Introduction
Stem cells are undifferentiated cells that possess
the ability to undergo self-renewal through cell division and to
differentiate into specialized cell types. As a result, individuals
develop a more specialized phenotype through the adoption of a
distinct genetic expression profile. Ultimately, these cells
differentiate into particular cell types that exhibit distinct and
defining characteristics (1).
Stem cells can be classified according to their
capacity for self-renewal and potency. Self-renewal refers to the
inherent capability of cells to undergo multiple rounds of cellular
division while preserving their undifferentiated form. On the other
hand, potency denotes the capacity of cells to differentiate into
distinct different types of cells (2). Stem cells can be categorized into
four distinct categories based on their potency. The initial
category consists of totipotent stem cells. The aforementioned
cells are generated from the union of a female egg and a male sperm
cell, and possess the ability to undergo differentiation into both
embryonic and extraembryonic cell lineages. The second category
encompasses pluripotent stem cells, which possess the capacity to
differentiate into cell types derived from all three primary germ
layers (endoderm, mesoderm and ectoderm), but do not possess the
ability to generate placental or extraembryonic cells. The third
category comprises multipotent stem cells, which possess the
capacity to undergo differentiation into a finite range of cell
types that are confined to a specific germinal layer. The fourth
category comprises unipotent cells, which undergo differentiation
leading to the formation of a singular cell type. These cells
appear in adult organisms (3).
According to Barky et al (4), stem cells can be categorized into
numerous types, including embryonic, fetal, adult,
amniotic-derived, cord blood-derived and induced pluripotent stem
cells. Embryonic stem cells are derived from embryos, as their name
suggests. Pluripotent cells possess the ability to remain in an
undifferentiated state or undergo differentiation into any cell
type found inside the human body (5).
The derivation of the initial embryonic line in 1998
marked the commencement of a prominent, impassioned and persistent
discourse within the realm of research ethics (5). In order to obtain embryonic stem
cells, it is imperative to terminate the preimplantation embryo at
the age of 5 days (6). Another
barrier to their potential medical use is the need for human
leukocyte antigen (HLA)-compatible human embryonic stem cell lines
(7). The limited understanding of
their biological characteristics serves as a constraining element
that diminishes their prospects for use. One further limitation
that poses a significant concern for their utilization in clinical
practice is the potential formation of teratomas and
teratocarcinomas (8).
Mesenchymal stem cells (MSCs) are currently
undergoing experimental testing in numerous biological systems and
clinical contexts in order to investigate potential therapeutic
effects for a wide range of reasons (9,10).
MSCs are distinguished and described by a number of key features.
Firstly, they exhibit the capacity to adhere to plastic culture
flasks. Additionally, they exhibit the positive expression of
specific membrane antigens, namely CD105, CD73, CD90 and CD44.
Conversely, they do not express certain antigens, such as CD45,
CD34, CD14 or CD11b, CD79 or CD19 and HLA-DR. Furthermore, they
possess the capacity to differentiate into osteoblasts, adipocytes
and chondroblasts under suitable conditions in vitro
(11). One of the major hurdles
for improving MSC transplantation efficacy is the lack of molecular
characterization (12).
This molecular characterization can be used to
replace current expensive and time-consuming characterization
protocols such as flow cytometry. The molecular characterization of
MSCs may provide a promising solution for easier and improved
characterization of MSCs, in order to make full use of stem cells
in regenerative medicine.
Materials and methods
Isolation of rat bone marrow-derived
MSCs (BM-MSCs)
BM-MSCs were isolated from the femurs and tibiae of
6-8-week-old female Sprague-Dawley rats. A total of 15 rats
(weighing 150-180 g) were obtained from the Medical Experimental
Research Center (Mansoura, Egypt) as previously described by Meurer
et al (13), with some
modifications. In the present study, rats were housed at a
temperature of 20-25˚C and a humidity of 30-50% in the animal house
of the Medical Experimental Research Center (MERC), Faculty of
Medicine, Mansoura University. They were conditioned in standard
metallic cages (6 rats per cage) with regular dark/light cycles.
They were acclimatized to the laboratory conditions, fed standard
rat chow and water was available ad libitum. The health
status of the rats their welfare condition were observed daily by a
veterinarian. The experimental protocol of the present study was
approved by the Local Ethics Committee, Faculty of Medicine,
Mansoura University in accordance with the Ethics Committee of the
National Research Center, Egypt with registration number (09/189),
which has been accepted by MU-ACUC.
Briefly, the rats were euthanized using an overdose
(5%) of halothane; the death of the rats was confirmed by the
absence of a corneal reflex, failure to detect respiration, and the
absence of a heart beat for a period >5 min. Following the
confirmation of death, the rats were soaked in 70% (v/v) alcohol
for 2 min and transferred to a surgery table in a class II
biological safety cabinet. The femur and tibia were dissected out.
The bilateral connection parts around the ankle, hip and knee
joints were cut to isolate the tibiae and femurs gently and
carefully. Any remaining muscle or tissue on the bones was removed
using sterile lint-free tissue paper (Kleenex; Kimberly-Clark
Worldwide, Inc.) and the bones were sprayed with 70% ethanol. The
proximal end of the femur and the distal end of the tibia were cut
using fine scissors. The needle of the syringe was filled with
proliferation medium (DMEM/F12, 10% FBS and 1%
penicillin/streptomycin; HyClone; Cytiva) and inserted into the
bone diaphysis, and all bone marrow was flushed into a sterile
50-ml conical tube. These flushed bone marrow cells were plated in
75-cm2 tissue culture flasks and the flasks were
incubated in a 5% CO2 incubator with a humidified
atmosphere containing 95% air at 37˚C.
The cells were allowed to adhere for 24 h, and the
non-adherent cells were transferred to another flask for further
culture. The culture medium was changed every 3 days. The adherent
cells were observed under an inverted microscope (Olympus
Corporation) and detached using 0.25% trypsin-EDTA when they became
70-90% confluent. At passage 3, the cells were divided into five
groups (A, B, C, D and E) according to their morphology.
Flow cytometry
Since there is no well-defined basis for determining
MSCs using their cellular surface markers, it was confirmed that
they were not of hematopoietic origin (CD45-), and
subsequently two selected markers that have been defined (11) as MSC markers
(CD29+/Entegrin β1 and CD105+/Endoglin; cat.
no. FMC003; R&D Systems, Inc.) were detected.
Following bone marrow extraction, the cells were
cultured in a T-75 flask as aforementioned and sub-cultured until
passage 3. Once the cells reached 80-90% confluency, the cells were
harvested using a trypsin/EDTA solution, and then washed twice with
PBS and resuspended in staining buffer at a concentration of
1x106 cells/ml. Each group was stained first for
CD45/CD29 and then for CD45/CD105. In brief, the cells were
labelled according to the manufacturer's instructions with
conjugated monoclonal antibodies:
Peridinin-chlorophyll-protein-conjugated mouse anti-rat CD45,
phycoerythrin-conjugated mouse anti-rat CD29/integrin β1 and
carboxyfluorescein-conjugated mouse Endoglin/CD105 (included with
the kit). The stained cells were then fixed with formaldehyde
buffer in PBS and analyzed using a flow cytometer (Invitrogen). The
results were analyzed using Attune NxT software v3,1,2 (Invitrogen;
Thermo Fisher Scientific, Inc.).
Gene expression analysis using reverse
transcription-quantitative PCR (RT-qPCR)
SOX-2, octamer-binding transcription factor 4
(Oct-4), Nanog and ES cell expressed Ras (Eras) mRNA expression was
assessed in the five categorized groups by total RNA extraction
followed by RT-qPCR. Briefly, RNA was extracted from the cells in
the five categorized groups using an RNeasy® Mini Kit
(Qiagen, Inc.). The isolated RNA was reverse-transcribed into cDNA
using the Maxima First Strand cDNA kit, and it was then kept at
-20˚C for further analysis. The qPCR reaction mixture mainly
contained 20 µl (total volume) triple-step SYBR-Green PCR master
mix (Thermo Fisher Scientific, Inc.), 2 µl (10 pmol/µl) forward and
reverse primers for the genes studied, and 2 µl cDNA template. The
amplification process for each reaction was conducted over a span
of 40 cycles. The primer designing tool [Primer designing tool
(nih.gov); https://www.ncbi.nlm.nih.gov/tools/primer-blast/]
was used to design primer sequences for the studied genes listed in
Table I. The cycle threshold (Cq)
value of the control gene, GAPDH, was used to normalize the Cq
values for each target gene. The determination of the expression of
several target genes was conducted using the comparative ΔCq
approach (14).
 | Table IPrimer sequences of the studied
genes. |
Table I
Primer sequences of the studied
genes.
| Gene | Sequence | Product size | Gene ID |
|---|
| Oct-4 | F:
CGAGAACCTTCAGGAGATATGC | 197 | NM_001009178.2 |
| | R:
TACAGAACCACACTCGAACC | | |
| Nanog | F:
CCTGAGCTATAAGCAGGTGAAGA | 144 | NM_001100781.1 |
| | R:
CTGCAATGGATGCTGGGATAC | | |
| SOX-2 | F:
AACCGTGATGCCGACTAGAA | 93 | NM_001109181.1 |
| | R:
CGCCTAACGTACCACTAGAAC | | |
| Eras | F:
CATCCTAACCCCCAACTGTCC | 186 | NM_001109375.1 |
| | R:
TGGCTCTCCTCTGGCGATCT | | |
Statistical analysis
The results were analyzed using one-way ANOVA with
GraphPad Prism 8.0 software (Dotmatics). To examine significant
differences between groups, Tukey's multiple comparison post hoc
test was applied following ANOVA. P<0.05 was considered to
indicate a statistically significant difference.
Results
Morphology of MSCs
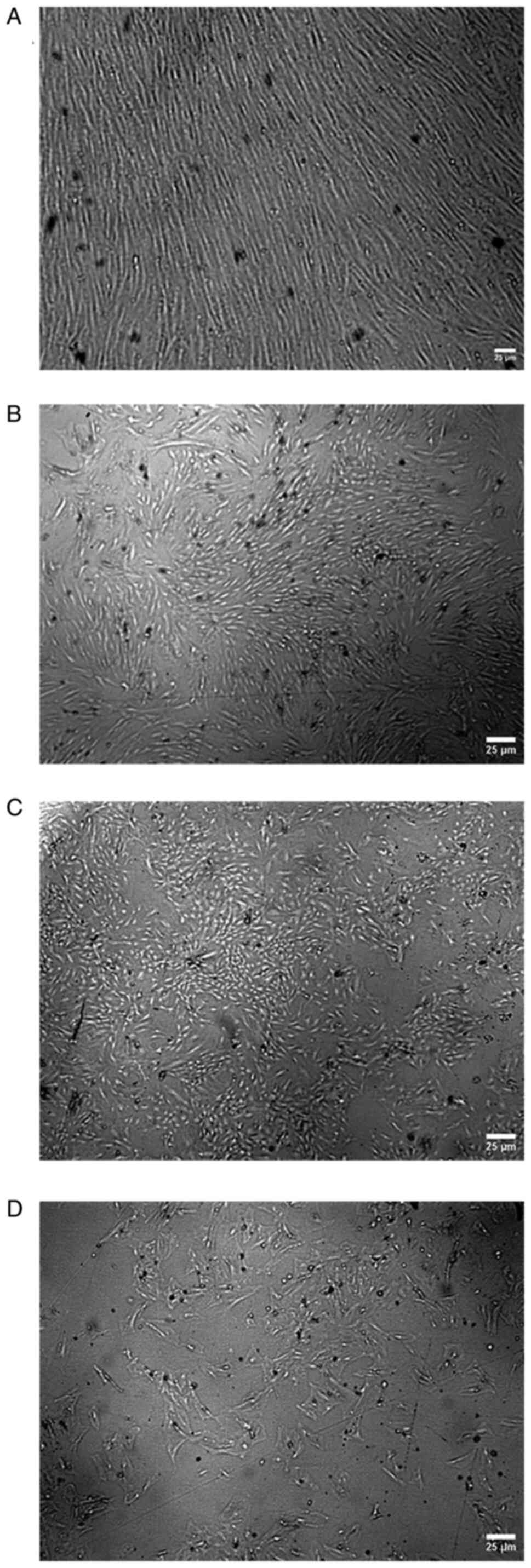
The morphology of the adhered BM-MSCs was examined
under an inverted microscope at passage 3, and then, according to
the shape of the cells and colonies, the cells were divided into
five groups (A, B, C, D and E). In group A, the cells had a spindle
fibroblast-like shape and formed a monolayer sheet, with cells
arranged in arrays without gaps between cells as shown in Fig. 1A. In group B, the cells still had a
spindle fibroblast-like shape, forming colonies with gaps between
cells, and cells could not form a monolayer sheet, as shown in
Fig. 1B. In group C, cells had the
ability to form colonies, but the cells in the colonies had
different shapes (ranging between spindle-shaped cells or flattened
cells), as shown in Fig. 1C. In
group D, the cells lost their ability to form colonies, and the
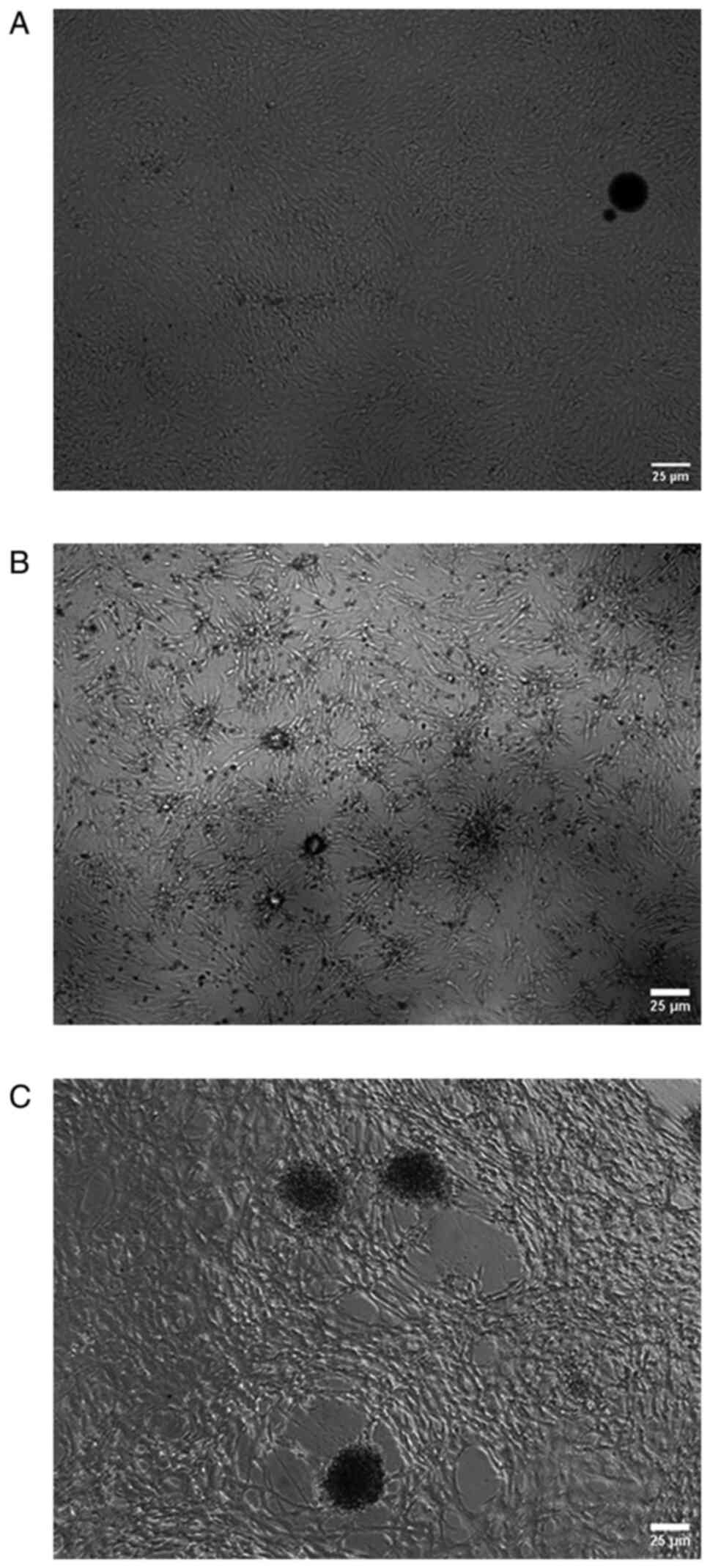
cells were fattened and large, as shown in Fig. 1D. In group E, the cells in the
monolayer sheet began to aggregate to form multilayer sheets, as
shown in Fig. 2A. Over time, the
cells collapsed, formed spheres and began to become enlarged, as
shown in Fig. 2B and C.
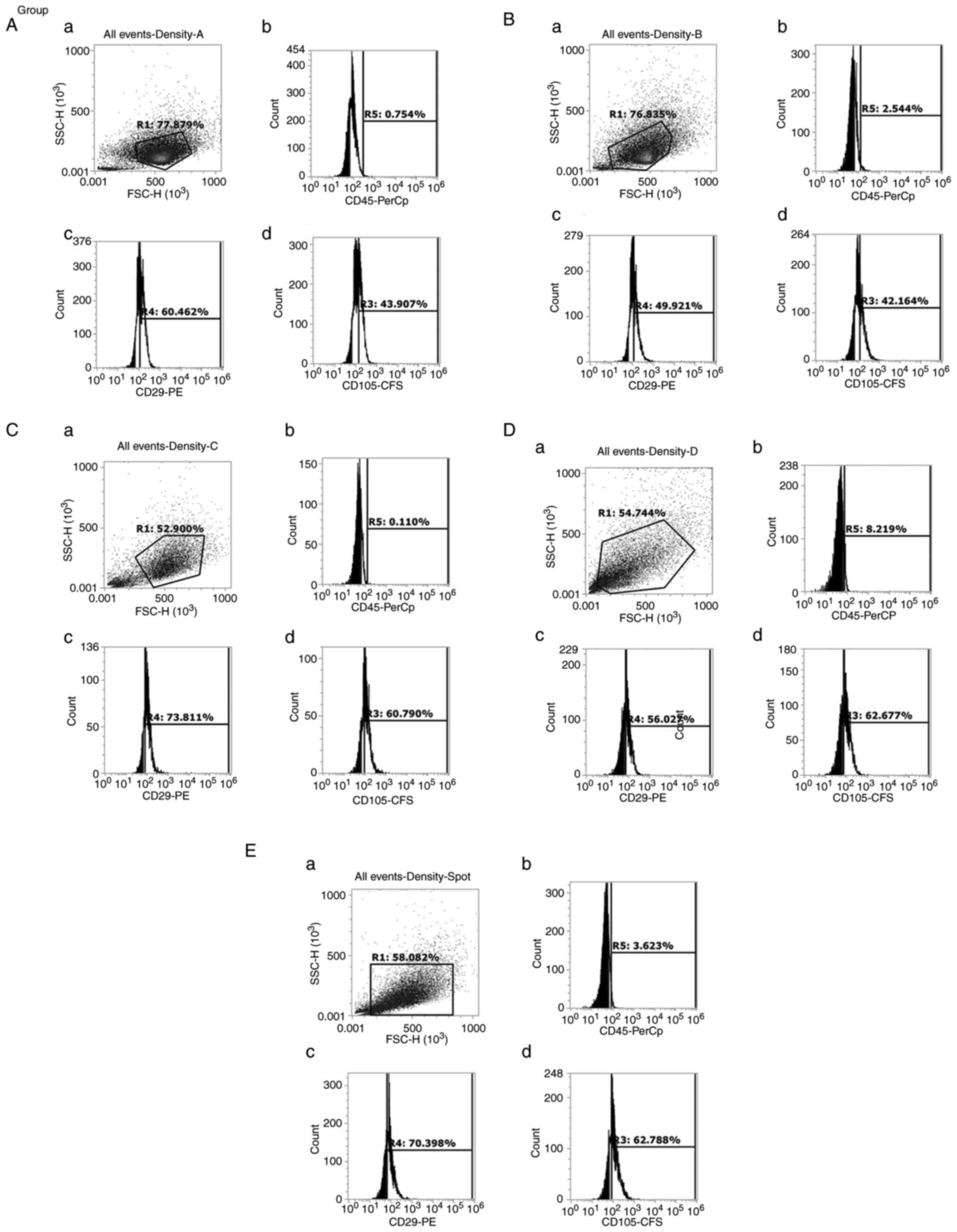
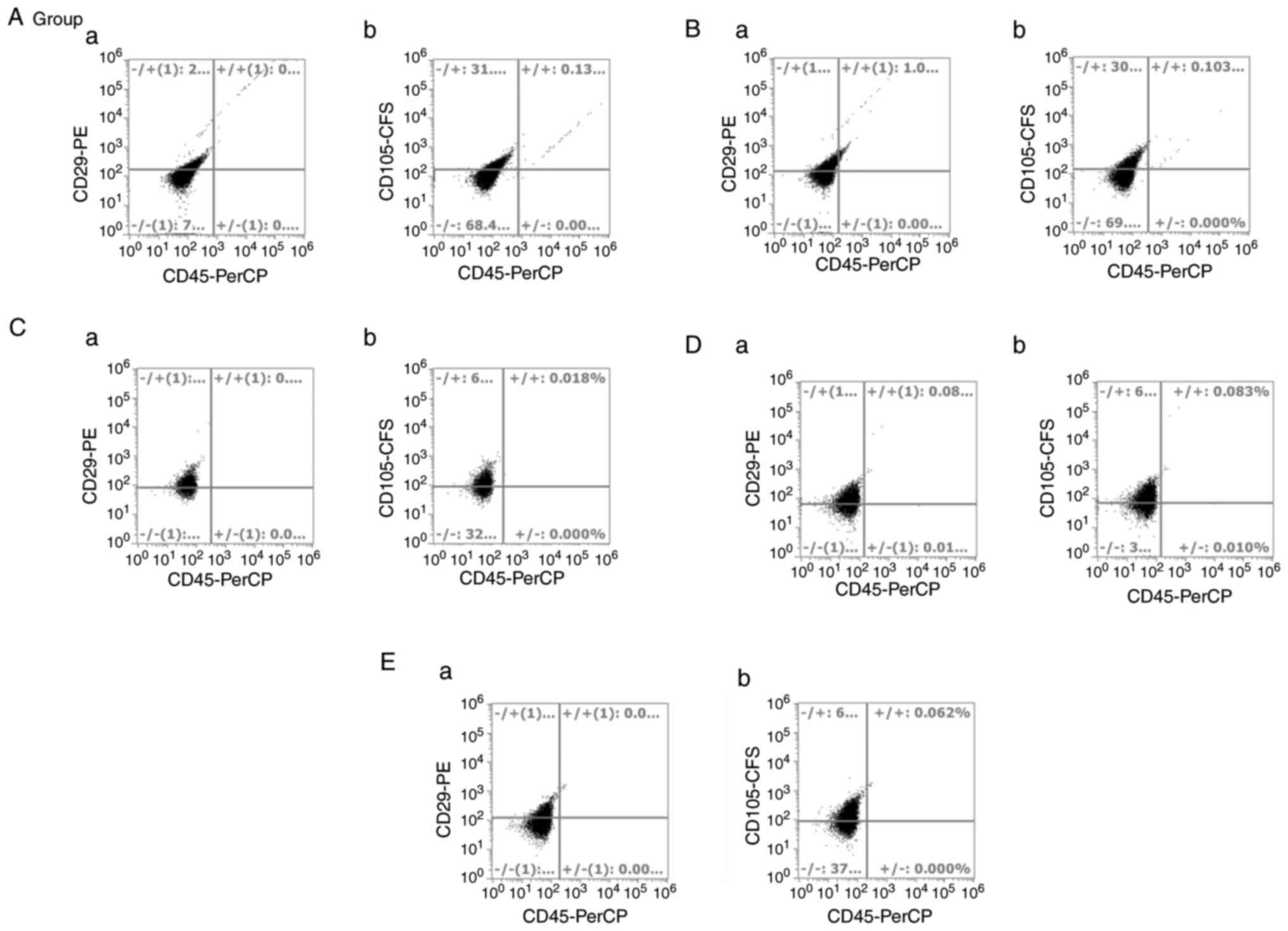
Flow cytometry
Once arrived at passage 3, the cells from the five
groups were harvested and stained as aforementioned. Flow
cytometric analysis of the five groups revealed that they met the
criteria of MSCs. They were all negative for CD45, and positive for
both mesenchymal markers, CD29 and CD105, as shown in Figs. 3 and 4. No differences in these markers were
observed among all groups, although they had different morphologies
and different self-renewal abilities.
Gene expression
The present study examined four genes (Nanog, Oct-4,
SOX-2 and Eras) that play a role in the pluripotency state of stem
cells, as previously reported by Takahashi and Yamanaka (15). In order to examine the expression
of different pluripotency genes in groups A, B, C, D and E, RT-qPCR
was performed on the cDNA of transcripts extracted from each group.
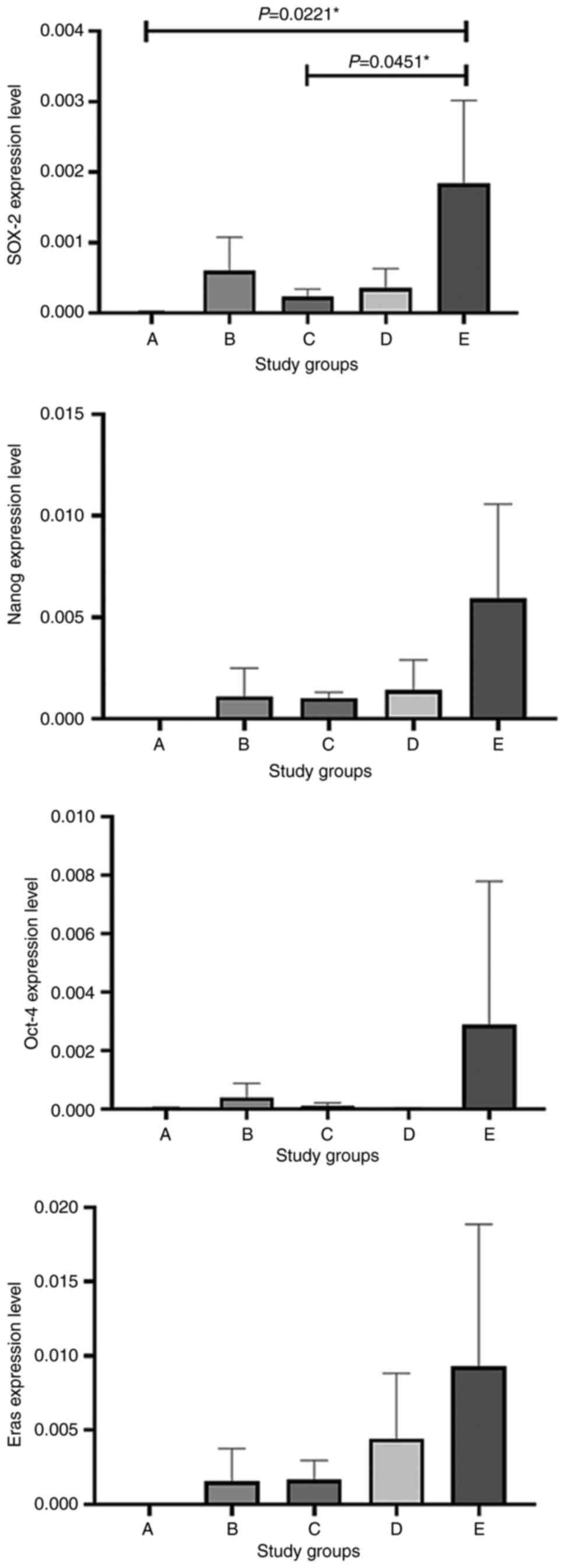
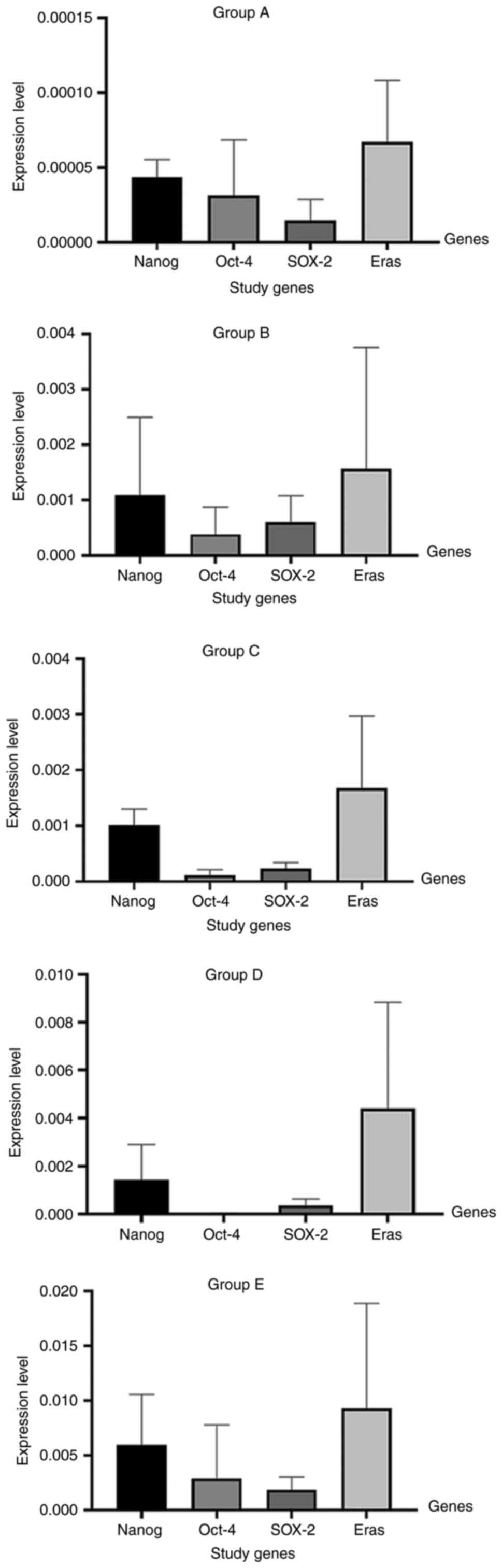
The highest expression levels of Nanog were observed in group E and
the lowest expression levels were observed in group A, while the
other groups exhibited different expression levels within the range
of those in groups A and E, as shown in Figs. 5 and 6. As regards Oct-4 and Eras expression,
the highest expression levels were also observed in group E, while
the lowest expression levels were observed in group A, as shown in
Figs. 5 and 6. The expression levels of SOX-2 were
significantly increased in group E compared with group A (P=0.0221)
and group C (P=0.0451), as shown in Figs. 5 and 6.
Discussion
In the present study, a total of 12 Sprague-Dawley
rats were used for breeding, and only the female rats aged 2 months
were selected for MSC isolation. The utilization of this particular
age criterion serves the purpose of mitigating the potential impact
of age-related disparities. It has been documented that the
functional attributes of BM-MSCs (16) and adipose-derived stem cells,
including yield, proliferation and differentiation capabilities,
are detrimentally influenced by an advancing age (17), which has also been confirmed at a
later date by another group (18).
In the present study, different shapes and
morphologies of MSCs were observed using the same methodology of
isolation and the same conditions in culture, including seeding
density, type of medium, percentage of serum and tissue culture
flasks.
In another study, different morphologies of cells
were observed when using six different types of culture media and
different seeding densities. Based on this, five different
morphologies were identified: i) Cells cultured in Rooster
Nourish-MSC XF media were spindle-shaped and elongated, and
aggregated; ii) cells cultured in Stem MACS-MSC XF and MSC
NutriStem XF were spindle-shaped and slender, and had a mat-like
appearance at higher confluency, as well as being shorter and
thicker; iii) cells cultured in PLTMax comparable to DMEM-KO, which
was used for control cultures, were spindle-shaped, elongated and
bright with tapering ends; and iv) cells cultured in StemXVivo MSC
SFM medium were highly elongated, with tapering ends at passage 4,
while at passage 5, few cells were aggregated and there was a
change in shape of the cells. However, the aforementioned
morphologies were medium-dependent and reflected the response of
cells to different environments rather than different types of stem
cells (19).
Another group from the University of Munich isolated
BM-MSCs, and then classified the cells into three sub-populations
according to their morphology: Rapidly self-renewing cells,
elongated fibroblast-like spindle-shaped cells and slowly
replicating, large, cuboidal or flattened cells. These
sub-populations exhibited distinct morphologies, proliferation
rates and differentiation properties (20).
In the present study, it was possible to easily and
clearly differentiate between five distinct groups of cells based
on the morphology and colony forming ability only after 1 week of
isolation. These different shapes were observed using the same
conditions, medium and methodology.
These different sub-populations of bone marrow stem
cells exhibited the same profile in flow cytometry analysis,
suggesting that they met the criteria of stem cells introduced by
the International Society for Cell & Gene Therapy. However,
these cells were not the same in terms of morphology, ability to
form colonies and doubling rate. Thus, it was determined which
sub-population was closer to the criteria of stem cells and the
pluripotency state in particular. Additionally, the results of flow
cytometry highlighted the inaccuracy of using cellular makers and
cytoplasmic markers to define and characterize stem cells.
The present study subsequently aimed to approach
this issue from another angle. Takahashi and Yamanaka (15) determined transcription factors that
controlled the internal cellular system and changed cells from the
quiescent state of being a fibroblast to the pluripotent state.
Thus, these factors should serve a crucial role in stem cells in
adult somatic tissues.
To better understand the heterogeneity among these
different morphologies of stem cells derived from the bone marrow,
the expression of four pluripotency genes was analyzed. This
investigation was motivated by the recognition that the
functionality of MSCs is regulated by distinct molecular profiles.
In the present study, the expression of SOX-2 was observed in the
isolated sub-populations, especially in group E, and this is a
factor implicated in the self-renewal of pluripotent stem cells and
the multipotency of BM-MSCs. This finding suggests that BM-MSCs
possess a more primitive state, which has been previously
acknowledged (21) and is in
accordance with the study by Heo et al (22), which revealed that SOX-2 expression
was higher in BM-MSCs than in adipose tissue-derived MSCs.
In the present study, the BM-MSCs also exhibited the
presence of considerable levels of a number of essential
transcription factors, including Oct-4 and Nanog. This was
confirmed by RT-qPCR, even in the absence of external stimuli. The
findings of the novel study by Labedz-Maslowska et al
(23) indicated a notable increase
in the mRNA concentration of two key transcription factors, Oct-4
and Nanog, in a specific subset of rat bone marrow cells.
Furthermore, it revealed a higher mRNA concentration of both
transcription factors regulating cell pluripotency (Oct-4 and
Nanog) in a purified
CD45-/Lin-/CD106+ population of
rat bone marrow cells compared with unfractionated bone marrow
cells (23). On the other hand,
the present study revealed low expression levels of Oct-4 in groups
A, C and D, and high expression levels in group E. Nanog expression
was higher than Oct-4 expression, but was still low in group A, and
different expression levels were observed in groups B, C and D,
while high expression levels were observed in group E.
In their novel study, Takahashi and Yamanaka
(15) reported that a number of
genes, including Stat3, Eras, c-myc, KLF transcription factor 4 and
β-catenin, which are commonly observed to be upregulated in
malignancies, play a role in sustaining the embryonic stem cell
phenotype over an extended period and promote the fast
proliferation of embryonic stem cells in a controlled environment.
This concurs with the results of the present study, which
demonstrated that Eras was highly expressed in group E rather than
other study groups.
In conclusion, the bone marrow is a rich source of
multiple types of cells that have adhesiveness properties as well
as a considerable ability to divide multiple times and
differentiate in some cases. Observations of the morphology and
colony-forming ability are more effective for the identification of
stem cells than flow cytometry and adhesiveness protocols. It would
be of great value to deeply study these 10 factors (Fbxo15, Nanog,
Eras, Dppa2, Oct3/4 (Pou5f1), Sox2, Tcl1, Klf4, β-catenin AND
c-Myc) mentioned in the study by Takahashi and Yamanaka (15) in adult stem cells, and to identify
the connection between pluripotent stem cells and stem cells in
adult tissues.
Acknowledgements
Not applicable.
Funding
Funding: The present work was funded by the Mansoura University
program for competitive research, Egypt.
Availability of data and materials
The datasets used and/or analyzed during this study
are available from the corresponding author on reasonable
request.
Authors' contributions
DASMA proposed the main hypothesis for the study,
designed the main experiments and participated in the writing of
the manuscript. MES proposed the main hypothesis for the study,
designed the main experiments, carried out many stages in the
experiments and participated in the writing of the manuscript. AAE
carried out many stages of the research protocol and prepared the
introduction section of the manuscript. SMF carried out many stages
of the research protocol, and prepared the figures and graphs for
the study, and participated in the writing of the manuscript. BHO
carried out the animal breading, animal care and bone marrow
extraction. SHH carried out the animal breading, animal care and
bone marrow extraction. All authors have read and approved the
final manuscript. MMS proposed the main hypothesis for the study,
designed the main experiments and participated in the writing of
the manuscript. DASMA and MES confirm the authenticity of all the
raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Animal care during the experimental procedures was
carried out according to the recommendations and following the
approval of the Mansoura University Animal Care and Use Committee
(MU-ACUC) (Decision no. MED.RP.23.06.02).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zakrzewski W, Dobrzyński M, Szymonowicz M
and Rybak Z: Stem cells: Past, present, and future. Stem Cell Res
Ther. 10(68)2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Pavlović M and Radotić K: Essential
characteristics of stem cells: Self-renewal, and plasticity. Animal
and plant stem cells: Concepts. Propagation and Engineering. 17–21.
2017.
|
|
3
|
Shah AA and Khan FA: Types and
classification of stem cells. Advances in application of stem
cells: From Bench to Clinics: 25-49, 2021.
|
|
4
|
Barky AR, Ali EMM and Mohamed TM: Stem
cells, classifications and their clinical applications. Am J
Pharmacol Ther. 1:001–007. 2017.
|
|
5
|
Khan FA, Almohazey D, Alomari M and
Almofty SA: Isolation, culture, and functional characterization of
human embryonic stem cells: Current trends and challenges. Stem
Cells Int. 2018(1429351)2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Volarevic V, Markovic BS, Gazdic M,
Volarevic A, Jovicic N, Arsenijevic N, Armstrong L, Djonov V, Lako
M and Stojkovic M: Ethical and safety issues of stem cell-based
therapy. Int J Med Sci. 15:36–45. 2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Kim A, Lee KG, Kwon Y, Lee KI, Yang HM,
Habib O, Kim J, Kim ST, Kim SJ, Kim JS and Hwang DY: Off-the-Shelf,
immune-compatible human embryonic stem cells generated via
CRISPR-mediated genome editing. Stem Cell Rev Rep. 17:1053–1067.
2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Doğan A: Embryonic stem cells in
development and regenerative medicine. Adv Exp Med Biol. 1079:1–15.
2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Margiana R, Markov A, Zekiy AO, Hamza MU,
Al-Dabbagh KA, Al-Zubaidi SH, Hameed NM, Ahmad I, Sivaraman R, Kzar
HH, et al: Clinical application of mesenchymal stem cell in
regenerative medicine: A narrative review. Stem Cell Res Ther.
13(366)2022.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Rehman A, Nigam A, Laino L, Russo D,
Todisco C, Esposito G, Svolacchia F, Giuzio F, Desiderio V and
Ferraro G: Mesenchymal stem cells in soft tissue regenerative
medicine: A comprehensive review. Medicina (Kaunas).
59(1449)2023.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Kobolak J, Dinnyes A, Memic A,
Khademhosseini A and Mobasheri A: Mesenchymal stem cells:
Identification, phenotypic characterization, biological properties
and potential for regenerative medicine through biomaterial
micro-engineering of their niche. Methods. 99:62–68.
2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Li Z, Zhang C, Weiner LP, Zhang Y and
Zhong JF: Molecular characterization of heterogeneous mesenchymal
stem cells with single-cell transcriptomes. Biotechnol Adv.
31:312–317. 2013.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Meurer SK, Neß M, Weiskirchen S, Kim P,
Tag CG, Kauffmann M, Huber M and Weiskirchen R: Isolation of mature
(Peritoneum-Derived) mast cells and immature (Bone Marrow-Derived)
mast cell precursors from mice. PLoS One.
11(e0158104)2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Takahashi K and Yamanaka S: Induction of
pluripotent stem cells from mouse embryonic and adult fibroblast
cultures by defined factors. Cell. 126:663–676. 2006.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Fafián-Labora J, Fernández-Pernas P,
Fuentes I, De Toro J, Oreiro N, Sangiao-Alvarellos S, Mateos J and
Arufe MC: Influence of age on rat bone-marrow mesenchymal stem
cells potential. Sci Rep. 5(16765)2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Jung HG, Ahn EK, Lee JH, Kim YH, Leem SH,
Heo J and Kim H: Effects of harvesting sites and ages on adipose
tissue-derived stem cells in rat. Tissue Engineering and
Regenerative Medicine. 11:137–142. 2014.
|
|
18
|
Siennicka K, Zołocińska A, Dębski T and
Pojda Z: Comparison of the donor age-dependent and in vitro
culture-dependent mesenchymal stem cell aging in rat model. Stem
Cells Int. 2021(6665358)2021.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Bhat S, Viswanathan P, Chandanala S,
Prasanna SJ and Seetharam RN: Expansion and characterization of
bone marrow derived human mesenchymal stromal cells in serum-free
conditions. Sci Rep. 11(3403)2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Haasters F, Prall WC, Anz D, Bourquin C,
Pautke C, Endres S, Mutschler W, Docheva D and Schieker M:
Morphological and immunocytochemical characteristics indicate the
yield of early progenitors and represent a quality control for
human mesenchymal stem cell culturing. J Anat. 214:759–767.
2009.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Yoon DS, Kim YH, Jung HS, Paik S and Lee
JW: Importance of Sox2 in maintenance of cell proliferation and
multipotency of mesenchymal stem cells in low-density culture. Cell
Prolif. 44:428–440. 2011.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Heo JS, Choi Y, Kim HS and Kim HO:
Comparison of molecular profiles of human mesenchymal stem cells
derived from bone marrow, umbilical cord blood, placenta and
adipose tissue. Int J Mol Med. 37:115–125. 2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Labedz-Maslowska A, Kamycka E,
Bobis-Wozowicz S, Madeja Z and Zuba-Surma EK: Identification of new
rat bone marrow-derived population of very small stem cell with
Oct-4A and Nanog expression by flow cytometric platforms. Stem
Cells Int. 2016(5069857)2016.PubMed/NCBI View Article : Google Scholar
|