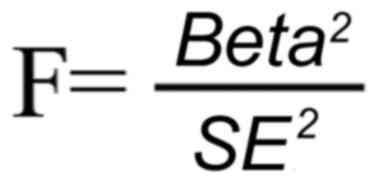Introduction
Melanoma is an invasive malignant tumor originating
from melanocytes (1). The
incidence of melanoma exhibits a linear increase in the age group
of 25 to 50 years (2). Although
melanoma is less common than other types of skin cancer, it
accounts for 73% of skin cancer-related mortality (3), with a 5-year survival rate of only
~18%. It is considered that the incidence and mortality rates of
melanoma will continue to increase over the next 10 years (4,5).
Regrettably, early surgical treatment and late-stage radiation
therapy and chemotherapy have not yielded satisfactory therapeutic
effects for patients with melanoma (6). Hence, the identification of the risk
factors involved in the occurrence of melanoma is of utmost
importance.
Studies have indicated a marked increase in the
expression and secretion of cathepsins in cancer, which play
recognized roles in cancer progression (7,8).
Within tumor cells, cathepsins and their messenger RNAs are
frequently upregulated, leading to the secretion of excess
proenzyme (unprocessed form) (7).
Extracellular cathepsins have been isolated from cancer cell
culture media, constituting 40% of total secreted proteins, and
have been observed in the serum of patients with cancer (9). Cathepsins play crucial roles in the
occurrence, development and metastasis of melanoma (8,10).
Secreted cathepsins promote melanoma invasion and metastasis by
directly and indirectly facilitating extracellular matrix
degradation (11,12). Saenger et al (13) utilized reverse
transcription-polymerase chain reaction to measure preprocessed
peripheral blood samples from 218 patients with melanoma,
demonstrating that the expression levels of cathepsins in
peripheral blood can predict the survival rates of patients with
melanoma. Elevated levels of cathepsins were shown to be associated
with the favorable prognosis of patients with melanoma (13). Kos et al (14) used quantitative enzyme-linked
immunosorbent assay to measure cathepsin B levels in the serum of
43 patients with metastatic melanoma, 54 patients with in
situ cutaneous melanoma (the absence of evidence of tumor
metastasis) and 30 healthy blood donors. The results of their study
revealed significantly higher levels of cathepsin B in the serum of
patients with metastatic melanoma compared with healthy
individuals, and patients with metastatic melanoma with high levels
of cathepsin B exhibited markedly lower overall survival rates than
those with low levels of cathepsin B (14). These associations suggest a link
between cathepsins and malignant melanoma; however, causal
associations cannot be established from observational studies due
to confounding and reverse causality. These studies may be
influenced by various confounding factors, such as the voluntary
treatment choices of patients and incomplete data records,
potentially resulting in the decreased internal validity of the
research findings. Furthermore, causal associations cannot be
established from these studies. Manipulating independent variables
through data collection and outcome observation does not allow for
the verification of causality.
In the present study, Mendelian randomization (MR)
was used to validate the causal associations in assessing the role
of cathepsins in the development of melanoma. With MR, the impact
of exposure (cathepsins) on the risk of developing melanoma is
assessed using genetic instrumental variables (IVs) (15). As IVs are randomly distributed in
conceptualization, they are not influenced by confounding factors.
In genome-wide association studies (GWASs), the common genetic
variants associated with cathepsins serve as IVs for measuring
cathepsins. The present study also discusses the causal risk of
melanoma development from a genetic perspective, providing a basis
for further prevention and diagnosis.
Materials and methods
Study design
A two-sample MR was conducted utilizing GWAS data.
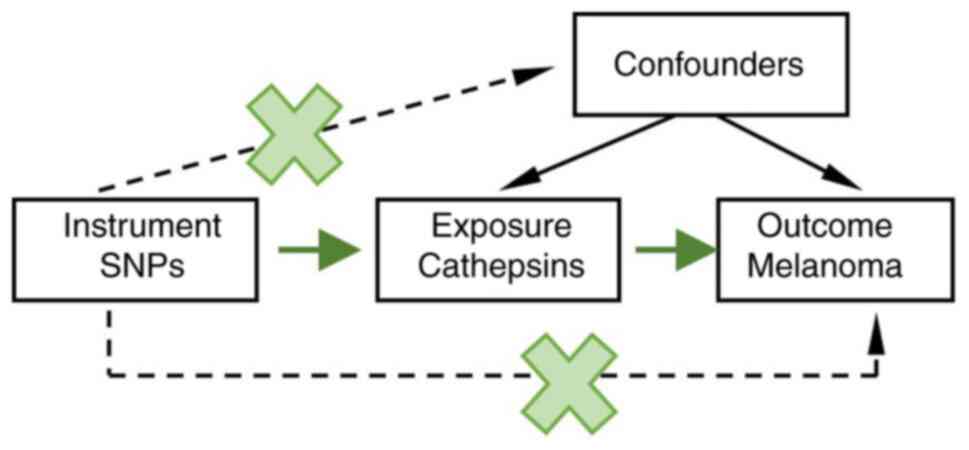
The MR method was based on the following three assumptions
(Fig. 1) (16,17):
i) The genetic variants selected as IVs are associated with
cathepsins; ii) the genetic variants are independent of any
unmeasured confounding factors; iii) the genetic variants are
associated with malignant melanoma solely through cathepsins and
not through other pathways.
Data sources
The GWAS involved in malignant melanoma and
cathepsins utilized data from GWAS databases. Specifically, the
GWAS data for malignant melanoma were sourced from the UK Biobank
(https://www.ukbiobank.ac.uk), and the
data for cathepsins were obtained from the GWAS Catalog (https://gwas.mrcieu.ac.uk/datasets). The
summarized levels of cathepsin S, cathepsin B, cathepsin O,
cathepsin E and cathepsin L2 in the European population were
extracted from the comprehensive summary data of GWAS protein-level
summaries described by Sun et al (18). As regards the outcome data details,
the melanoma data from the UK Biobank included 3,751 cases and
372,016 controls, covering a total of 11,396,019 single nucleotide
polymorphisms (SNPs). All melanoma cases met the diagnostic
criteria for melanoma: immunohistochemical staining revealed
positivity for characteristic markers of melanocytes, including
S100, Melan-A, and HMB45(19). The
study design, quality control procedures, phenotype definition and
inference methods for GWAS were described in the previous
publication where sample collection was reported (20). Participants in these data sources
were of European ancestry. The intentional selection of European
participants helps ensure sample population homogeneity, reduce
confounding factors and enhance result reliability. As the data
included in the present study have been previously published,
ethical approval or informed consent were not required (20). Detailed information regarding the
data sources is provided in Table
I, along with corresponding links to the specific data sources
(Table SI).
 | Table IMendelian randomization results for
the causal effects of cathepsins on melanoma. |
Table I
Mendelian randomization results for
the causal effects of cathepsins on melanoma.
| | IVW | MR-Egger | Weighted mode | Weighted
median |
IVW-Heterogeneity | Pleiotropy |
|---|
| Trait | SNPs | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | I2 | P-value | Egger
intercept | P-value |
|---|
| Cathepsin S | 23 | 1.000
(0.999-1.001) | 0.943 | 1.000
(0.999-1.001) | 0.924 | 1.000
(0.999-1.001) | 0.836 | 1.000
(0.999-1.001) | 0.8 | 0.198 | 0.196 | -9.30E-06 | 0.947 |
| Cathepsin B | 17 | 1.000
(0.999-1.001) | 0.763 | 1.000
(0.998-1.002) | 0.797 | 0.999
(0.998-1.001) | 0.34 | 0.999
(0.998-1.001) | 0.32 | 0.158 | 0.268 | -0.0001118 | 0.673 |
| Cathepsin O | 10 | 1.000
(0.999-1.001) | 0.646 | 0.998
(0.996-1.001) | 0.294 | 1.000
(0.997-1.003) | 0.95 | 1.000
(0.998-1.002) | 0.927 | 0.742 | 0.820 | 0.0003042 | 0.335 |
| Cathepsin E | 11 | 0.999
(0.998-1.001) | 0.375 | 1.000
(0.997-1.003) | 0.809 | 1.000
(0.998-1.002) | 0.906 | 1.000
(0.998-1.001) | 0.857 | 1.448 | 0.943 | -0.0001668 | 0.489 |
| Cathepsin L2 | 11 | 1.000
(0.999-1.001) | 0.872 | 1.001
(0.998-1.004) | 0.404 | 1.001
(0.998-1.004) | 0.447 | 1.000
(0.998-1.004) | 0.323 | 0.212 | 0.242 | -0.0002378 | 0.394 |
Genetic variants
As regards the exposure data, genetic variants
exceeding the genome-wide association thresholds
(P<5.0x10-6) were selected as IVs, and a clumping
algorithm (r2 threshold=0.001; kb=10,000 mB) was used to
eliminate linkage disequilibrium. Additionally, the strength of the
instrument was assessed using the F-statistic (F-value calculation
formula below), which is approximated by the squared SNP-phenotype
association divided by its variance. A potential weak instrument is
indicated by an F-statistic <10(21). Therefore, strongly correlated IVs,
based on an F-statistic >10, were selected to meet the
assumption of correlation in MR analysis as follows:
where beta represents the effect size of SNP
exposure, and SE is the standard error of SNP exposure. A total of
23 SNPs were found to be associated with cathepsin S, 17 SNPs with
cathepsin B, 11 SNPs with cathepsin O, 11 SNPs with cathepsin E and
11 SNPs with cathepsin L2. The effect sizes of changes in
cathepsins for each additional effect allele are presented in
Table SII, Table SIII, Table SIV, Table SV and Table SVI. No linkage disequilibrium was
observed among the SNPs.
Two-sample MR analysis
MR analysis, a prominent tool for determining the
causal effect of exposure variables on outcomes via genetic
variation, was used to confirm the causal association between
cathepsins and melanoma (22,23).
Various MR techniques, including inverse variance weighted (IVW),
MR-Egger, weighted median (WM) and weighted mode, were utilized.
Notably, the IVW and MR-Egger techniques, commonly utilized as
fundamental methodologies in MR analysis worldwide, play essential
roles. An IVW analysis was conducted, utilizing multiplicative
random effects assuming balanced pleiotropy, with SNP-specific Wald
estimates, i.e., SNP-outcome association divided by SNP-exposure
association, as the main analysis. Additionally, sensitivity
analyses were performed using WM and MR-Egger (24). The WM assumes that 50% of the
weight comes from valid SNPs. Furthermore, MR-Egger exhibited
robustness to genetically invalid instruments due to instrument
strength independent of direct effect, meaning the instruments do
not confound the exposure of the outcomes. A zero MR-Egger
intercept indicated no evidence for this genetic pleiotropy. IVW,
serving as the main tool, determines causal links between exposure
variables and outcomes in MR analysis, with a result deemed
significant when the P-value of IVW is <0.05. Additionally,
under the condition of IVW, the direction of MR-Egger and the WM
method must align with that of IVW. To eliminate bias, MR-PRESSO
(https://github.com/rondolab/MR-PRESSO) was used to
detect and correct potentially pleiotropic outliers (SNPs) for all
reported results. Heterogeneity, quantified using Cochran Q
statistics and I² statistics, increased with larger I² values.
Further analysis included leave-one-out analysis conducted by
removing each SNP to assess the stability and reliability of the MR
results. The final results, depicted using forest plots, scatter
plots and funnel plots, showcased the effectiveness of the MR
analysis methods. These methods were implemented using the
‘Two-SampleMR’ (https://mrcieu.github.io/TwoSampleMR) and ‘MR-PRESSO’
(https://github.com/rondolab/MR-PRESSO) R packages in R
(version 4.0.3).
Statistical analysis
IVW: IVW is a method used in MR for the
meta-analysis of the effects of multiple SNPs across different
locus. The prerequisite for applying IVW is that all SNPs serve as
valid IVs and are completely independent of each other. WM: WM
represents the median of the distribution function obtained by
sorting the individual SNP effect values according to weights. When
at least 50% of the information comes from valid IVs, WM can
provide robust estimates. Weighted mode: The weighted mode method
evaluates the combined effects of different genotypes on the
phenotype by calculating the weighted average of each genotype.
This approach can better control the impact of genotype frequency
differences on the analysis results, thereby improving the
robustness and accuracy of the analysis. MR-Egger method: MR-Egger
does not enforce the regression line to pass through the origin,
allowing for the inclusion of IVs with directional pleiotropy. The
presence of genetic pleiotropy is indicated when the regression
intercept is non-zero and P<0.05. MR-PRESSO method: MR-PRESSO
can be used to obtain more accurate estimates by identifying and
excluding specific SNPs as outliers, thus reducing the influence of
potential confounding factors in MR analysis.
Results
Selection of IVs
Following an extensive quality control review, 72
SNPs associated with five cathepsins were identified as IVs for
melanoma. Notably, the cathepsin S levels were linked to 23 SNPs,
cathepsin B to 17 SNPs, cathepsin O to 11 SNPs, cathepsin E to 11
SNPs and cathepsin L2 to 11 SNPs (Tables I and SII, SIII, SIV, SV
and SVI).
Causal role of cathepsins in
melanoma
In the analysis, IVW was used to provide estimates
of the causal association between cathepsins and melanoma,
indicating that cathepsin S [odds ratio (OR), 1.000; 95% confidence
interval (CI) 0.999-1.001; P=0.943], cathepsin B (OR, 1.000; 95%
CI, 0.999-1.001; P=0.763), cathepsin O (OR, 1.000; 95% CI,
0.999-1.001; P=0.646), cathepsin E (OR, 0.999; 95% CI, 0.998-1.001;
P=0.375) and cathepsin L2 (OR, 1.101; 95% CI 0.831-1.458; P=0.503)
were not significantly associated with the risk of developing
melanoma. The overall estimates of the MR Egger and WM methods were
consistent with the IVW analysis (Table I).
Sensitivity analyses
For cathepsin S, neither the IVW test (Q=27.421,
P=0.196) nor the MR-Egger test (Q=27.415, P=0.158) revealed
significant heterogeneity (P>0.05). Similarly, for cathepsin B
(IVW test: Q=19.013, P=0.268; MR-Egger test: Q=18.782, P=0.224),
cathepsin O (IVW test: Q=5.165, P=0.820; MR-Egger test: Q=4.114,
P=0.847), cathepsin E (IVW test: Q=4.086, p = 0.943; MR-Egger test:
Q=3.566, P=0.938) and cathepsin L2 (IVW test: Q=12.683, P=0.242;
MR-Egger test: Q=11.644, P=0.234), no apparent heterogeneity was
observed (Table SVII).
Additionally, the MR-Egger intercept test indicated the absence of
horizontal pleiotropy (P>0.05) (Table SVIII). Scatter plots demonstrated
the genetic association between cathepsins and melanoma (Fig. S1). Furthermore, funnel plots
revealed no significant asymmetry, suggesting negligible
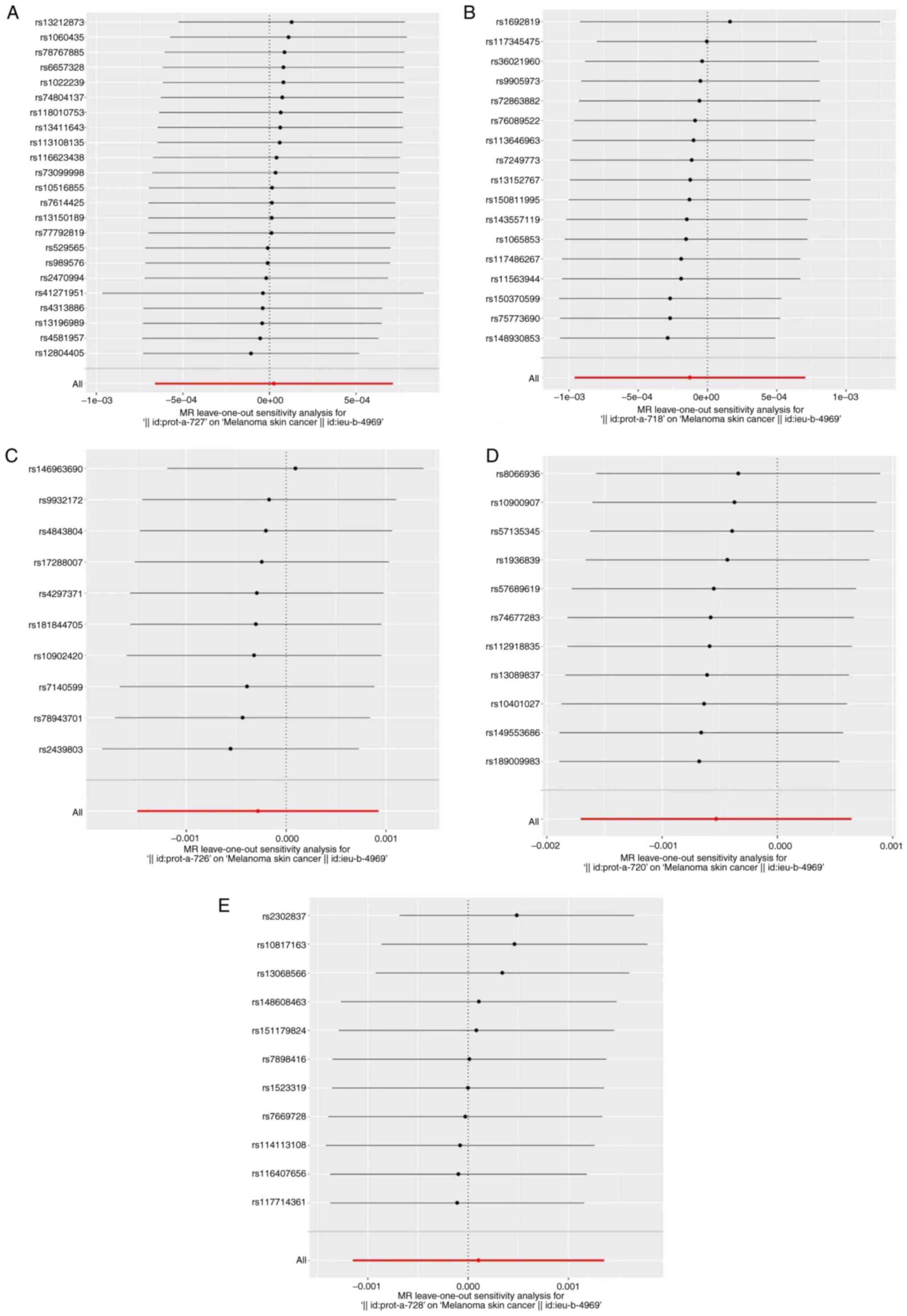
publication bias and directional horizontal pleiotropy (Fig. S2). Following a systematic
elimination of SNPs through leave-one-out analysis, the outcomes
showed minimal alteration, indicating that no single variant
influenced the association between cathepsins (cathepsin S,
cathepsin B, cathepsin O, cathepsin E and cathepsin L2) and
melanoma (Fig. 2).
Discussion
In the present study, genetic variants were utilized
to assess whether a risk factor has a causal effect on the results
of MR studies. To the best of our knowledge, this is the first MR
study aimed at evaluating the impact of cathepsins on the
occurrence of melanoma. The present study found no causal
association between several cathepsins (cathepsin S, cathepsin B,
cathepsin O, cathepsin E and cathepsin L2) and melanoma among
Europeans. In other words, alterations in cathepsins levels are not
associated with the occurrence of melanoma.
Cathepsins are a group of enzymes involved in
determining the metastatic potential of cancer cells (10), including cysteine cathepsins
(cathepsin B and cathepsin L) and aspartyl protease (cathepsin D),
which are typically present in the form of inactive zymogens in
lysosomes. Once released into the extracellular space, cathepsins
contribute to the enhancement of tumor metastatic potential by
promoting cell migration and invasion capabilities (25,26).
However, the current understanding of the association between
cathepsins and melanoma is still at a laboratory experiment phase.
Only a limited number of clinical observational studies are
currently available; however, due to limitations in sample size,
the results of these observational studies are not yet reliable
(13,14,27).
In the present study, not all capabilities were selected as
exposure factors for MR analysis, as other capabilities are rarely
reported to be closely associated with cutaneous melanoma. Previous
studies have indicated that various cathepsins are overexpressed in
melanoma . Furthermore, cathepsin B co-localizes with
HMB-45-positive cells in malignant melanoma (28). It is noteworthy that the levels of
cathepsin B and H are significantly elevated in metastatic melanoma
patient groups compared to healthy control groups (14). The transition of human melanoma
cells from a non-metastatic to a highly metastatic phenotype is
directly associated with the secretion of cathepsin L (29). The precursor of cathepsin-L is a
protease capable of cleaving human C3 (the third component of
complement), imparting high tumorigenicity and metastatic potential
to human melanoma cells (29).
Cathepsin S is a single-chain non-glycosylated enzyme, the only
member among 11 cathepsins (Cat B, C, F, H, L, K, O, S, V, W, X/Z)
possessing protein hydrolysis activity at neutral pH (30). Research has also revealed increased
expression of cathepsin S in certain types of cancer, such as
colorectal, gastric and breast cancer (31-33),
with its crucial role demonstrated in tumor invasion and metastasis
through inducing tumor angiogenesis and degradation of the
extracellular matrix (34,35). The inhibition of cathepsin S
expression has been shown to be associated with the inhibition of
malignant phenotypes in cancer cells and improved clinical outcomes
in patients with breast cancer (33,36).
Additionally, specific cathepsin S activity in primary choroidal
melanoma is the strongest predictor of tumor metastatic behavior
(37). Notably, unlike other
cathepsins, cathepsin E has been found to be associated with
antitumor activity in vitro, as research using mice carrying
human and murine tumor xenografts has demonstrated the antitumor
activity of cathepsin E (38). In
addition, the injection of purified cathepsin E into nude mice
carrying human tumor xenografts has been shown to induce tumor cell
apoptosis in a dose-dependent manner and to inhibit tumor growth
(38).
Although some of the aforementioned studies strongly
suggest that cathepsins in serum can serve as serum markers for the
early diagnosis and prognostic prediction of melanoma, the present
study did not provide sufficient evidence to support this
conclusion. Serum markers possess advantages, such as easy
detection, minimal patient trauma and a low cost, rendering them
markedly advantageous for the early screening and follow-up care of
patients with cancer, and they are also a current focus and
priority of research (39,40). Tumor markers for melanoma lack
specificity; hence, broad-spectrum tumor markers, such as CA50,
CA199, etc., can serve as melanoma tumor markers. Since these tumor
markers are also highly expressed in patients with other types of
cancer, there is a need to discover sensitive and specific
biomarkers for cutaneous malignant melanoma (41). The cathepsins hold value for the
early diagnosis and prognosis assessment of patients with melanoma;
however, the results are typically derived from clinical studies
with small sample sizes, lacking authenticity and reliability
(14,27). In conclusion, diagnosing melanoma
using cathepsins remains challenging; therefore, further evaluation
using large sample sizes, multicenter, and rigorously designed
studies are required to validate their authenticity. Further
prospective studies are warranted to verify their effectiveness,
and concrete steps should be taken to establish standards for
guiding the effective use of diagnostic and prognostic assessment
tools.
In addition, the present study aimed to increase the
number of SNPs to alleviate bias caused by limited IVs; therefore,
relevant literature on MR of cathepsins was examined and a
significance threshold with a P-value <5x10-6 was
established. However, caution is required in interpreting the
results. Third, the MR analysis method is a theoretical causal
analysis method that requires further validation through animal
experiments to establish causality. On the whole, the findings in
the present study may aid in the understanding of the complex
mechanisms linking cathepsins and melanoma.
In conclusion, the present study found no evidence
of a causal association between cathepsins and melanoma, which
contradicts the results of the majority of previous observational
studies, as aforementioned. Prior research has suggested a
potential association between melanoma and cathepsins (42); however, confounding factors or
reverse causality may also be at play. Further investigations are
required to fully elucidate the association between melanoma and
cathepsins.
Supplementary Material
Scatter plots for the causal
association between cathepsins and melanoma. (A) Associations
between cathepsin S and melanoma. (B) Associations between
cathepsin B and melanoma. (C) Associations between cathepsin O and
melanoma. (D) Associations between cathepsin E and melanoma. (E)
Associations between cathepsin L2 and melanoma.
Funnel plots of cathepsins. (A) Funnel
plots of cathepsin S. (B) Funnel plots of cathepsin B. (C) Funnel
plots of cathepsin O. (D) Funnel plots of cathepsin E. (E) Funnel
plots of cathepsin L2.
Data sources of the Mendelian
randomization study.
Summary statistics utilized in the
Mendelian randomization study of cathepsin S in melanoma.
Summary statistics utilized in the
Mendelian randomization study of cathepsin B in melanoma.
Summary statistics utilized in the
Mendelian randomization study of cathepsin O in melanoma.
Summary statistics utilized in the
Mendelian randomization study of cathepsin E in melanoma.
Summary statistics utilized in the
Mendelian randomization study of cathepsin L2 in melanoma.
Heterogeneity tests of cathepsins in
melanoma.
Test for the directional horizontal
pleiotropy of cathepsins in melanoma.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Author's contributions
WW contributed to the conceptualization and
methodology of the study. JL and WW were involved in the data
analysis and visualization of the results. WW participated in the
drafting and reviewing of the main manuscript. JL and WW have
reviewed and approved the final manuscript. JL and WW confirm the
authenticity of all the raw data
Ethics approval and consent to
participate
Two-sample MR was conducted using GWAS data. The
study did not require ethical approval since all data were derived
from summary statistics from previously published GWASs.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hasan N, Nadaf A, Imran M, Jiba U, Sheikh
A, Almalki WH, Almujri SS, Mohammed YH, Kesharwani P and Ahmad FJ:
Skin cancer: Understanding the journey of transformation from
conventional to advanced treatment approaches. Mol Cancer.
22(168)2023.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Carr S, Smith C and Wernberg J:
Epidemiology and risk factors of melanoma. Surg Clin North Am.
100:1–12. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Welch HG, Mazer BL and Adamson AS: The
rapid rise in cutaneous melanoma diagnoses. N Engl J Med.
384:72–79. 2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Whiteman DC, Green AC and Olsen CM: The
growing burden of invasive melanoma: Projections of incidence rates
and numbers of new cases in six susceptible populations through
2031. J Invest Dermatol. 136:1161–1171. 2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Yuan J, Li X and Yu S: Global, Regional,
and National incidence trend analysis of malignant skin melanoma
between 1990 and. 2019, and projections until 2034. Cancer Control.
31(10732748241227340)2024.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Oya K, Nakamura Y, Shen LT, Ishizuki S,
Matsusaka S and Fujisawa Y: Soluble PD-L1 predicts tumor response
and immune-related adverse events in patients with advanced
melanoma treated with anti-PD-1 antibodies. J Dermatol. 51:807–815.
2024.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Olson OC and Joyce JA: Cysteine cathepsin
proteases: Regulators of cancer progression and therapeutic
response. Nat Rev Cancer. 15:712–729. 2015.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Palermo C and Joyce JA: Cysteine cathepsin
proteases as pharmacological targets in cancer. Trends Pharmacol
Sci. 29:22–28. 2008.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Sudhan DR and Siemann DW: Cathepsin L
targeting in cancer treatment. Pharmacol Ther. 155:105–116.
2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Mohamed MM and Sloane BF: Cysteine
cathepsins: Multifunctional enzymes in cancer. Nat Rev Cancer.
6:764–775. 2006.PubMed/NCBI View
Article : Google Scholar
|
|
11
|
Tripathi R and Plattner R: EnABLing
cathepsin-driven melanoma metastasis. Molr Cell Oncol.
5(e1458016)2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Tripathi R, Fiore LS, Richards DL, Yang Y,
Liu J, Wang C and Plattner R: Abl and Arg mediate cysteine
cathepsin secretion to facilitate melanoma invasion and metastasis.
Sci Signal. 11(eaao0422)2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Saenger Y, Magidson J, Liaw B, de Moll E,
Harcharik S, Fu Y, Wassmann K, Fisher D, Kirkwood J, Oh WK and
Friedlander P: Blood mRNA expression profiling predicts survival in
patients treated with tremelimumab. Clin Cancer Res. 20:3310–3318.
2014.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Kos J, Stabuc B, Schweiger A, Krasovec M,
Cimerman N, Kopitar-Jerala N and Vrhovec I: Cathepsins B, H, and L
and their inhibitors stefin A and cystatin C in sera of melanoma
patients. Clin Cancer Res. 3:1815–1822. 1997.PubMed/NCBI
|
|
15
|
Guo W, Zhao L, Huang W, Chen J, Zhong T,
Yan S, Hu W, Zeng F, Peng C and Yan H: Sodium-glucose cotransporter
2 inhibitors, inflammation, and heart failure: A two-sample
Mendelian randomization study. Cardiovasc Diabetol.
23(118)2024.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Bowden J, Davey Smith G and Burgess S:
Mendelian randomization with invalid instruments: Effect estimation
and bias detection through Egger regression. Int J Epidemiol.
44:512–525. 2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Yang C, Long C, Zhang Q, He D, Bi H and
Liu X: Mendelian randomization study of the relationship between
serum matrix metalloproteinases and the occurrence of sepsis. Int
Care Res. 3:215–220. 2023.
|
|
18
|
Sun BB, Maranville JC, Peters JE, Stacey
D, Staley JR, Blackshaw J, Burgess S, Jiang T, Paige E, Surendran
P, et al: Genomic atlas of the human plasma proteome. Nature.
558:73–79. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Swetter SM, Thompson JA, Albertini MR,
Barker CA, Baumgartner J, Boland G, Chmielowski B, DiMaio D, Durham
A, Fields RC, et al: NCCN Guidelines® Insights:
Melanoma: Cutaneous, Version 2.2021. J Natl Compr Canc Netw.
19:364–376. 2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Lou J, Cui S, Li J, Jin G, Fan Y and Huang
N: Causal relationship between the gut microbiome and basal cell
carcinoma, melanoma skin cancer, ease of skin tanning: Evidence
from three two-sample mendelian randomisation studies. Front
Immunol. 15(1279680)2024.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Chan II, Kwok MK and Schooling CM: The
total and direct effects of systolic and diastolic blood pressure
on cardiovascular disease and longevity using Mendelian
randomisation. Sci Rep. 11(21799)2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Agarwal I, Fuller ZL, Myers SR and
Przeworski M: Relating pathogenic loss-of-function mutations in
humans to their evolutionary fitness costs. Elife.
12(e83172)2023.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Burgess S and Thompson SG: Avoiding bias
from weak instruments in Mendelian randomization studies. Int J
Epidemiol. 40:755–764. 2011.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zhao G, Lu Z, Liao Y, Sun Y, Zhang Y, Kang
Z, Feng X, Sun J and Yue W: Association of intestinal
anti-inflammatory drug target genes with psychiatric Disorders: A
Mendelian randomization study. J Adv Res: S2090-S1232, 2024 doi:
10.1016/j.jare.2024.05.002 (Epub ahead of print).
|
|
25
|
Matarrese P, Ascione B, Ciarlo L, Vona R,
Leonetti C, Scarsella M, Mileo AM, Catricalà C, Paggi MG and
Malorni W: Cathepsin B inhibition interferes with metastatic
potential of human melanoma: An in vitro and in vivo study. Mol
Cancer. 9(207)2010.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Mijanović O, Branković A, Panin AN,
Savchuk S, Timashev P, Ulasov I and Lesniak MS: Cathepsin B: A
sellsword of cancer progression. Cancer Lett. 449:207–214.
2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Yadati T, Houben T, Bitorina A and
Shiri-Sverdlov R: The ins and outs of cathepsins: Physiological
function and role in disease management. Cells.
9(1679)2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Fröhlich E, Mack AF, Garbe C and Klessen
C: Distribution and colocalization of markers for proliferation,
invasion, motility and neoangiogenesis in benign melanocytic naevi
and malignant melanomas. Br J Dermatol. 153:1159–1165.
2005.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Frade R, Rodrigues-Lima F, Huang S, Xie K,
Guillaume N and Bar-Eli M: Procathepsin-L, a proteinase that
cleaves human C3 (the third component of complement), confers high
tumorigenic and metastatic properties to human melanoma cells.
Cancer Res. 58:2733–2736. 1998.PubMed/NCBI
|
|
30
|
Yuan L, Sheng L, He W, Zou C, Hu B, Liu J,
Ge W, Liu Y, Wang J and Ma E: Discovery of novel cathepsin
inhibitors with potent anti-metastatic effects in breast cancer
cells. Bioorg Chem. 81:672–680. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Gormley JA, Hegarty SM, O'Grady A,
Stevenson MR, Burden RE, Barrett HL, Scott CJ, Johnston JA, Wilson
RH, Kay EW, et al: The role of Cathepsin S as a marker of prognosis
and predictor of chemotherapy benefit in adjuvant CRC: A pilot
study. Br J Cancer. 105:1487–1494. 2011.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Yang Y, Lim SK, Choong LY, Lee H, Chen Y,
Chong PK, Ashktorab H, Wang TT, Salto-Tellez M, Yeoh KG and Lim YP:
Cathepsin S mediates gastric cancer cell migration and invasion via
a putative network of metastasis-associated proteins. J Proteome
Res. 9:4767–4778. 2010.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Gautam J, Bae YK and Kim JA: Up-regulation
of cathepsin S expression by HSP90 and 5-HT7 receptor-dependent
serotonin signaling correlates with triple negativity of human
breast cancer. Breast Cancer Res Treat. 161:29–40. 2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Small DM, Burden RE, Jaworski J, Hegarty
SM, Spence S, Burrows JF, McFarlane C, Kissenpfennig A, McCarthy
HO, Johnston JA, et al: Cathepsin S from both tumor and
tumor-associated cells promote cancer growth and
neovascularization. Int J Cancer. 133:2102–2112. 2013.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Arnlöv J: Cathepsin S as a biomarker:
Where are we now and what are the future challenges? Biomark Med.
6:9–11. 2012.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Wilkinson RDA, Burden RE, McDowell SH,
McArt DG, McQuaid S, Bingham V, Williams R, Cox ÓT, O'Connor R,
McCabe N, et al: A novel role for cathepsin S as a potential
biomarker in triple negative breast cancer. J Oncol.
2019(3980273)2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Sangster AB, Chang-McDonald B, Patel J,
Bockett N, Paterson E, Davis PF and Tan ST: Expression of
cathepsins B and D by cancer stem cells in head and neck metastatic
malignant melanoma. Melanoma Res. 31:426–438. 2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Kawakubo T, Okamoto K, Iwata J, Shin M,
Okamoto Y, Yasukochi A, Nakayama KI, Kadowaki T, Tsukuba T and
Yamamoto K: Cathepsin E prevents tumor growth and metastasis by
catalyzing the proteolytic release of soluble TRAIL from tumor cell
surface. Cancer Res. 67:10869–10878. 2007.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Smith-Byrne K, Hedman Å, Dimitriou M,
Desai T, Sokolov AV, Schioth HB, Koprulu M, Pietzner M, Langenberg
C, Atkins J, et al: Identifying therapeutic targets for cancer
among 2074 circulating proteins and risk of nine cancers. Nat
Commun. 15(3621)2024.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Versluis JM, Blankenstein SA, Dimitriadis
P, Wilmott JS, Elens R, Blokx WAM, van Houdt W, Menzies AM, Schrage
YM, Wouters MWJM, et al: Interferon-gamma signature as prognostic
and predictive marker in macroscopic stage III melanoma. J
Immunother Cancer. 12(e008125)2024.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Liu X, Feng F, Wang T, Qin J, Yin X, Meng
G, Yan C, Xing Z, Duan J, Liu C and Liu J: Laparoscopic
pancreaticoduodenectomy for metastatic pancreatic melanoma: A case
report. Medicine (Baltimore). 97(e12940)2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Quintanilla-Dieck MJ, Codriansky K, Keady
M, Bhawan J and Rünger TM: Cathepsin K in melanoma invasion. J
Invest Dermatol. 128:2281–2288. 2008.PubMed/NCBI View Article : Google Scholar
|