and angiogenesis
(6). Mammals have 39 HOX genes
arranged into four clusters: A, B, C and D (7,8).
Previous research showed that aberrant HOX gene expression has a
role in the progression of certain types of cancer, such as breast
cancer (9), lung cancer (10) and glioblastoma (11). HOXD, located on chromosome 2q31, is
a subfamily of HOX genes (12) and
includes HOXD1, D3, D4, D8, D9, D10, D11, D12 and D13 genes
(13). In lung cancer research, the
HOXD3 pro-oncogene enhances lung adenocarcinoma cell metastasis by
downregulating E-cadherin and upregulating N-cadherin, as well as
facilitating tumor invasion and angiogenesis by influencing
urokinase-type plasminogen activator and MMP-2 expression (14). Similarly, in A549 cells, a loss of
microRNA (miR)-520a-3p causes overexpression of HOXD8, which
enhances cell proliferation and cancer cell stemness (15). Overexpressed HOXD9 has also been
reported to target the
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 promoter
region, which may lead to malignant behavior in NSCLC (16). Moreover, miR-224 can participate in
NSCLC progression via regulating HOXD10 expression (17). In summary, dysregulation of the HOXD
genes is associated with the occurrence and progression of lung
cancer. However, to the best of our knowledge, there have been no
reports regarding the role of HOXD1 in the occurrence and
development of lung cancer to date.
Epigenetic control is the most prevalent method in
HOX gene regulation (18), and
abnormal DNA methylation levels of certain HOX gene promoters have
been identified in cancer studies (8). For example, peripheral blood DNA
methylation profiles showed hypermethylation of the HOX gene CpG
region in non-Hodgkin lymphoma (19). Furthermore, the promoters of HOXB5
and HOXB7 have been shown to exhibit hypomethylation levels, which
increases the metastasis of small cell lung cancer (20). Between 60–90% of the CpGs in the
genome are methylated, and the unmethylated CpGs are clustered into
CpG islands in the promoter and exon regions of structural genes
(21). Therefore, an abnormal
methylation status in the CpG islands can lead to the onset of
cancer. The hypermethylation of the tumor suppressor gene CpG
islands inhibits gene transcription, resulting in the loss of an
anti-cancer effect (21,22). Hypermethylation of various genes in
the HOXD family, including HOXD3, HOXD10 and HOXD13, has also been
identified in lung cancer (8).
There are three main catalytic DNA methyltransferases (DNMTs),
DNMT1, DNMT3A and DNMT3B, involved in the DNA methylation process
(23). It has been reported that
normal DNA methylation levels are impacted by DNMT dysregulation. A
previous study reported that DNMT1/3A/3B are overexpressed in oral
squamous cell carcinoma, which triggers promoter hypermethylation
silencing of >40 cancer-suppressor genes (24). In addition, overexpression of
DNMT1/3A in prostate cancer accelerates cancer progression by
epigenetic silencing of Claudin-1 (25). In summary, DNA methylation of
certain HOX genes has been reported to occur in lung cancer and to
contribute to the development of cancer. However, reports on the
mechanism of DNA methylation control of HOXD1 in LUAD re currently
limited.
In the present study, the objective was to
investigate the function of HOXD1 and its regulation mechanisms in
the initiation and development of LUAD. Through the exploration,
this study aimed to reveal the potential of HOXD 1 as a molecular
target for clinical treatment of LUAD, providing theoretical basis
for early diagnosis and treatment of LUAD. The present findings
promoted the promise of novel and impactful therapeutic strategies
for LUAD.
Materials and methods
Differential expression gene analysis
and prognostic value in LUAD
The expression of nine HOX genes of the HOXD cluster
in LUAD tissues and normal tissues were examined using The
University of Alabama at Birmingham Cancer data analysis Portal
(UALCAN) database (http://ualcan.path.uab.edu/analysis-prot.html)
(26). The prognostic value of
HOXD1 expression for overall survival (OS), first progression (FP)
and post-progression survival (PPS) was analyzed using the
Kaplan-Meier plotter (http://kmplot.com) (27).
Cell culture
The human normal lung epithelial cell line (BEAS-2B)
and LUAD cell lines (A549, H1299, H1650 and H1975) were purchased
from Cell Bank/Stem Cell Bank, Chinese Academy of Sciences. BEAS-2B
and A549 were cultured in DMEM (Shanghai VivaCell Biosciences,
Ltd.) supplemented with 1% penicillin/streptomycin (Beijing
Solarbio Science & Technology Co., Ltd.) and 10% FBS (CellMax).
H1299, H1650 and H1975 cells were cultured in RPMI 1640 (Shanghai
VivaCell Biosciences, Ltd.) supplemented with 1%
penicillin/streptomycin (Beijing Solarbio Science & Technology
Co., Ltd.) and 10% FBS. All cells were cultured at 37°C and 5%
CO2.
Virus production and infection
The lentiviruses used in this study were produced by
Shanghai GeneChem Co., Ltd. Lentiviruses expressing HOXD1 were
produced using the lentiviral vector GV492 (Shanghai GeneChem Co.,
Ltd.). The virus transfection process was performed according to
manufacturer's protocol. Briefly, A549 or H1299 cells were seeded
at 3–5×104 cells/ml in 6-well plates. Lentiviruses
containing the negative control (LV-NC; empty lentiviral vector
GV-492) or HOXD1 vector (LV-HOXD1) were transfected into A549 or
H1299 cells (multiplicity of infection, 10). The volume of culture
medium was 1 ml/well in 6-well plates and 1X HitransG P (Shanghai
GeneChem Co., Ltd.) was added to each well and incubate for 14–26 h
at 37°C and 5% CO2, the medium was replaced with fresh
complete culture medium. Puromycin (6 µg/ml) selection was then
used for 2 days and 3 µg/ml puromycin was used for maintenance to
produce stably transfected cells.
Xenograft mouse model
A total of 16 female BALB/cA-nu mice (weight, 18–20
g; age, 4–5-weeks; Beijing HFK Bioscience Co., Ltd.,) were housed
under specific pathogen-free conditions. The mice had ad
libitum access to food/water and were housed under a 12 h
light/dark cycle. The temperature was maintained at 24±2°C and a
relative humidity range of 50–60% was maintained. All procedures
were approved by the Animal Use and Care Committee at Shandong
Provincial Hospital Affiliated with Shandong First Medical
University (approval no. 2021-622; Jinan, China). A549 cells
transfected with either the LV-NC or LV-HOXD1 plasmid vector via
lentiviruses were used in the tumor formation assay. The nude mice
were divided into two groups (8 mice/group): LV-NC and LV-HOXD1.
All mice were injected subcutaneously in the right hind limb with
3×106 A549 LV-NC or LV-HOXD1 cells in 150 µl PBS. The
experimental duration of the present study was 5 weeks, with the
first week allocated for adaptive feeding. The experiment commenced
at the beginning of the second week, with the subcutaneous
injection of cells to establish a xenograft tumor animal model.
Animal health was observed daily and the body weight of the mice
and tumor diameters were measured weekly for 4 weeks. If any humane
endpoints were reached, the animals were sacrificed. These included
a tumor diameter >20 mm, weight loss >20% of body weight, the
animal exhibited cachexia or wasting syndrome or the size of the
solid tumor >10% of body weight. Notably, none of the mice
succumbed to humane endpoints during the experimental process. The
mice were euthanized by cervical dislocation. Tumors were then
dissected and tumor weights were measured.
Small interfering (si)RNA
transfection
The siRNAs (siR) used in the present study were
constructed by external companies (siR-DNMTs, Guangzhou RiboBio
Co., Ltd.; siR-NC, Shanghai GenePharma Co., Ltd.). A549 cells were
incubated with 10 µM siR-DNMTs (Guangzhou RiboBio Co., Ltd.) and
siR-negative control (siR-NC) (Shanghai GenePharma Co., Ltd.) using
Lipofectamine® RNAiMAX Reagent (cat. no. 13778-150;
Invitrogen; Thermo Fisher Scientific, Inc.) (Table SI). An equal concentration of
siR-NC was added to A549 cells as a control. Cells were seeded in
6-well plates until they reached 40–50% confluence. To prepare the
transfection mix according to the manufacturer's protocol, 9 µl
Lipofectamine® RNAiMAX was diluted in 150 µl Opti-MEM
Medium (Gibco; Thermo Fisher Scientific, Inc.) and mixed.
Additionally, 3 µl siRNA was diluted in 150 µl Opti-MEM Medium and
mixed separately. The diluted siRNA was then mixed with the diluted
Lipofectamine® RNAiMAX in a 1:1 ratio to form a
siRNA-lipid complex. This complex was incubated for 5 min at room
temperature before being added to the cultured cells. Following
transfection, the cells were incubated at 37°C. After 48 h of
transfection, the medium was removed from the wells. Cells were
washed 3 times with cold PBS and then 500 µl RNAiso Plus reagent
(Takara Bio, Inc.) was added to thoroughly lyse cells using
RNase-Free Pipette Tips.
RT-qPCR
Total RNA was extracted from BEAS-2B cells and the
LUAD cell lines, A549 cells transfected with siR-NC or with
siR-DNMTs, and A549 and H1299 cells transfected with lentiviruses
containing the negative control (LV-NC) or HOXD1 (LV-HOXD1) vector
using RNAiso Plus reagent (Takara Bio, Inc.) according to the
manufacturer's instructions. The RNA pellet was recovered in
RNase-free water and the RNA concentration and purity were measured
using the NanoDrop 1000 (Thermo Fisher Scientific, Inc.). cDNA was
synthesized using the Evo M-MLV RT Premix (Hunan Accurate
Bio-Medical Technology Co., Ltd.) according to the manufacturer's
protocol. The cDNA was used as the template for RT-qPCR using the
Taq SYBR Green qPCR Premix (Jiangsu Best-Enzymes Biotechnology Co.,
Ltd.). The following thermocycling conditions were used for qPCR:
Initial denaturation at 95°C for 10 sec; 40 cycles of 60°C for 10
sec and 72°C for 30 sec. The final results were analyzed as
previously described (28) and the
relative quantification of target genes were analyzed using the
2−ΔΔCq method (29)
after normalization to β-actin or GAPDH expression levels. The
primers used are listed in Table
SII.
Western blotting
Cells were lysed on ice using RIPA lysis buffer (New
Cell & Molecular Biotech Co., Ltd.) containing Protease
Inhibitor Cocktail (New Cell & Molecular Biotech Co., Ltd.).
Protein concentrations in the samples were quantified using the
NanoDrop 1000 (Thermo Fisher Scientific, Inc.). A total of 75 µg
protein/lane were separated by SDS-PAGE electrophoresis using a
10–12.5% gel, followed by transfer to PVDF membranes (cat. no.
IPVH00010; MilliporeSigma). After blocking in 5% skim milk for 2 h
at room temperature, the membranes were incubated with primary
antibodies against HOXD1 (1:500; cat. no. ab220856; Abcam), Flag M2
(1:5,000; cat. no. m20008m; Abmart Pharmaceutical Technology Co.,
Ltd.) and GAPDH (1:50.000; cat. no. 60004-I–Ig; Proteintech Group,
Inc.) overnight at 4°C. After washing three times with TBST
containing 0.1% Tween-20, the membranes were incubated with
anti-mouse IgG HRP-linked antibodies (1:20,000; cat. no. AB0102;
Shanghai Abways Biotechnology Co., Ltd.) and anti-rabbit IgG
HRP-linked antibodies (1:50,000; cat. no. AB0101; Shanghai Abways
Biotechnology Co., Ltd.) at room temperature for 1 h and an ECL kit
(cat. no. WBKLS0500; MilliporeSigma) were used for visualization.
The chemiluminescence imaging system (ChemiDoc M, Bio-Rad
Laboratories, Inc.) was used for imaging and ImageJ software
(version 1.51j8; National Institutes of Health,) was used for
semi-quantitation of protein levels. GAPDH was used as a reference
control.
Methylation analysis
The UALCAN database was used to analyze HOXD1
promoter methylation levels (26).
CpG islands in the HOXD1 promoter sequence were predicted using
MethPrimer (http://www.urogene.org/methprimer/) (30). TBS was performed by Igenebook
Biotechnology Co., Ltd. Briefly, bisulfite sequencing PCR primers
targeting the HOXD1 promoter region were designed using MethPrimer
(Table SIII). The HOXD1 promoter
region was selected from upstream 2,000 bp to downstream 1,000 bp
of the transcription start site (TSS) and located on the National
Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/) nucleotide sequence
starting from 176,186,579 and ending at 176,189,579 (accession no.
NC_000002). Genomic DNA from BEAS-2B, A549 and H1299 cells were
extracted and bisulfite treatment was performed using the EZ DNA
Methylation-Gold™ Kit (cat. no. D5005; Zymo Research
Corp.). DNA quality was detected using the Qubit 4.0.
Bisulfite-treated templates were subjected to bisulfite sequencing
PCR (BSP) amplification by high-fidelity U-base-resistant DNA
polymerase BSP amplification. BSP amplification products from the
same sample were mixed and labeled primers were amplified to obtain
a bisulfite-converted DNA library. The library from each sample was
pooled and sequenced using the Illumina NovaSeq6000 platform
(Illumina, Inc.) using the paired end 150 bp method as previously
described (31,32). A library concentration of 13.1 ng/µl
was measured using Qubit 4.0. Trimmomatic (version 0.36), BSMAP
(version v2.7.3) and FastQC (version 0.11.7; http://www.bioinformatics.babraham.ac.uk/projects/fastqc/)
were used for data analysis (33,34).
Decitabine treatment
A549 cells at a density of 70–80% were treated with
0.1, 1.0, 5.0 or 10.0 µM decitabine [5-aza-2′-deoxycytidine (DAC);
cat. no. ID0120; Beijing Solarbio Science & Technology Co.,
Ltd.] and 10 µM DMSO (cat. no. D8371; Beijing Solarbio Science
& Technology Co., Ltd.) for 96 h at 37°C and 5% CO2.
The cells were then collected for RNA extraction and RT-qPCR
analysis.
Cell proliferation assay and colony
formation assay
Cell proliferation was measured using crystal violet
staining, Cell Counting Kit-8 (CCK-8) and colony formation assays.
For the crystal violet staining assay, cells were seeded in a
12-well culture plate at 1×105 cells/well and incubated
for 3 days. After washing twice with cold PBS, cells were fixed
using 5% glacial acetic acid for 15 min at room temperature and
then stained with 0.1% crystal violet for 10 min at room
temperature. Cells were imaged using a camera. For the CCK-8 assay,
it was performed according to manufacturer's protocol of CCK-8
reagent (cat. no. BS350B; Biosharp Life Sciences). Briefly, cells
were seeded in a 96-well culture plate at a density of
2–3×103 cells/well. After the cells were adherent and
were cultured for different time periods (0, 24, 48 and 72 h), 10
µl/well of CCK-8 reagent was added, and then the cells were
incubated for 2 h at 37°C and 5% CO2. Absorbance at 450
nm was measured using a microplate reader (SpectraMax 190;
Molecular Devices, LLC). For the colony formation assay, cells were
seeded at a density of 1×103 cells/well in a 6-well
culture plate and incubated for 7 days until a single cell
proliferated to form a visible cluster which was defined as a
colony. After discarding the culture medium and washing twice with
PBS, 4% paraformaldehyde was added to each well to fix the cells
for 30 min at room temperature. Then, cells were stained with 2 ml
of 0.1% crystal violet for 3 min at room temperature. Visible
colonies were counted using Image J software.
Wound healing and Transwell
assays
For the wound-healing assay, 1×106 A549
cells were seeded in a 6-well plate and then scratched with a 10 µl
pipette tip. The cells were washed 3 times with PBS to remove the
scratched cells and then serum-free medium (DMEM or RPMI 1640) was
used to culture the cells. Cells were cultured at 37°C and 5%
CO2. The migration of cells at the indicated time points
of 0 and 96 h was observed using an inverted fluorescence
microscope (Olympus IX73; Olympus Corporation). The width of the
wound was measured using Photoshop (version 2017.1.6; Adobe
Systems, Inc.). For the Transwell experiments, Transwell inserts
were used with or without a Matrigel coating (cat. no. HY-K6001;
MedChemExpress) for the cell invasion and migration experiments.
Matrigel was melted at 4°C overnight and diluted with pre-cooled
serum-free medium at 4°C to a final concentration of 1 mg/ml and
maintained on ice. Then, 100 µl of diluted Matrigel was added to
the center of the bottom of the upper chamber and incubated at 37°C
for 4–5 h to dry. Control cells (A549 LV-NC and H1299 LV-NC) and
HOXD1-overexpression cells (A549 LV-HOXD1 and H1299 LV-HOXD1) were
seeded at a density of 2×105 cells in the upper chamber
in serum-free medium. Regular culture medium containing 10% FBS was
added to the lower chamber. Cells were then incubated at 37°C for
24 h. Migratory and invasive cells in the lower chambers were
stained using 0.1% crystal violet for 2 min at room temperature.
Finally, cells were imaged using a light microscope (Olympus IX73;
Olympus Corporation). Cells were counted and photographed in three
randomly selected fields of view and quantified using Image J
software.
ChIP-seq and ChIP-qPCR
The ChIP assays were conducted by Igenebook
Biotechnology Co., Ltd. Briefly, ~3×107 A549 cells were
washed twice in cold PBS, cross-linked with 1% formaldehyde for 10
min at room temperature, quenched with glycine for 5 min at room
temperature and then washed twice with cold PBS at room
temperature. Cells were collected by centrifugation at 1,000 × g
for 5 min at 4°C. Samples were lysed using 50 mM Tris-HCl (pH 8.0),
10 mM EDTA, 1% SDS, 1X protease inhibitor cocktail (cat. no.
5056489001; MilliporeSigma) and chromatin on ice. The chromatin was
sheared into an average DNA fragment length of 200–500 bp.
Additionally, 20 µl of chromatin was stored at −20°C for input DNA
and 100 µl chromatin was incubated at 4°C overnight with antibodies
against Flag M2 (1:50; cat. no. 14793; Cell Signaling Technology),
DNMT1 (1:100; cat. no. 24206-1-AP; Proteintech), DNMT3A (1:100;
cat. no. 20954-1-AP; Proteintech), DNMT3B (1:50; cat. no.
26971-1-AP; Proteintech) or IgG (1:100; cat. no. 2729S; Cell
Signaling Technology, Inc.) as a negative control for
immunoprecipitation. Then, 30 µl of protein beads
(Dynabeads™ protein G; cat. no. 10004D; Thermo Fisher
Scientific, Inc.) were added and the samples were further incubated
for 3 h at 4°C. The beads were then washed once with 20 mM Tris/HCL
(pH 8.1), 50 mM NaCl, 2 mM EDTA, 1% Triton X-100 and 0.1% SDS,
washed twice with 10 mM Tris/HCL (pH 8.1), 250 mM LiCl, 1 mM EDTA,
1% NP-40 and 1% deoxycholic acid and twice with 1X TE buffer (10 mM
Tris-Cl at pH 7.5 and 1 mM EDTA). Bound material was then eluted
from the beads using 300 µl of elution buffer (100 mM NaHCO3 and 1%
SDS), treated with RNase A (8 µg/ml; cat. no. EN0531; Thermo Fisher
Scientific, Inc.) for 6 h at 65°C and then with proteinase K (345
µg/ml; cat. no. P6556; MilliporeSigma) overnight at 45°C. DNA
concentration and purity were detected using the Q-bit (Qbit 4.0;
Invitrogen; Thermo Fisher Scientific, Inc.). The purified products
were used to construct sequencing libraries following the protocol
provided by the I NEXTFLEX® ChIP-Seq Library Prep Kit
for Illumina® Sequencing (cat. no. NOVA-5143-02, Bioo
Scientific). The concentration of the libraries were assayed using
the Qubit 4.0 and the fragment size determined using the QSep400
(Bioptic). The library concentration of 22.5 nM was determined by
qPCR. Then, the purified products were sequenced on an Illumina
NovaSeq using the paired end 150 bp method (35). Trimmomatic (version 0.36) was used
to filter out low-quality reads. Next, clean reads were mapped to
the human genome using BWA (version 0.7.15-r1140). Samtools
(version 1.3.1) was used to remove potential PCR duplicates. MACS2
software (version 2.1.1.20160309) was adopted to screen peaks
(bandwidth 300 bp; model fold 5, 50; q value 0.05). Peaks were
assigned to genes if their midpoint was closest to the TSS of a
single gene (36). HOMER (version
3) was used to predict motif occurrences within peaks with default
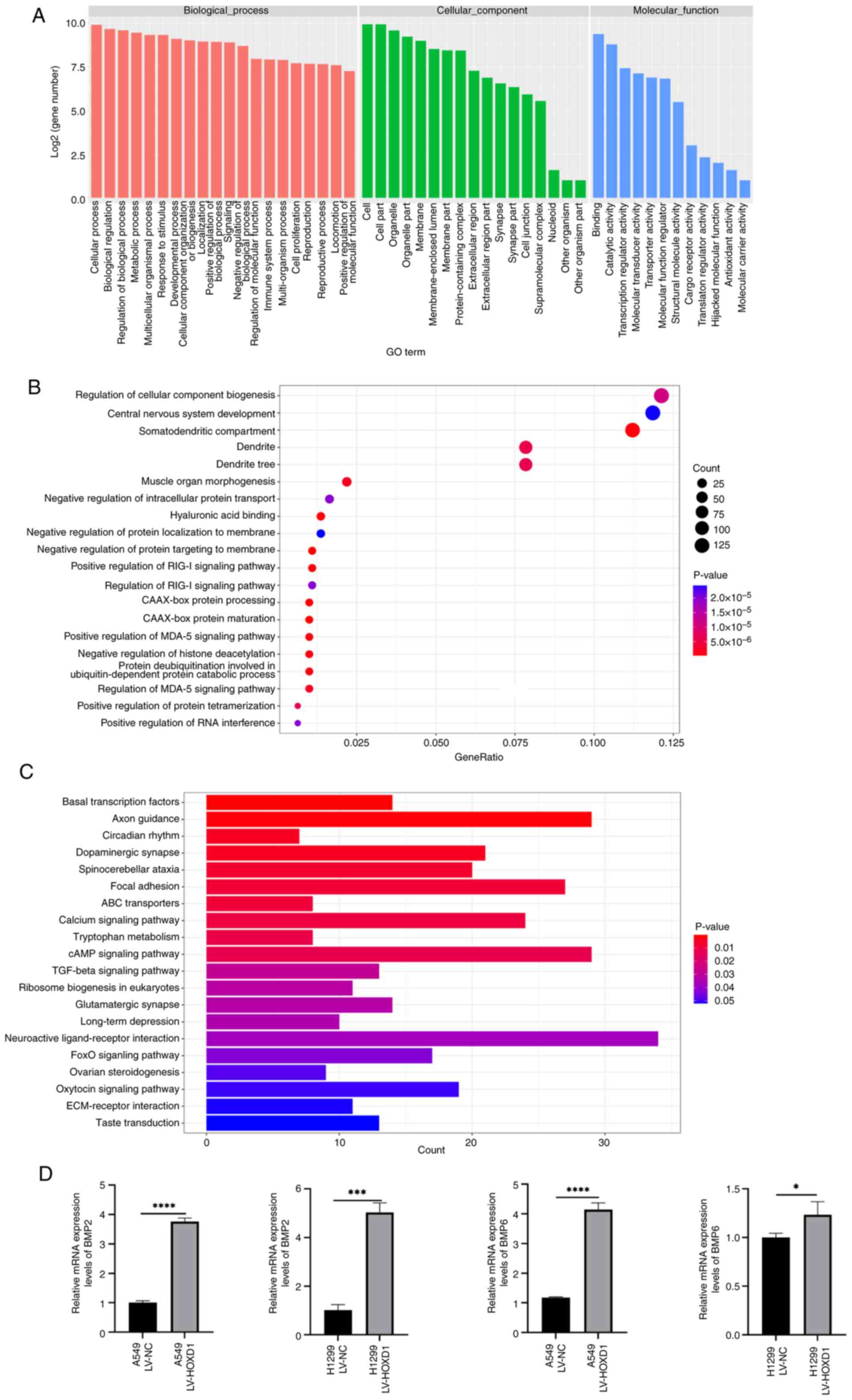
settings for a maximum motif length of 12 bp (37). The ClusterProfiler (http://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html)
R package (38) was employed to
perform Gene Ontology (GO; http://geneontology.org/) (39) and Kyoto Encyclopedia of Genes and
Genomes (KEGG; http://www.genome.jp/kegg/) (40) enrichment analyses. The GO and KEGG
enrichment analyses were calculated using hypergeometric
distribution with a q value cutoff of 0.05.
For ChIP-qPCR, the extracted DNA fragments were used
as templates for qPCR analysis using the ChamQ SYBR Color qPCR
Master Mix (cat. no. Q411; Vazyme Biotech Co., Ltd.). The 2,000 bp
region upstream of the TSS in the HOXD1 promoter region was divided
into seven segments (F1, F2, F3, F4, F5, F6 and F7), and primers
were designed for each segment. Primers are listed in Table SIV. The following thermocycling
conditions were used for qPCR: Initial denaturation at 95°C for 30
sec; 40 cycles of 95°C for 10 sec and 60°C for 30 sec. The data
were normalized to the input. The final results were analyzed as
previously described (28) and the
relative quantification of target genes were analyzed using the
2−ΔΔCq method (29).
Statistical analysis
Differential expression data for the HOXD family in
lung adenocarcinoma derived from the UALCAN database were analyzed
using Welch's t-test. Survival analyses from the Kaplan-Meier
Plotter database were conducted using Cox regression analysis. All
experiments were repeated at least three times. GraphPad Prism
software (version 8.0.1; Dotmatics) was used for all statistical
analyses. A student's t-test was used to analyze the colony
formation, Transwell and xenograft tumor assays and the
differential expression of BMP2 and BMP6. The CCK-8 assay data were
analyzed using a two-way ANOVA followed by Bonferroni's post hoc
test. HOXD1 expression in lung adenocarcinoma cell lines and data
from the DAC treatment experiments were analyzed using one-way
ANOVA followed by Dunnett's post hoc test. The ChIP-qPCR results
were analyzed using a two-tailed unpaired Student's t-test. Data
were presented as mean ± SD. P<0.05 was considered to indicate a
statistically significant difference.
Results
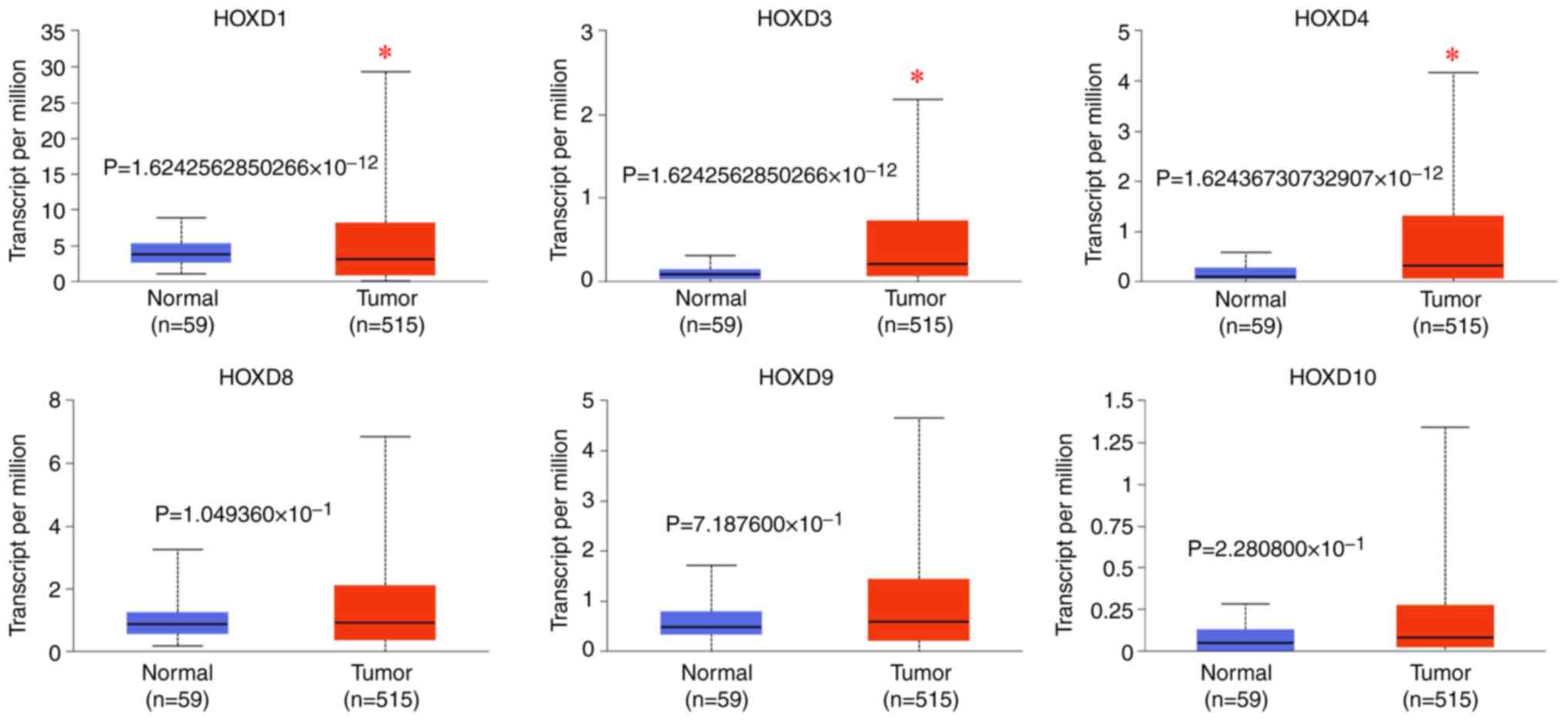
HOXD1 was downregulated in LUAD and
low expression levels predicted poor patient prognosis
The UALCAN database was used to examine the
expression levels of nine HOX genes from the HOXD cluster in LUAD
(Fig. 1). It was demonstrated that
HOXD1 expression level in the tumor group (median level, 3.125) was
significantly lower compared with that in the normal group (median
level, 3.735) (P<0.05), while HOXD3 (median level of tumor
group, 0.211; median level of normal group, 0.084) and HOXD4
(median level of tumor group, 0.317; median level of normal group,
0.097) expression levels were significantly increased in the tumor
groups compared with the normal groups (P<0.05). There were no
significant differences in HOXD8 (median level of tumor group,
0.903; median level of normal group, 0.864) (P>0.05), HOXD9
(median level of tumor group, 0.582; median level of normal group,
0.47) (P>0.05) and HOXD10 (median level of tumor group, 0.082;
median level of normal group, 0.049) (P>0.05) expression levels
between the tumor and normal groups. There were no data for the
expression of HOXD11, HOXD12 and HOXD13 in the samples analyzed. In
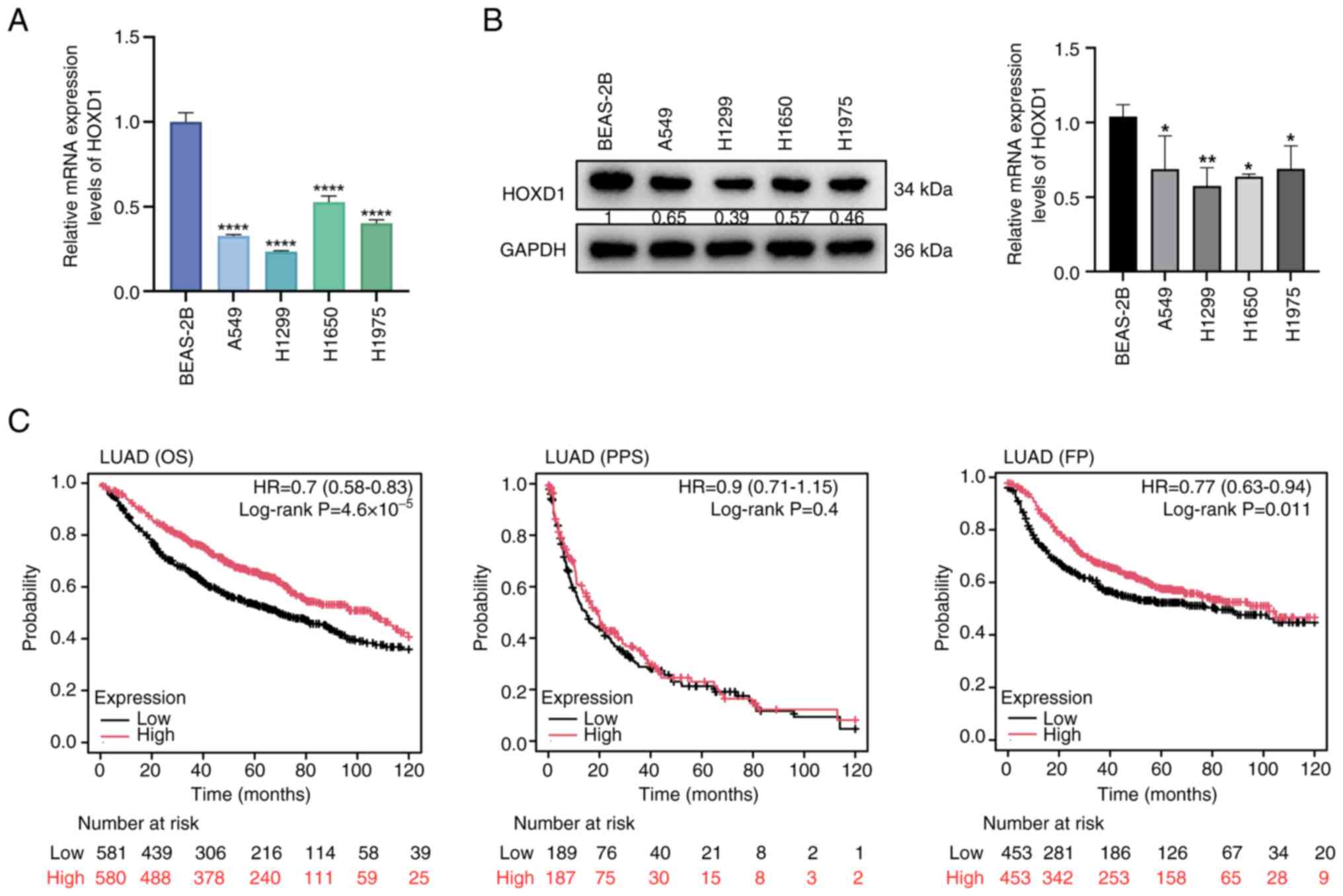
addition, HOXD1 expression levels were demonstrated to be low in
LUAD cell lines compared with a normal lung epithelial cell line
(Fig. 2A and B).
The prognostic values of the HOXD1 gene in patients
with LUAD were analyzed using Kaplan-Meier plotter. It was
demonstrated that patients with LUAD who had lower expression
levels of HOXD1 had a poorer predicted OS (P=0.0014) and FP
(P=0.00023) compared with patients with high expression levels of
HOXD1, whereas there was no difference in PPS between the two
patient groups (P=0.4) (Fig. 2C).
These data indicated that HOXD1 expression was downregulated in
LUAD in comparison to the normal lung, and low HOXD1 expression
levels may be associated with a poor prognosis in patients with
LUAD.
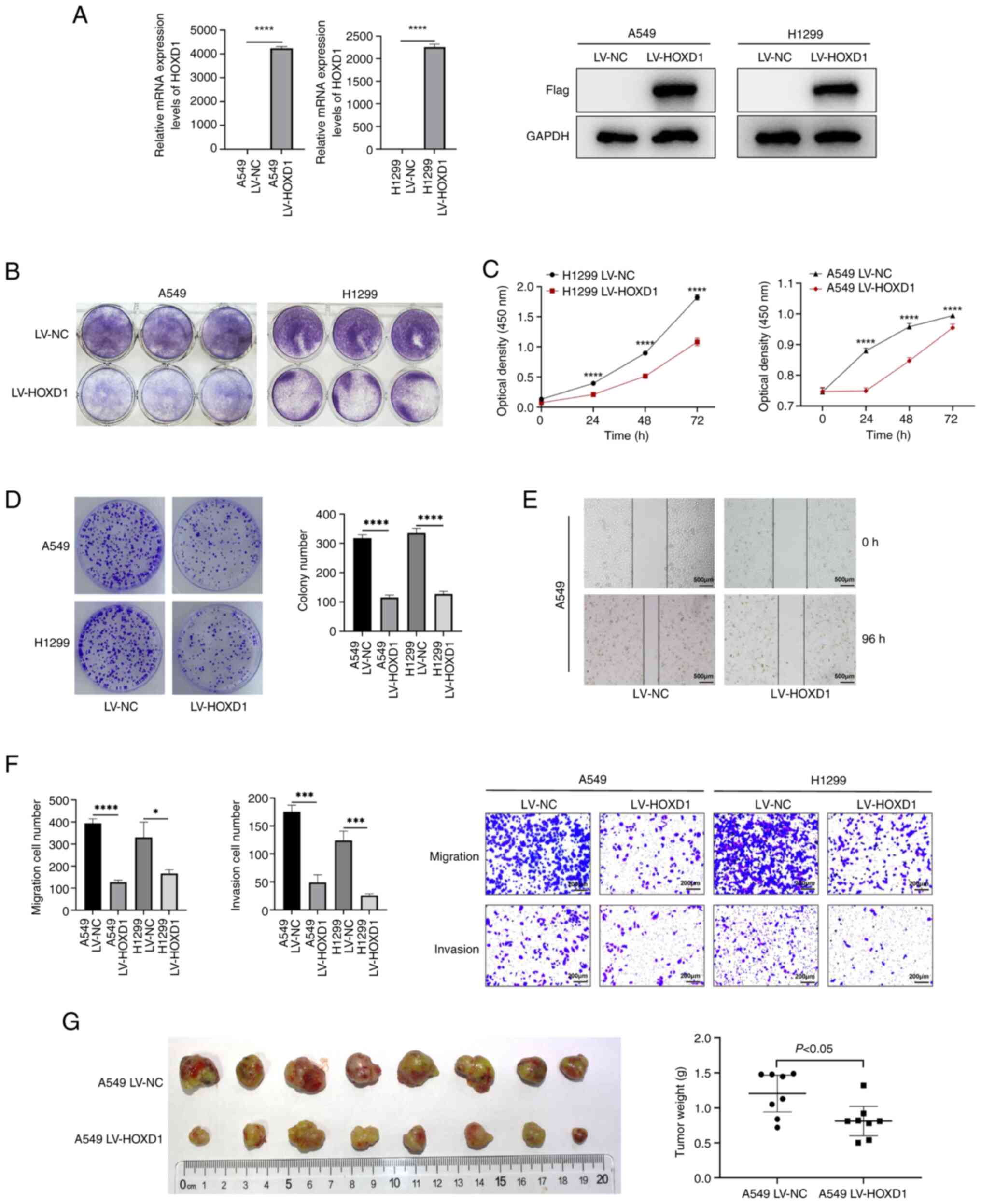
HOXD1 suppressed LUAD progression in
vivo and in vitro
To explore the function of HOXD1 in LUAD, two stably
transfected cell lines (A549 and H1299) overexpressing HOXD1 were
established. A549 and H1299 cells were transfected with LV-NC and
LV-HOXD1 to establish HOXD1 overexpression cell lines (Fig. 3A). Crystal violet staining, CCK-8
assays and colony formation assays demonstrated that the
upregulation of HOXD1 significantly inhibited cell proliferation
compared with control cells (P<0.05; Fig. 3B-D). Additionally, to determine cell
motility, a wound healing assay was performed. These results
demonstrated that HOXD1 overexpression in LUAD cells was associated
with a slower wound closure (Fig.
3E). Moreover, Transwell assays showed that the migration and
invasion of LUAD cells were inhibited when HOXD1 was upregulated
compared with control cells (Fig.
3F).
A549 LV-NC and A549 LV-HOXD1 cells were
subcutaneously injected into nude mice and after 28 days, the mice
were sacrificed and tumors were dissected. It was demonstrated that
the weight of subcutaneous tumors in the HOXD1-upregulated group
was lower compared with that in the control group (Fig. 3G). These assays indicated that HOXD1
may potentially have the ability to suppress the progression of
LUAD.
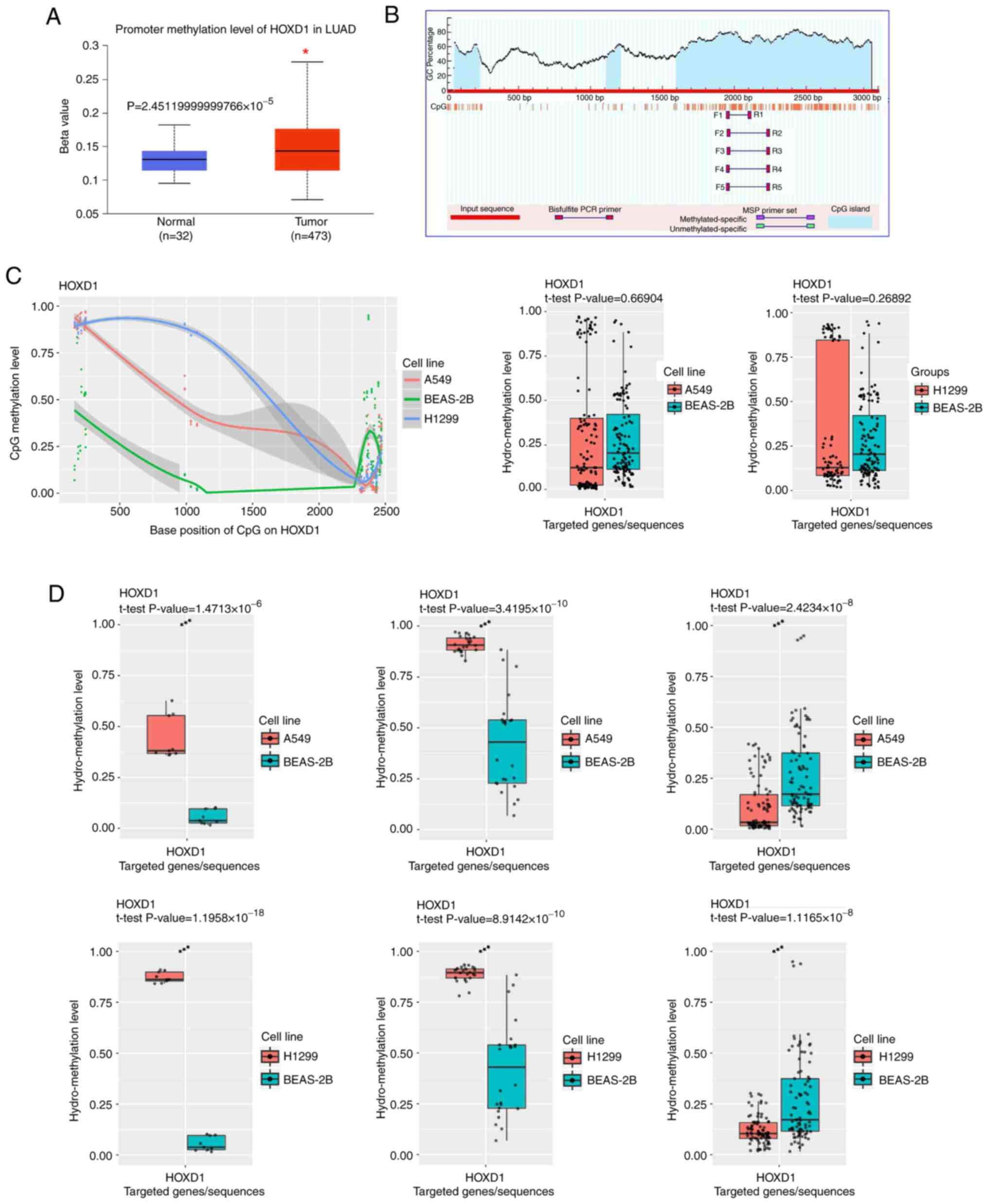
HOXD1 promoter undergoes DNA
methylation and is associated with the regulation of DNMTs
To investigate the molecular mechanism of low HOXD1
expression levels in LUAD cells, the UALCAN database was used to
analyze the methylation level of the HOXD1 promoter region in LUAD
samples and normal samples. These results showed that the
methylation level of LUAD samples was significantly higher compared
with that of normal samples (Fig.
4A). This indicated that the expression of HOXD1 may be
regulated by DNA methylation. CpG islands in the promoter region of
the HOXD1 gene were predicted using MethPrimer. These results
showed three CpG islands in the upstream 2,000 bp to downstream
1,000 bp of the TSS (Fig. 4B).
Furthermore, the methylation level of the HOXD1 promoter in
BEAS-2B, A549 and H1299 cells was measured, and the TBS results
showed that the total level of methylation was not significantly
different in A549 and H1299 cells compared with BEAS-2B cells
(Fig. 4C). It is noteworthy that
one CpG island in the posterior segment was significantly less
methylated, and two CpG islands in the anterior section of this
promoter region were significantly more methylated in LUAD cells
compared with BEAS-2B cells (Fig.
4D). These findings suggested that in LUAD, the HOXD1 promoter
region was regulated by DNA methylation.
The regulation mechanism of HOXD1 hypermethylation
was investigated as it was hypothesized that DNMTs were involved in
regulating HOXD1 expression in LUAD cells. To determine whether
DNMTs were upstream regulators of HOXD1 expression in LUAD cells,
A549 cells were treated with the DNMT inhibitor decitabine for 96
h. The mRNA expression level of the HOXD1 gene was detected using
RT-qPCR, which demonstrated that DAC significantly increased HOXD1
expression levels in a dose-dependent manner (Fig. 5A). In addition, A549 cells were
transfected with siRNAs targeting DNMT1, DNMT3A or DNMT3B and a
significant increase in HOXD1 expression levels were demonstrated
compared with negative controls (Fig.
5B). Furthermore, the 2,000 bp region upstream of the TSS in
the HOXD1 promoter region was divided into seven segments, and
primers were designed for each segment (Fig. 5C). ChIP-qPCR assays using A549 cells
showed that DNMTs bound to the HOXD1 promoter region. DNMT1
combined with F1, F5 and F7 segments of the HOXD1 promoter
(Fig. 5D). DNMT3A combined with F1,
F2, F3, F4, F5, F6 and F7 segments of the HOXD1 promoter (Fig. 5E). DNMT3B combined with F1, F2, F3,
F4, F5, F6 and F7 segments of the HOXD1 promoter (Fig. 5F). These ChIP-qPCR analyses
suggested that the HOXD1 promoter region may be enriched with DNMT
protein binding. These results suggested that DNMTs target the
HOXD1 promoter region and could potentially contribute to local
hypermethylation, inducing the dysregulation of HOXD1 in LUAD.
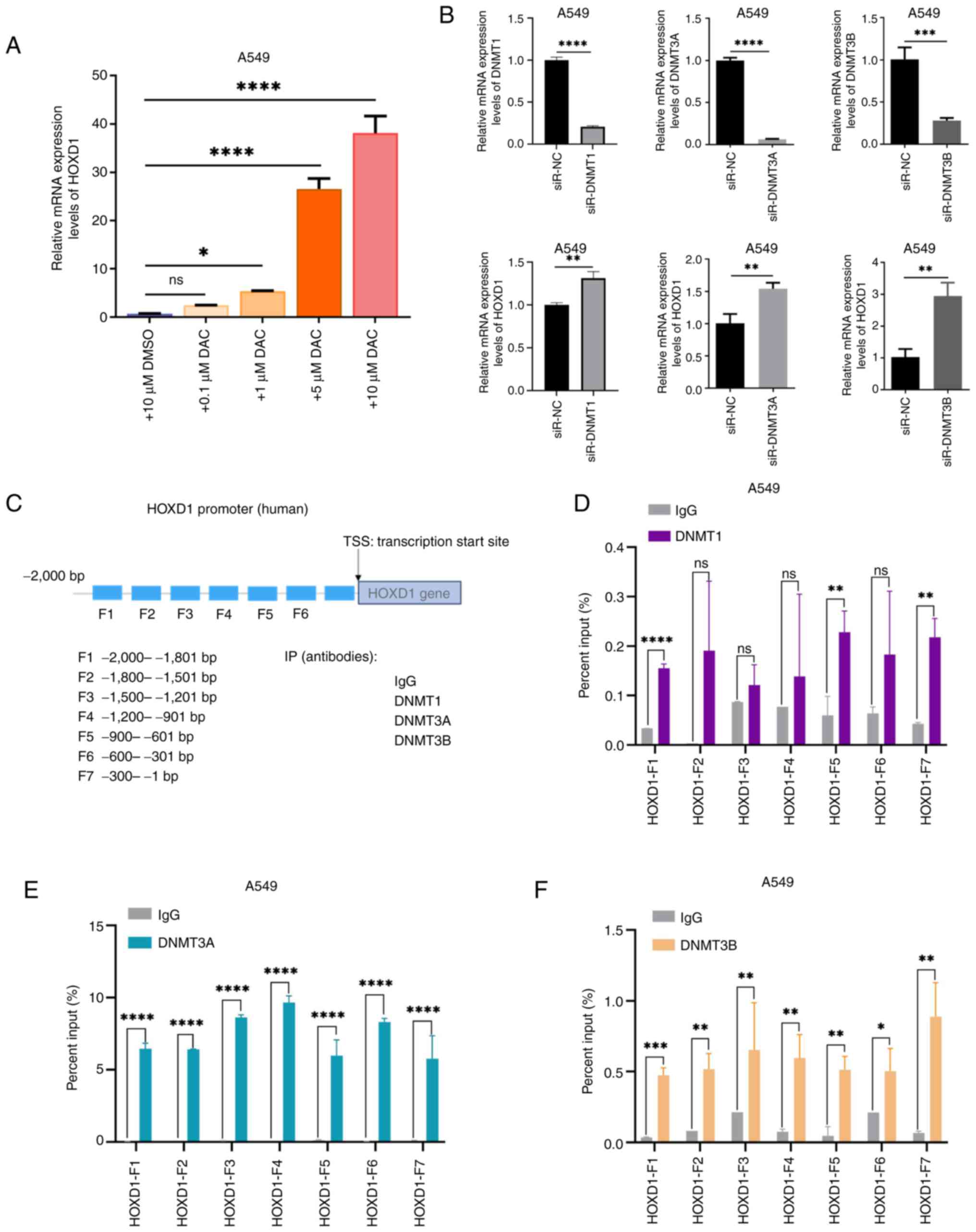 | Figure 5.DNMTs targeted the HOXD1 promoter
region and led to local hypermethylation. (A) A549 cells were
treated with different concentrations of DAC and the mRNA
expression levels of HOXD1 were detected using reverse
transcription-quantitative PCR. (B) HOXD1 expression was rescued
following the transfection of si-DNMTs in A549 cells. (C) Schematic
representation of the HOXD1 promoter and ChIP-qPCR primer design.
The binding sites of (D) DNMT1, (E) DNMT3A and (F) DNMT3B to the
HOXD1 promoter were determined using ChIP-qPCR. Data were presented
as mean ± SD. *P<0.05, **P<0.01, ***P<0.001,
****P<0.0001. HOX, homeobox; DAC, decitabine; si, small
interfering RNA; DNMT, DNA methyltransferase; TSS transcription
start site; ChIP-qPCR, chromatin immunoprecipitation-quantitative
PCR; ns, not significant. |
Upregulated HOXD1 increases BMP2 and
BMP6 transcriptional expression
To better identify the downstream target gene
potentially regulated by HOXD1 as a transcription factor in LUAD,
ChIP-seq was performed on HOXD1 overexpressing A549 cells to
analyze the downstream regulatory network of HOXD1. GO annotation
analysis demonstrated that the target sequences of HOXD1 were most
enriched in biological processes (Fig.
6A). After the GO enrichment data were sorted according to the
P-value, from low to high, the top 20 GO-enriched functions were
presented as a bubble graph, which reflected the number of genes
and the degree of enrichment of HOXD1-enriched annotations to GO
functions. (Fig. 6B). Moreover,
following a descending order of P-value for all KEGG enrichment
data, bar graphs representing the top 20 KEGG-enriched signaling
pathways were created to show the enrichment of HOXD1 in various
metabolic pathways (Fig. 6C).
Based on the results of ChIP-seq analysis, 24 genes
associated with cellular processes were screened. Then, the
expression of these genes was confirmed using RT-qPCR. These
results showed that overexpression of HOXD1 elevated the mRNA
expression levels of BMP2 and BMP6 (Fig. 6D).
Discussion
The HOX gene family encodes transcription factors
that serve a vital role in embryonic development. Studies have also
reported a connection between cancer and the abnormal expression of
HOX genes (41–43). A subfamily of HOX genes, HOXD,
functions as an oncogene or a tumor suppressor that participates in
the growth of tumors. Tan et al (44) reported that HOXD11 accelerated the
development of penile squamous cell carcinoma through degrading the
extracellular matrix and promoting epithelial-mesenchymal
transition via the fibronectin 1/MMP2/MMP9 pathway. In malignant
glioma, decreased miR-7156-3p expression promotes HOXD13
upregulation and induces glioma cell stemness and invasiveness
(45). Furthermore, as a tumor
suppressor, HOXD8 upregulates serine hydroxymethyltransferase 1
expression and inhibits renal cell carcinoma tumor progression
(46). HOXD1 is also recognized as
a tumor suppressor gene as it inhibits the progression of renal
clear cell carcinoma (47). Hence,
the different roles of HOXD in cancer vary depending on the
specific cancer type. In the present study, RT-qPCR and western
blotting analyses showed that downregulation of HOXD1 was observed
in LUAD cell lines. Additionally, low expression levels of HOXD1
were positively correlated with a poor prognosis in patients with
LUAD. Moreover, the overexpression of HOXD1 suppressed cancer cell
proliferation, migration and invasion in LUAD cells and an animal
model. In this regard, HOXD1 could potentially serve as a tumor
suppressor gene in LUAD and may be a future promising therapeutic
target for anticancer treatment.
A previous study reported that DNA methylation of
HOX genes is a common event in cancer (48). The HOXD family is frequently
regulated by upstream DNA methylation (49). The methylation level of HOXD3 has
been linked to prostate cancer pathology and HOXD3 hypermethylation
indicated a poorer prognosis (50).
Additionally, hypermethylation of HOXD8 can serve as a biomarker
for the identification of biliary tract cancer (51). Furthermore, methylation of CpG
islands at the HOXD locus has also been reported in LUAD (52). Therefore, the present study aimed to
explore the upstream regulatory mechanism of HOXD1 in LUAD
development. It was demonstrated that DNMTs bind to the HOXD1
promoter region and repress its expression, as shown through
bioinformatic analysis and ChIP-qPCR. It was demonstrated that
HOXD1 was regulated by DNMTs, through inhibiting or overexpressing
DNMTs. Following DNMT inhibition, there was a considerable increase
in HOXD1 expression. This suggested that DNMTs over accumulate in
the HOXD1 promoter region, thereby repressing HOXD1 expression in
LUAD. It is currently unknown how DNMTs regulate the
transcriptional expression of HOXD1, and whether other factors are
involved in the interactions between DNMTs and HOXD1. Long
non-coding RNAs (lncRNAs) have been proposed to serve a significant
role in DNA methylation-mediated transcriptional expression
regulation of downstream genes (53–55).
The potential involvement of lncRNAs in the DNMT-mediated
regulation of HOXD1 expression should be analyzed in future
studies.
Preliminary downstream mechanistic studies suggested
that HOXD1 positively regulates the mRNA expression levels of BMP2
and BMP6, which belong to the TGF-β superfamily (56). Increased TGF-β expression levels in
certain types of cancer has been linked to tumor progression and
enhanced stem cell characteristics, which allows cancer cells to
form tumors and develop resistance to immunotherapy (57). The involvement of BMP in cellular
biological processes is primarily mediated through the SMAD and
MAPK pathways (58). Previous
research on NSCLC has demonstrated that BMP2 is highly expressed in
LUAD, while BMP6 expression was not significantly different in
NSCLC tissue compared with healthy lung tissue (59). Mechanistically, BMP2 targets
downstream PNMA family member 5 to enhance cancer cell migration
and invasion (60). Vora et
al (61) reported that
increased BMP2 expression levels cause metabolic dysregulation by
suppressing AMP-activated protein kinase expression and
upregulating PI3K expression. Moreover, BMP2 can enhance SMAD1/5
phosphorylation, which promotes lung cancer metastasis (62). Furthermore, it has been reported
that methylation regulates BMP6 in NSCLC, resulting in epigenetic
dysregulation (63). Additionally,
it has been shown that deficiency of BMP6 expression results from
increased myosin heavy chain 16 in the advancement of LUAD
(64). In the present study, HOXD1,
as a transcription factor, regulated the expression levels of BMP2
and BMP6 at the transcriptional level. However, other factors may
also be involved in this regulatory progression. Therefore,
exploring the binding mechanism between HOXD1 and BMP2 and BMP6, as
well as their detailed regulatory relationship with downstream
signaling pathways, should be pursued in future research.
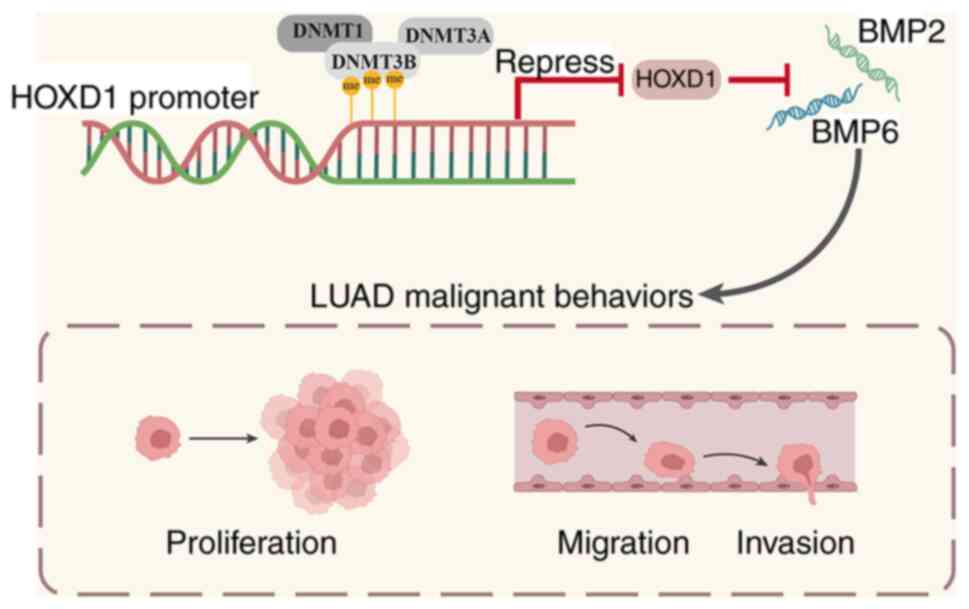
In conclusion, the present study demonstrated that
HOXD1 was significantly downregulated and HOXD1 functioned as a
tumor suppressor in LUAD. DNA methylation-regulated HOXD1 inhibited
LUAD progression by increasing BMP2 and BMP6 mRNA expression levels
(Fig. 7). These results could
potentially provide a theoretical basis for future studies into the
early diagnosis and development of novel treatment strategies for
NSCLC.
Supplementary Material
Supporting Data
Acknowledgments
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant no. 82103131).
Availability of data and materials
The ChIP sequencing data and target bisulfite
sequencing data generated in the present study can be found in the
GEO database under accession numbers GSE277213 and GSE277214,
respectively, or at the following URLs: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE277213
and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE277214,
respectively.
Authors' contributions
XH, SZ, XZ, HL and YD performed the experiments,
analyzed the data and reviewed and edited the manuscript. LL
contributed to study conceptualization, methodology, supervision,
validation, reviewing and editing. XH and LL confirmed the
authenticity of all the raw data. All authors read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
All animal procedures were approved by the Animal
Use and Care Committee at Shandong Provincial Hospital Affiliated
with Shandong First Medical University (approval no. 2021-622).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
HOX
|
homeobox
|
|
BMP
|
bone morphogenetic protein
|
|
NSCLC
|
non-small cell lung cancer
|
|
LUAD
|
lung adenocarcinoma
|
|
DNMTs
|
DNA methyltransferases
|
|
TBS
|
targeted bisulfite sequencing
|
|
ChIP-qPCR
|
chromatin
immunoprecipitation-quantitative PCR
|
|
OS
|
overall survival
|
|
FP
|
first progression
|
|
PPS
|
post-progression survival
|
|
siRNA
|
small interfering RNA
|
|
DAC
|
decitabine
|
|
CCK-8
|
cell counting kit-8
|
|
TSS
|
transcription start site
|
|
ChIP-seq
|
ChIP-sequencing
|
|
lncRNAs
|
long non-coding RNAs
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
References
|
1
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Giaquinto AN and Jemal A:
Cancer statistics, 2024. CA Cancer J Clin. 74:12–49. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bourreau C, Treps L, Faure S, Fradin D and
Clere N: Therapeutic strategies for non-small cell lung cancer:
Experimental models and emerging biomarkers to monitor drug
efficacies. Pharmacol Ther. 242:1083472023. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ettinger DS, Wood DE, Aisner DL, Akerley
W, Bauman JR, Bharat A, Bruno DS, Chang JY, Chirieac LR, D'Amico
TA, et al: Non-small cell lung cancer, version 3.2022, NCCN
clinical practice guidelines in oncology. J Natl Compr Canc Netw.
20:497–530. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Riely GJ, Wood DE, Ettinger DS, Aisner DL,
Akerley W, Bauman JR, Bharat A, Bruno DS, Chang JY, Chirieac LR, et
al: Non-small cell lung cancer, version 4.2024, NCCN clinical
practice guidelines in oncology. J Natl Compr Canc Netw.
22:249–274. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Steens J and Klein D: HOX genes in stem
cells: Maintaining cellular identity and regulation of
differentiation. Front Cell Dev Biol. 10:10029092022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jonkers J, Pai P and Sukumar S: Multiple
roles of HOX proteins in metastasis: Let me count the ways. Cancer
Metastasis Rev. 39:661–679. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Paço A, de Bessa Garcia SA and Freitas R:
Methylation in HOX clusters and its applications in cancer therapy.
Cells. 9:16132020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Belpaire M, Taminiau A, Geerts D and
Rezsohazy R: HOXA1, a breast cancer oncogene. Biochim Biophys Acta
Rev Cancer. 1877:1887472022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li L, Zhang X, Liu Q, Yin H, Diao Y, Zhang
Z, Wang Y, Gao Y, Ren X, Li J, et al: Emerging role of HOX genes
and their related long noncoding RNAs in lung cancer. Crit Rev
Oncol Hematol. 139:1–6. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang R, Zhang G, Dong Z, Wang S, Li Y,
Lian F, Liu X, Li H, Wei X and Cui H: Homeobox A3 and KDM6A
cooperate in transcriptional control of aerobic glycolysis and
glioblastoma progression. Neuro Oncol. 25:635–647. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bolt CC, Lopez-Delisle L, Mascrez B and
Duboule D: Mesomelic dysplasias associated with the HOXD locus are
caused by regulatory reallocations. Nat Commun. 12:50132021.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu M, Zhan J and Zhang H: HOX family
transcription factors: Related signaling pathways and
post-translational modifications in cancer. Cell Signal.
66:1094692020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hamada J, Omatsu T, Okada F, Furuuchi K,
Okubo Y, Takahashi Y, Tada M, Miyazaki YJ, Taniguchi Y, Shirato H,
et al: Overexpression of homeobox gene HOXD3 induces coordinate
expression of metastasis-related genes in human lung cancer cells.
Int J Cancer. 93:516–525. 2001. View Article : Google Scholar
|
|
15
|
Liu Y, Miao L, Ni R, Zhang H, Li L, Wang
X, Li X and Wang J: microRNA-520a-3p inhibits proliferation and
cancer stem cell phenotype by targeting HOXD8 in non-small cell
lung cancer. Oncol Rep. 36:3529–3535. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wan K, Shao J, Liu X, Cai Y, Xu Y, Li L,
Xiong L and Liang S: HOXD9 contributes to the Warburg effect and
tumor metastasis in non-small cell lung cancer via transcriptional
activation of PFKFB3. Exp Cell Res. 427:1135832023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li S, Zhang J, Zhao Y, Wang F, Chen Y and
Fei X: miR-224 enhances invasion and metastasis by targeting HOXD10
in non-small cell lung cancer cells. Oncol Lett. 15:7069–7075.
2018.PubMed/NCBI
|
|
18
|
Shah N and Sukumar S: The Hox genes and
their roles in oncogenesis. Nat Rev Cancer. 10:361–371. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Espín-Pérez A, Brennan K, Ediriwickrema
AS, Gevaert O, Lossos IS and Gentles AJ: Peripheral blood DNA
methylation profiles predict future development of B-cell
Non-Hodgkin Lymphoma. NPJ Precis Oncol. 6:532022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Na F, Pan X, Chen J, Chen X, Wang M, Chi
P, You L, Zhang L, Zhong A, Zhao L, et al: KMT2C deficiency
promotes small cell lung cancer metastasis through DNMT3A-mediated
epigenetic reprogramming. Nat Cancer. 3:753–767. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mancarella D and Plass C: Epigenetic
signatures in cancer: Proper controls, current challenges and the
potential for clinical translation. Genome Med. 13:232021.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang J, Yang J, Li D and Li J:
Technologies for targeting DNA methylation modifications: Basic
mechanism and potential application in cancer. Biochim Biophys Acta
Rev Cancer. 1875:1884542021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen Z and Zhang Y: Role of mammalian DNA
methyltransferases in development. Annu Rev Biochem. 89:135–158.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Flausino CS, Daniel FI and Modolo F: DNA
methylation in oral squamous cell carcinoma: From its role in
carcinogenesis to potential inhibitor drugs. Crit Rev Oncol
Hematol. 164:1033992021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhu L, Tang N, Hang H, Zhou Y, Dong J,
Yang Y, Mao L, Qiu Y, Fu X and Cao W: Loss of claudin-1 incurred by
DNMT aberration promotes pancreatic cancer progression. Cancer
Lett. 586:2166112024. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chandrashekar DS, Karthikeyan SK, Korla
PK, Patel H, Shovon AR, Athar M, Netto GJ, Qin ZS, Kumar S, Manne
U, et al: UALCAN: An update to the integrated cancer data analysis
platform. Neoplasia. 25:18–27. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Győrffy B: Integrated analysis of public
datasets for the discovery and validation of survival-associated
genes in solid tumors. Innovation (Camb). 5:1006252024.PubMed/NCBI
|
|
28
|
Li A, Xie J, Lv L, Zheng Z, Yang W, Zhuo
W, Yang S, Cai D, Duan J, Liu P, et al: RPL9 acts as an oncogene by
shuttling miRNAs through exosomes in human hepatocellular carcinoma
cells. Int J Oncol. 64:582024. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li LC and Dahiya R: MethPrimer: Designing
primers for methylation PCRs. Bioinformatics. 18:1427–1431. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zuo Y, Zhong J, Bai H, Xu B, Wang Z, Li W,
Chen Y, Jin S, Wang S, Wang X, et al: Genomic and epigenomic
profiles distinguish pulmonary enteric adenocarcinoma from lung
metastatic colorectal cancer. EBioMedicine. 82:1041652022.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pu W, Qian F, Liu J, Shao K, Xiao F, Jin
Q, Liu Q, Jiang S, Zhang R, Zhang J, et al: Targeted bisulfite
sequencing reveals dna methylation changes in zinc finger family
genes associated with KRAS mutated colorectal cancer. Front Cell
Dev Biol. 9:7598132021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bolger AM, Lohse M and Usadel B:
Trimmomatic: A flexible trimmer for Illumina sequence data.
Bioinformatics. 30:2114–2120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xi Y and Li W: BSMAP: Whole genome
bisulfite sequence MAPping program. BMC Bioinformatics. 10:2322009.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yachida S, Mizutani S, Shiroma H, Shiba S,
Nakajima T, Sakamoto T, Watanabe H, Masuda K, Nishimoto Y, Kubo M,
et al: Metagenomic and metabolomic analyses reveal distinct
stage-specific phenotypes of the gut microbiota in colorectal
cancer. Nat Med. 25:968–976. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Salmon-Divon M, Dvinge H, Tammoja K and
Bertone P: PeakAnalyzer: Genome-wide annotation of chromatin
binding and modification loci. BMC Bioinformatics. 11:4152010.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hull RP, Srivastava PK, D'Souza Z, Atanur
SS, Mechta-Grigoriou F, Game L, Petretto E, Cook HT, Aitman TJ and
Behmoaras J: Combined ChIP-Seq and transcriptome analysis
identifies AP-1/JunD as a primary regulator of oxidative stress and
IL-1β synthesis in macrophages. BMC Genomics. 14:922013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yadav C, Yadav R, Nanda S, Ranga S, Ahuja
P and Tanwar M: Role of HOX genes in cancer progression and their
therapeutical aspects. Gene. 919:1485012024. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Morgan R, Hunter K and Pandha HS:
Downstream of the HOX genes: Explaining conflicting tumour
suppressor and oncogenic functions in cancer. Int J Cancer.
150:1919–1932. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Paco A, Aparecida de Bessa Garcia S,
Leitao Castro J, Costa-Pinto AR and Freitas R: Roles of the HOX
proteins in cancer invasion and metastasis. Cancers (Basel).
13:102020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tan X, Liu Z, Wang Y, Wu Z, Zou Y, Luo S,
Tang Y, Chen D, Yuan G and Yao K: miR-138-5p-mediated HOXD11
promotes cell invasion and metastasis by activating the
FN1/MMP2/MMP9 pathway and predicts poor prognosis in penile
squamous cell carcinoma. Cell Death Dis. 13:8162022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang J, Deng M, Tong H, Xue W, Guo Y,
Wang J, Chen L and Wang S: A novel miR-7156-3p-HOXD13 axis
modulates glioma progression by regulating tumor cell stemness. Int
J Biol Sci. 16:3200–3209. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yang Y, Zhang M, Zhao Y, Deng T, Zhou X,
Qian H, Wang M, Zhang C, Huo Z, Mao Z, et al: HOXD8 suppresses
renal cell carcinoma growth by upregulating SHMT1 expression.
Cancer Sci. 114:4583–4595. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang L, Wang X, Sun H, Wang W and Cao L: A
pan-cancer analysis of the role of HOXD1, HOXD3, and HOXD4 and
validation in renal cell carcinoma. Aging (Albany NY).
15:10746–10766. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hu X, Wang Y, Zhang X, Li C, Zhang X, Yang
D, Liu Y and Li L: DNA methylation of HOX genes and its clinical
implications in cancer. Exp Mol Pathol. 134:1048712023. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang L, Qiao C, Cao L, Cai S, Ma X, Song
X, Jiang Q, Huang C and Wang J: Significance of HOXD transcription
factors family in progression, migration and angiogenesis of
cancer. Crit Rev Oncol Hematol. 179:1038092022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kron KJ, Liu L, Pethe VV, Demetrashvili N,
Nesbitt ME, Trachtenberg J, Ozcelik H, Fleshner NE, Briollais L,
van der Kwast TH and Bapat B: DNA methylation of HOXD3 as a marker
of prostate cancer progression. Lab Invest. 90:1060–1067. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Loi E, Zavattari C, Tommasi A, Moi L,
Canale M, Po A, Sabato C, Vega-Benedetti AF, Ziranu P, Puzzoni M,
et al: HOXD8 hypermethylation as a fully sensitive and specific
biomarker for biliary tract cancer detectable in tissue and bile
samples. Br J Cancer. 126:1783–1794. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Shiraishi M, Sekiguchi A, Oates AJ, Terry
MJ and Miyamoto Y: HOX gene clusters are hotspots of de novo
methylation in CpG islands of human lung adenocarcinomas. Oncogene.
21:3659–3662. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Huang W, Li H, Yu Q, Xiao W and Wang DO:
LncRNA-mediated DNA methylation: An emerging mechanism in cancer
and beyond. J Exp Clin Cancer Res. 41:1002022. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Geng X, Zhao J, Huang J, Li S, Chu W, Wang
WS, Chen ZJ and Du Y: lnc-MAP3K13-7:1 inhibits ovarian GC
proliferation in PCOS via DNMT1 downregulation-mediated CDKN1A
promoter hypomethylation. Mol Ther. 29:1279–1293. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Xu SF, Zheng Y, Zhang L, Wang P, Niu CM,
Wu T, Tian Q, Yin XB, Shi SS, Zheng L and Gao LM: Long non-coding
RNA LINC00628 interacts epigenetically with the LAMA3 promoter and
contributes to lung adenocarcinoma. Mol Ther Nucleic Acids.
18:166–182. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wakefield LM and Hill CS: Beyond TGFβ:
Roles of other TGFβ superfamily members in cancer. Nat Rev Cancer.
13:328–341. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Derynck R, Turley SJ and Akhurst RJ: TGFβ
biology in cancer progression and immunotherapy. Nat Rev Clin
Oncol. 18:9–34. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Massagué J and Sheppard D: TGF-β signaling
in health and disease. Cell. 186:4007–4037. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Langenfeld EM, Bojnowski J, Perone J and
Langenfeld J: Expression of bone morphogenetic proteins in human
lung carcinomas. Ann Thorac Surg. 80:1028–1032. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Huang F, Cao Y, Wang C, Lan R, Wu B, Xie
X, Hong J, Fu L and Wu G: PNMA5 promotes bone metastasis of
non-small-cell lung cancer as a target of BMP2 signaling. Front
Cell Dev Biol. 9:6789312021. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Vora M, Mondal A, Jia D, Gaddipati P, Akel
M, Gilleran J, Roberge J, Rongo C and Langenfeld J: Bone
morphogenetic protein signaling regulation of AMPK and PI3K in lung
cancer cells and C. elegans. Cell Biosci. 12:762022. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Wu CK, Wei MT, Wu HC, Wu CL, Wu CJ, Liaw H
and Su WP: BMP2 promotes lung adenocarcinoma metastasis through BMP
receptor 2-mediated SMAD1/5 activation. Sci Rep. 12:163102022.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Kraunz KS, Nelson HH, Liu M, Wiencke JK
and Kelsey KT: Interaction between the bone morphogenetic proteins
and Ras/MAP-kinase signalling pathways in lung cancer. Br J Cancer.
93:949–952. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhang L, Liu J, Wang H, Xu Z, Wang Y, Chen
Y and Peng H: MYH16 upregulation is associated with lung
adenocarcinoma aggressiveness and immune infiltration. J Biochem
Mol Toxicol. 37:e234902023. View Article : Google Scholar : PubMed/NCBI
|