Introduction
Breast cancer (BC) is among the most common
malignancies affecting women and is expected to cause 42,250
related deaths and 310,000 new cases in the United states in
2024(1). Based on the expression
of the hormone receptors, progesterone receptor (PR), estrogen
receptor (ER) and human epidermal growth factor 2 (HER2),
currently, the most widely used and accepted approach for
categorizing BC is immunohistochemistry. Consequently, the four
most well-known subtypes of BC are luminal A, luminal B,
HER2-positive and triple-negative BC (TNBC) (2,3). The
TNBC subtype, an aggressive disease, accounts for 10 to 20% of all
BC cases (4). TNBC is
characterized by the absence of the expression of all three
receptors, high relapse rates, resistance to current endocrine
therapies and short survival rates (5); therefore, the treatment of this
subtype is challenging. TNBC arises from the complex interactions
between targets and factors (6).
This heterogeneity is associated with the reduced efficacy of
therapies, a decreased overall survival, and a more aggressive
disease course (5,7). Furthermore, ER and PR are present in
70% of luminal A malignancies, which also lack HER2, exhibit low
levels of the cell proliferation marker Ki-67 (<20%) and lack
HER2. In terms of clinical characteristics, these tumors are
low-grade, slow growing and are associated with the optimal
prognosis, a greater survival rate and a lower recurrence rate. In
addition, the treatment efficiency of the most prevalent subtype of
BC (ER+) has improved over the past few years following
the introduction of endocrine-targeted therapies, such as
tamoxifen. However, approximately one third of patients may suffer
from metastasis and relapse due to the development of resistance to
endocrine therapies (8,9).
Kinesin superfamily proteins (KIFs) are motor
proteins that hydrolyze adenosine triphosphate as an energy source
and mediate the intracellular transportation of macromolecules,
such as chromosomes, organelles, RNA, vesicles and proteins
(10). Kinesin family member
(KIF15), also known as kinesin 12, plays an essential role in cell
division (mitosis), cytoskeleton structure and the transportation
of cellular molecules. KIF15 is a microtubule-protein complex that
maintains the bipolar geometry of the mitotic spindle and
facilitates chromosome segregation in collaboration with KIF11
(11,12). Recently, multiple studies have
shown that KIF15 is highly expressed in various malignancies, such
as BC, gallbladder cancer and gastric cancer, suggesting that KIF15
is a biomarker that could be exploited therapeutically (13-16).
Over the past two decades, the development of
various selective kinesin spindle inhibitors has become an
attractive avenue (17). Several
studies have investigated the role of KIF15 in carcinogenesis. Wang
et al (18) reported that
KIF15 was overexpressed in patients with pancreatic cancer and that
KIF15 silencing was associated with a reduced proliferation both
in vitro and in vivo. Similarly, another research
group suggested that KIF15 is a candidate biomarker for glioma and
could be used as a therapeutic target (19). More importantly, KIF15 has been
identified as a risk factor for TNBC, and its downregulation by
lentiviral infection has been shown to be associated with the
induction of apoptosis (20).
Notably, scientists have identified 10 candidate kinesin
small-molecule chemical inhibitors for clinical evaluation, two of
which are the most promising KIF11 inhibitors: Filanesib (Arry-520)
and ispinesib (SB-715992) (21).
In addition, a recent study utilized the small-molecule KIF15
inhibitor, Kif15-IN-1, as a combination therapy with ispinesib in
gastric cancer cells. This combination was found to exert
synergistic effects both in vitro and in vivo
(12).
Previous molecular and functional analyses have
suggested that KIF15 motor proteins contribute to cell division
and, consequently, malignant growth (22). Therefore, targeted therapies are
warranted to provide effective treatments with increased efficacy
and low toxicity. The present study aimed to investigate the impact
of the KIF15 inhibitor, Kif15-IN-1, on the viability, apoptosis
induction, and migration of BC cell lines with different statuses
of ER expression.
Materials and methods
Cells, cell culture and drug
preparation
In the present study, two human breast
adenocarcinoma cell lines were used: MCF7, which expresses ER
(ER+) and MDA-MB231, which lacks ER (ER-) and
belongs to the TNBC subtype (23,24).
In addition, non-transformed rat embryo fibroblasts (REF) were used
as a normal control (25). The
MDA-MB231 (ATCC HTB-26) cells were kindly supplied by the Faculty
of Science, Baghdad University, and the MCF7 (ATCC HTB-22) cells
were purchased from ATCC. In addition, REF were kindly provided by
the Biotechnology Research Center at Al-Nahrain University. REF
were originally established by the Experimental Therapy Department
of the Iraqi Center for Cancer and Medical Genetic Research
(ICCMGR), which is affiliated with Mustansiriyah University in
Baghdad, Iraq. As described in a previous study by the authors
(26), the cells were cultured in
Roswell Park Memorial Institute-1640 medium (RPMI-1640)
supplemented with L-glutamine (Capricorn Scientific GmbH). To
create a complete medium, 1% penicillin/streptomycin solution
(100X; Euroclone S.p.A.) and 10% fetal bovine serum (FBS) were
added to the RPMI-1640 medium. The cells were kept in an incubator
with 5% CO2 and 95% humidity at 37˚C, as previously
described (27).
The potent KIF15 inhibitor, Kif15-IN-1, was
purchased from MedChemExpress (cat. no. HY-15948). For in
vitro assays, a stock solution of 20 mM was created by
dissolving it in 100% dimethyl sulfoxide (DMSO). The required
concentrations for subsequent experiments were prepared in complete
medium from the stock solution.
Cytotoxicity assay
Cytotoxic effects were assessed using an in
vitro 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium
bromide (MTT) assay (28,29). A total of 10,000 cells were seeded
in each well of a 96-well plate and incubated overnight to ensure
cell adhesion. MDA-MB231, MCF7 and REF cell lines were then exposed
to increasing concentrations of Kif15-IN-1 (10-100 µM) three
replicate wells were used for each treatment. Following incubation
(24, 48 h for MDA-MB231 and MCF7 and 24, 48 and 96 h for REF), the
medium was removed from the plate, and 20 µl MTT solution (5 mg/ml)
(Shanghai Macklin Biochemical Co., Ltd.) was added to each well and
incubated for 3 h at 37˚C in the dark. To dissolve the MTT, 50 µl
DMSO (Bio Basic Inc.) was added, followed by 10 min of shaking
(30). A microplate reader (BioTek
Instruments, Inc.) was used to measure the absorbance at 490 nm.
The following equation was used to determine the percentage of
viable cells from the raw absorbance data:
where ‘A’ represents the absorbance. The
dose-response curve was generated using GraphPad Prism software
version 6 (Dotmatics), and the growth inhibitory concentration that
reduces viability by 50% (GI50) was determined via the
same curve.
Morphological analysis
The present study investigated the effects of
Kif15-IN-1 treatment on cell morphology. The cell lines (MDA-MB231,
MCF7 and REF), in a 24-well plate, were seeded at a seeding density
of 26x103 cells per well. The cell lines were treated
with their relevant GI50 doses of Kif15-IN-1 for 24 h at
37˚C in 5% CO2 and 95% humidified air. The cells were
then analyzed at a magnification of x200 using an inverted
microscope (Meiji Techno). Subsequently, images were captured using
digital camera (Canon, Inc.), as previously described (31).
Scratch assay
The MDA-MB231 cells were seeded in 24-well plates
under the same conditions used in the morphological study. Although
serum starvation is necessary for scratch assay experiments, serum
starvation has been shown to significantly influence the migration,
proliferation and migration-associated genes of MDA-MB-231 breast
cancer cells (32). Moreover,
serum deprivation has been demonstrated to affect the cell cycle
(33,34), which may subsequently influence the
efficacy of the Kif15 inhibitor. Therefore, the present study
utilized complete medium (containing 10% FBS) to reduce the
possible effects of serum deprivation. In addition, we aimed to
perform this test in more physiological settings. Therefore, the
MDA-MB231 cells were cultured using complete medium (10% FBS) for
24 h until they became nearly confluent, and then a scratch was
made using a pipette tip in the middle of the well before washing
with PBS. Fresh complete medium (10% FBS) was replaced with the
relevant wells containing DMSO (Control) or Kif15-IN-1 (58 µM).
Images were captured using an inverted microscope (MEIJI, Japan)
(x100 magnification) with a digital camera (Canon, Japan) before
the plate was returned to a humidified, warm incubator (37˚C).
Following incubation for 48 h, images were obtained. In addition,
ImageJ™ software version 1.46r was used for the analysis
of wound healing images.
Apoptosis detection by fluorescence
microscopy
As described in a previous study by the authors
(31), apoptosis analysis was
performed after 25x103 MDA-MB231 cells were exposed to
the control (DMSO) or Kif15-IN-1 (58 µM) for 48 h in a 24-well
plate. The cells were then trypsinized, rinsed with PBS, and
collected in 1.5 ml tubes (Eppendorf Germany). A total of 9 µl of
the cell suspension was mixed with 1 µl of acridine orange-ethidium
bromide (AO/EB) staining dye [100 µg/ml AO and 100 µg/ml EB (Fluka
Chemie GmbH)]. Subsequently, 100 cells were scored for different
stages of apoptosis using a fluorescence microscope (Human
Diagnostics), and images of randomly selected representative fields
were obtained.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the MDA-MB231 and MCF7
cells using TRIzol™ reagent 2023 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. The
ProtoScript® First Strand complementary DNA (cDNA)
Synthesis kit (E6300S) from New England Biolabs was used according
to the vendor's instructions, and cDNA was obtained by the reverse
transcription of total RNA. This assay is highly specific for
double-stranded DNA (dsDNA) over RNA. To measure the mRNA levels,
Luna Universal qPCR Master Mix (M3003S) from New England Biolabs
(SYBR®/FAM channel) was used. In addition, the
thermocycling conditions were as follows: 30 sec of denaturation at
94˚C, 40 cycles of denaturation at 94˚C for 5 sec, 15 sec of
annealing at 58˚C, and 10 sec of extension at 72˚C. KIF15
gene expression was measured in the treated and untreated cells
using RT-qPCR and relative cycle threshold (2-ΔΔCq)
methodology (35). GAPDH
was used as the internal control (housekeeping gene). The primers
used were as follows: KIF15 forward,
5'-CTGCCTGGGCCAAGTGATTA-3' and reverse, 5'-CGGGATTCCTTGTGGAGCTT-3';
and GAPDH forward, 5'-GGTGTGAACCATGAGAAGTATGA-3' and
reverse, 5'-GAGTCCTTCCACGATACCAAAG-3'.
Statistical analysis
The data were analyzed using GraphPad Prism software
(version 6.0.0; Dotmatics) and Microsoft Excel 2019. The majority
of the experiments were conducted at least twice. The unpaired
t-test was used to compare two means. The data were analyzed
according to the standard error of the mean (SEM). A P-value
<0.05 was considered to indicate a statistically significant
difference.
Results
Kif15-IN-1 inhibits BC cell
proliferation in a concentration-dependent manner regardless of the
ER status
The present study investigated the cytotoxic effects
of a KIF15 inhibitor (Kif15-IN-1) on BC cells. The MDA-MB231
(ER-) and MCF7 (ER+) cell lines were treated
with increasing concentrations of Kif15-IN-1 for 24 and 48 h
(Fig. 1A and B). In addition, a normal cell line (REF)
was also incubated with Kif15-IN-1 for 24, 48 and 96 h.
Subsequently, cell viability was evaluated using an MTT assay. Of
note, the MDA-MB231 cells exposed for 48 h were more sensitive to
Kif15-IN-1 (mean GI50=57.95±3.99 µM) than were the cells
incubated for 24 h (mean GI50=85.94±4.75 µM) (Fig. 1A). The MCF7 cells were less
sensitive following incubation for 24 h (GI50=93.17±3.82
µM), but were more sensitive after 48 h (mean
GI50=61.9±0.8 µM) (Fig.
1B). Notably, the REF were resistant (GI50 >100
µM) to Kif15-IN-1 at the different time points examined (Fig. 1C).
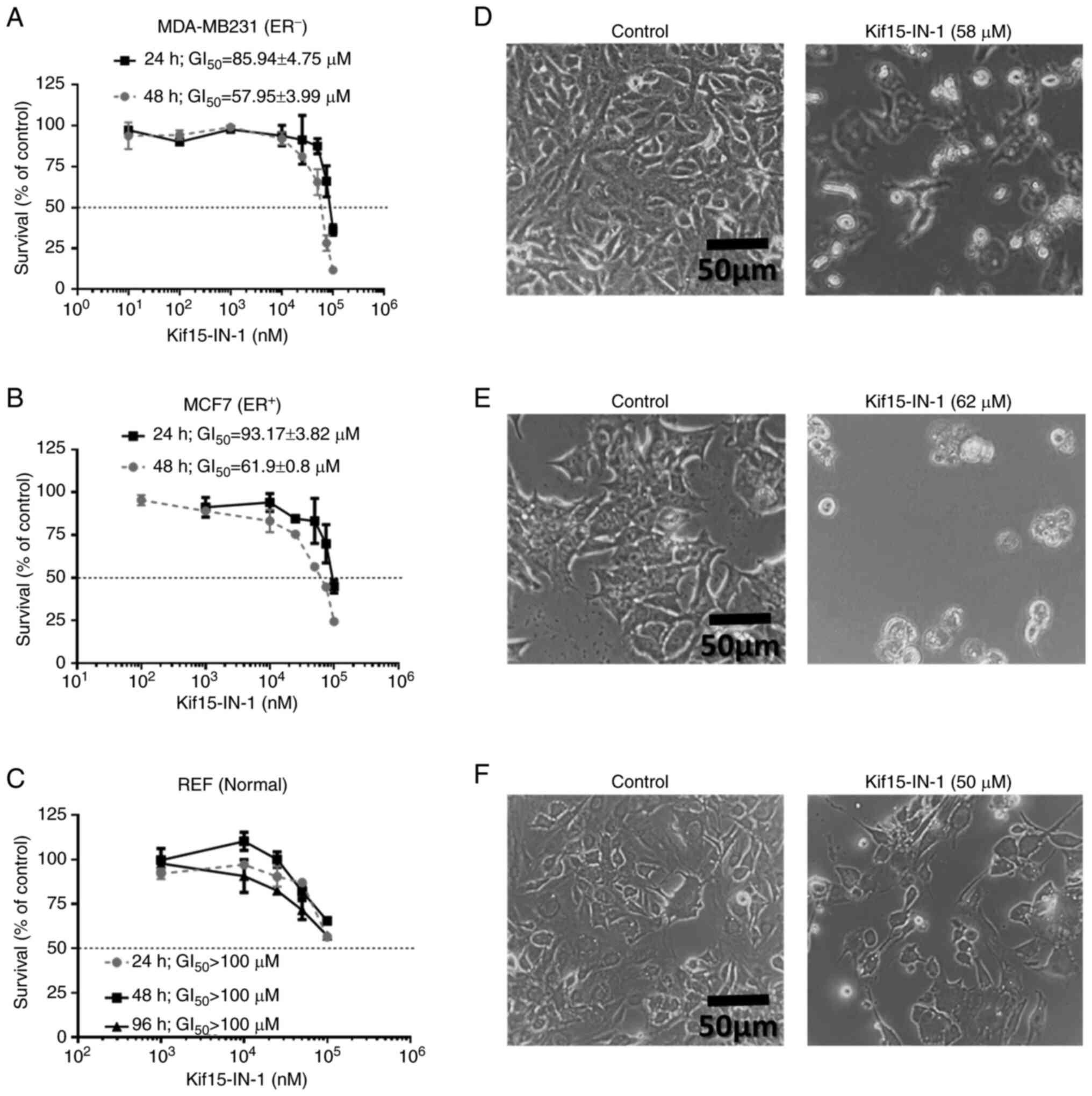 | Figure 1Kif15-IN-1 inhibits the proliferative
activity and alters the morphology of BC cells. (A) MDA-MB231, (B)
MCF7, and (C) REF were exposed to increasing concentrations of
Kif15-IN-1 for the indicated durations; only the BC cell lines were
relatively sensitive to 48 h of incubation. Compared with those of
the control cells, the data are presented as the mean percentage
survival ± SEM, and at least two replicates were performed with
triplicate measurements, which produced the findings. The
GI50 values are indicated. In addition, (D) MDA-MB231,
(E) MCF7, and (F) REF were incubated with the indicated treatments
for 24 h, and images of morphological alterations were obtained
using an inverted microscope at x200 magnification; the images are
representative of two replicates. BC, breast cancer; KIF15, kinesin
family member 15; REF, rat embryo fibroblasts; GI50,
growth inhibitory concentration that reduces viability by 50%. |
Kif15-IN-1 treatment alters the
morphology and suppresses the migratory potential of BC cells
To further examine the effects of Kif15-IN-1, both
MDA-MB231 and MCF7 cells were exposed to their relevant
GI50 concentrations, and after 24 h, the morphology of
the cells was examined under an inverted microscope. Of note, in
the MDA-MB231 cells, the impact on viability was associated with a
decrease in cell number, and the size of the treated cells was
greater than that of the control cells. In addition, some cells
floated in the medium, which may indicate that these cells were
dead (Fig. 1D). However, in the
MCF7 cells, the treatment was associated with a decreased cell
number and size; all treated cells were rounded compared with those
in the control group (Fig. 1E).
Notably, no obvious morphological changes were observed in the REF
treated with Kif15-IN-1 compared with the untreated cells (Fig. 1F).
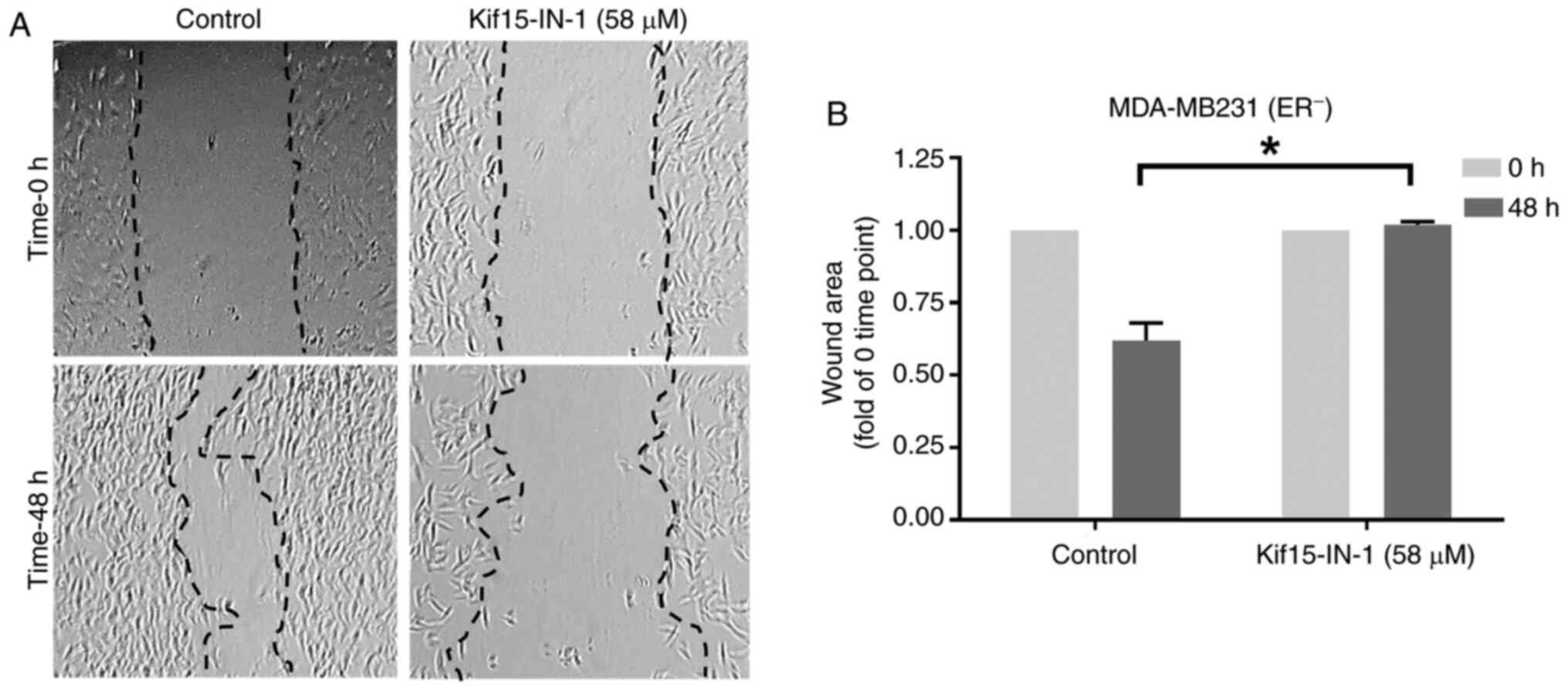
Additionally, the effects of Kif15-IN-1 on the
migratory potential of MDA-MB231 cells were examined using a
scratch assay (Fig. 2). As
illustrated in Fig. 2A, there was
less wound healing in the Kif15-IN-1-treated cells than in the
control-treated cells, and the bar chart indicates a significant
decrease of ~39% (P=0.011) after 48 h of exposure (Fig. 2B). Of note, small gaps are present
in the cells treated with the KIF15 inhibitor in the wound healing
image at 48 h. As indicated in Fig.
3, Kif15-IN-1 promotes apoptosis. This may explain the tiny
gaps observed in the cell monolayer of the Kif15-IN-1-treated cells
(Fig. 2A).
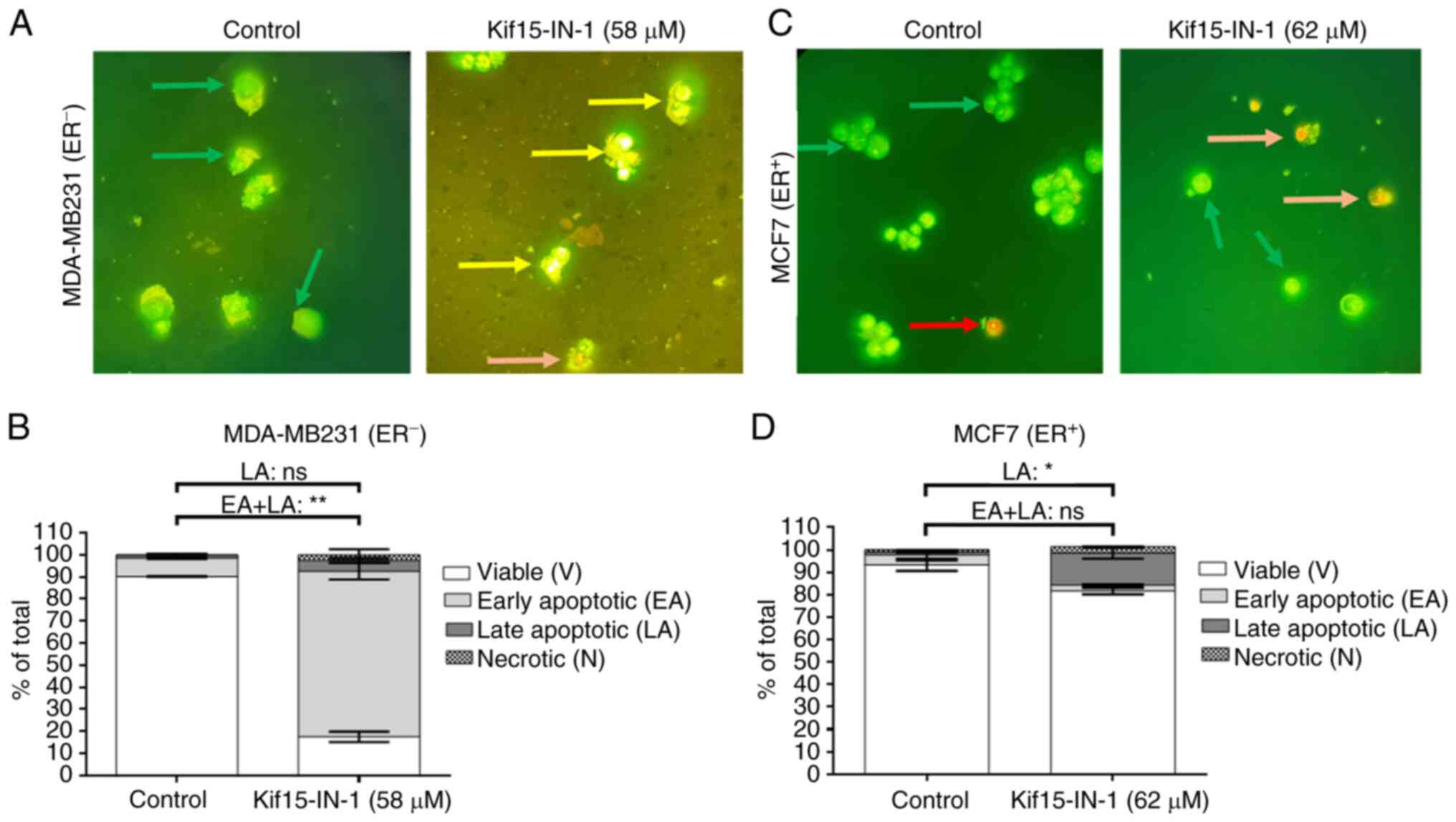
Kif15-IN-1 induces BC cell apoptosis
and significantly reduces the expression of KIF15
To better understand the effects of Kif15-IN-1, the
induction of the apoptosis of MDA-MB231 and MCF7 cells was
estimated using AO/EB staining to confirm the cytotoxic ability of
Kif15-IN-1. BC cells were co-cultured with the indicated treatments
for 48 h before apoptosis was detected (Fig. 3). As shown in Fig. 3A and B, the majority of the MDA-MB231 cells
underwent early apoptosis (~75%), and ~80% of the cells treated
with the GI50 dose of Kif15-IN-1 were apoptotic (early
and late apoptosis). In addition, the counts of MCF7 cells treated
with Kif15-IN-1 at the GI50 dose revealed that ~15% of
the cells were apoptotic at the late stage; however, no significant
difference in the total number of apoptotic cells (early and late
apoptosis) was observed (Fig. 3C
and D).
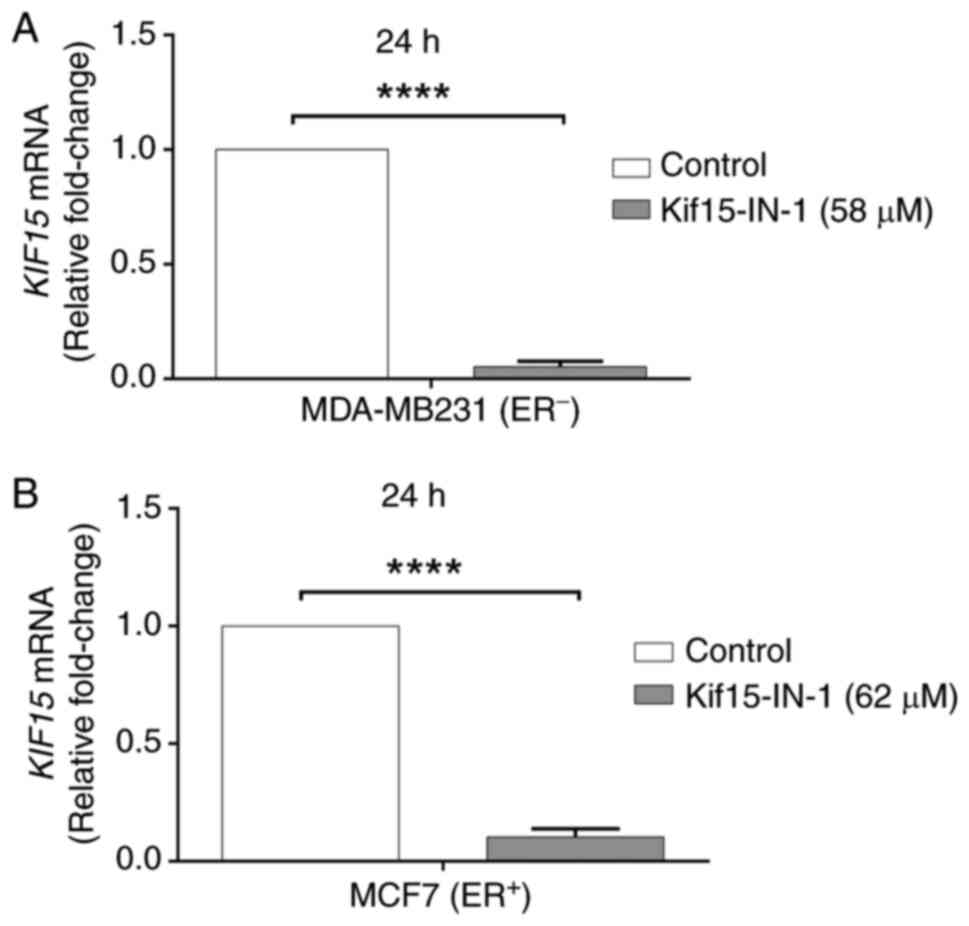
In addition, RT-qPCR was used to investigate the
effects of Kif15-IN-1 on KIF15 mRNA levels. Notably, it was
found that, compared with the control, Kif15-IN-1 significantly
reduced the expression of the KIF15 gene in both the
MDA-MB-231 (18.8-fold) (Fig. 4A)
and MCF7 (9.7-fold) cells after 24 h (Fig. 4B).
Discussion
BC is the leading cause of cancer-related mortality
among women worldwide; several subtypes of BC, such as TNBC, are
challenging to treat (36). In
addition, the emergence of resistance to current therapeutics is a
matter of concern; therefore, novel therapies targeting aberrant
cellular mechanisms are warranted to overcome resistance and
improve treatment efficacy. Multiple studies have emphasized the
role of mitotic motor proteins in the carcinogenesis of various
malignancies, including BC (16,18,20,37).
Of note, recent reports have investigated the function and
structure of the motor kinesin protein KIF15 in several tumors,
suggesting that this protein could be exploited therapeutically
(12,15,22).
Recent studies have shown that KIF15 expression is
upregulated in patients with BC and is abnormal in accelerating
cell cycle progression (13,20).
In addition, Alfarsi et al (38) reported that KIF18A was highly
expressed in >2,000 patients with ER+ BC and that it
was a significant predictor of a poor response to endocrine
treatment. Of note, multiple attempts to downregulate KIF15 protein
expression via RNA interference techniques in different types of
cancer have yielded promising results in vitro and in
vivo (16,20). However, despite the potential
therapeutic advantages of siRNAs, several issues have made it
difficult to employ siRNA-based antiviral treatments, including
off-target effects, siRNA instability, poor long-term protein
expression, drug resistance and immunological reactions (39). Therefore, the present study aimed
to explore the impact of a small-molecule KIF15 inhibitor
(Kif15-IN-1) on BC cell lines derived from different subtypes
(ER+ and TNBC) in an attempt to identify a
chemical-targeted therapy that may enhance the treatment efficacy
of existing therapies for BC and overcome resistance.
To explore the effects of KIF15 inhibition in BC
cells, MDA-MB231 and MCF7 cells were treated with Kif15-IN-1
monotherapy in vitro. Interestingly, viability was
inhibited, and the KIF15 inhibitor was cytotoxic in both cell
lines, regardless of mutational status and subtype. Considering
that KIF15 is highly expressed in BC cells (13,20),
it was hypothesized that Kif15-IN-1 would inhibit the target
protein KIF15, as evidenced by the significant downregulation of
KIF15 in both cell lines (MDA-MB231 and MCF7). Additionally,
the normal cells used, REF, were resistant to Kif15-IN-1, unlike
the BC cell lines, which further supports the assumption of target
(KIF15) inhibition. The downregulation of the KIF15 gene
expression may be attributed to the off-target effect of the KIF15
inhibitor or the mechanism of a negative feedback network between
protein and gene expression through the binding of proteins to RNA
regulatory motifs of mRNAs (40).
In addition, cytotoxicity is associated with the
induction of programmed cell death and a decreased migratory
potential. Of note, a similar effect was previously observed
following KIF15 knockdown in BC (13,20),
gallbladder cancer (15),
pancreatic cancer (18) and
Burkitt lymphoma (41).
Additionally, bioinformatics and experimental studies have revealed
that KIF15 knockdown induces programmed cell death in
ovarian cancer by triggering the interaction of several pathways
(42). Additionally, recent
research in prostate cancer cells has revealed that the knockdown
of KIF15 was associated with the inhibition of proliferation
and migration potential from one side and the induction of
programmed cell death by decreasing the activity of the PI3K/Akt
pathway (43). Another recent
study revealed that KIF15 may modulate apoptotic pathways to reduce
the production of seven anti-apoptotic proteins in gastric cancer
(16).
KIF15 plays a vital role in clustering microtubules
into bundles to mediate the movement of cells (44); therefore, the suppression of
migration is a consequence of KIF15 inhibition. In addition,
PI3K/Akt activity is associated with the regulation of cell
movement (43,45). Notably, in addition to the
cytotoxic effect, herein, the microscopic examination of BC cells
via both inverted light and fluorescence microscopy revealed
greater enlargement in cells treated with Kif15-IN-1 than in the
control-treated cells. This result may indicate that KIF15
inhibition is associated with the inhibition of both chromosomal
segregation and cell cycle progression (20,46).
KIF15 is vital in driving centrosome separation and promotes
bipolar spindle assembly during cell division. In addition, the
change in cell shape may support the assumption of KIF15 protein
inhibition, as KIF15 is involved in microtubule generation and
maintaining cell shape and the skeleton (47).
Of note, Dumas et al (48) assumed that KIF15 may partially
compensate for the effect of mitotic spindle inhibitors (e.g., Eg5
inhibitors) and thereby induce resistance. Therefore, to overcome
this resistance, researchers (49,50)
have explored the addition of KIF15 inhibitors to mitotic spindle
inhibitors and obtained successful combinations. However, further
experimentation is required for a more in-depth understanding of
the effect mechanisms, the primary limitation of which is the lack
of financial support and instrumentation.
In conclusion, in the present study, KIF15
expression was found to be upregulated in BC. Of note, the present
study revealed that the small-molecule inhibitor, Kif15-IN-1, was
cytotoxic to ER+ and TNBC BC cell lines. This resulted
in a significant decrease in the ability to migrate, along with
morphological alterations and the activation of apoptosis. The
effects were linked mechanistically to the downregulation of the
KIF15 gene. Therefore, further in vivo studies are
warranted to confirm these findings, and exploring the combination
of Kif15-IN-1 with existing chemotherapies in BC is advisable.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets generated during the current study are
available from the corresponding author upon reasonable
request.
Authors' contributions
AHA designed and executed the experiments (MTT, cell
culture, apoptosis analysis, scratch, morphological assays and
statistical analysis). AHA was also involved in the
conceptualization of the study, in material collection, in the
writing, reviewing and editing of the original draft of the
manuscript, in study supervision, and in project administration.
SGA was involved in the conceptualization of the study, in the
study methodology, in the validation of the RT-qPCR results, in
data curation, and in study supervision. SGA was also involved in
the writing of the original draft of the manuscript and in material
collection. SIAJ was involved in the conceptualization and design
of the study, study execution, and in the analysis of the RT-qPCR
results. SIAJ was also involved in the writing of the original
draft of the manuscript and in material collection. SAM maintained
the cells and participated in the experiments (MTT, cell culture,
apoptosis, scratch, and morphological assays and material
collection). AHA and SGA confirm the authenticity of all the raw
data. All the authors have read and approved the final version of
the manuscript for publication.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Giaquinto AN and Jemal A:
Cancer statistics, 2024. CA Cancer J Clin. 74:12–49.
2024.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Al-Juboori SI, Vadakekolathu J, Idri S,
Wagner S, Zafeiris D, Pearson JR, Almshayakhchi R, Caraglia M,
Desiderio V, Miles AK, et al: PYK2 promotes HER2-positive breast
cancer invasion. J Exp Clin Cancer Res. 38(210)2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Łukasiewicz S, Czeczelewski M, Forma A,
Baj J, Sitarz R and Stanisławek A: Breast cancer-epidemiology, risk
factors, classification, prognostic markers, and current treatment
Strategies-an updated review. Cancers (Basel).
13(4287)2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Larkin L: Breast cancer genetics and risk
assessment: An overview for the clinician. Climacteric. 26:229–234.
2023.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Park JH, Jonas SF, Bataillon G,
Criscitiello C, Salgado R, Loi S, Viale G, Lee HJ, Dieci MV, Kim
SB, et al: Prognostic value of tumor-infiltrating lymphocytes in
patients with early-stage triple-negative breast cancers (TNBC) who
did not receive adjuvant chemotherapy. Ann Oncol. 30:1941–1949.
2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Aine M, Boyaci C, Hartman J, Häkkinen J,
Mitra S, Campos AB, Nimeus E, Ehinger A, Vallon-Christersson J,
Borg Å, et al: Molecular analyses of triple-negative breast cancer
in the young and elderly. Breast Cancer Res. 23(20)2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Lehmann BD, Pietenpol JA and Tan AR:
Triple-negative breast cancer: Molecular subtypes and new targets
for therapy. Am Soc Clin Oncol Educ Book. e31–e39. 2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zboril EK, Grible JM, Boyd DC, Hairr NS,
Leftwich TJ, Esquivel MF, Duong AK, Turner SA, Ferreira-Gonzalez A,
Olex AL, et al: Stratification of tamoxifen synergistic
combinations for the treatment of ER+ breast cancer. Cancers
(Basel). 15(3179)2023.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Höller A, Nguyen-Sträuli BD,
Frauchiger-Heuer H and Ring A: ‘Diagnostic and prognostic
biomarkers of luminal breast cancer: Where are We Now?’. Breast
Cancer (Dove Med Press). 15:525–540. 2023.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Hirokawa N, Noda Y, Tanaka Y and Niwa S:
Kinesin superfamily motor proteins and intracellular transport. Nat
Rev Mol Cell Biol. 10:682–696. 2009.PubMed/NCBI View
Article : Google Scholar
|
|
11
|
Miki H, Okada Y and Hirokawa N: Analysis
of the kinesin superfamily: Insights into structure and function.
Trends Cell Biol. 15:467–476. 2005.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Sun RF, He N, Zhang GY, Yu ZY, Li LS, Ma
ZJ and Jiao ZY: Combined inhibition of KIF11 and KIF15 as an
effective therapeutic strategy for gastric cancer. Curr Cancer Drug
Targets. 23:293–306. 2023.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zeng H, Li T, Zhai D, Bi J, Kuang X, Lu S,
Shan Z and Lin Y: ZNF367-induced transcriptional activation of
KIF15 accelerates the progression of breast cancer. Int J Biol Sci.
16:2084–2093. 2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Gao X, Zhu L, Lu X, Wang Y, Li R and Jiang
G: KIF15 contributes to cell proliferation and migration in breast
cancer. Hum Cell. 33:1218–1228. 2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wang J, Wang D, Fei Z, Feng D, Zhang B,
Gao P, Hu G, Li W, Huang X, Chen D, et al: KIF15 knockdown
suppresses gallbladder cancer development. Eur J Cell Biol.
100(151182)2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ding L, Li B, Yu X, Li Z, Li X, Dang S, Lv
Q, Wei J, Sun H, Chen H, et al: KIF15 facilitates gastric cancer
via enhancing proliferation, inhibiting apoptosis, and predict poor
prognosis. Cancer Cell Int. 20(125)2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Al Kufi SGJH, Emmerson J, Rosenqvist H,
Garcia CMM, Rios-Szwed DO and Wiese M: Absence of DEATH kinesin is
fatal for Leishmania mexicana amastigotes. Sci Rep.
12(3266)2022.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wang J, Guo X, Xie C and Jiang J: KIF15
promotes pancreatic cancer proliferation via the MEK-ERK signalling
pathway. Br J Cancer. 117:245–255. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Wang Q, Han B, Huang W, Qi C and Liu F:
Identification of KIF15 as a potential therapeutic target and
prognostic factor for glioma. Oncol Rep. 43:1035–1044.
2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Sheng J, Li C, Dong M and Jiang K:
Identification by comprehensive bioinformatics analysis of KIF15 as
a candidate risk gene for Triple-negative breast cancer. Cancer
Manag Res. 12:12337–12348. 2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Shahin R and Aljamal S: Kinesin spindle
protein inhibitors in cancer: From high throughput screening to
novel therapeutic strategies. Future Sci OA.
8(FSO778)2022.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Gibbs B, Douglas J, Wates R, McDonald P,
Whitaker A, Ndi C, Pathak H, Harned L, Neuenswander S, Broward M,
et al: Abstract 5334: Targeting the KIF15-TPX2 PPI to overcome
KIF11 inhibitor resistance in epithelial ovarian cancer. Cancer
Res. 83:5334. 2023.
|
|
23
|
Holliday DL and Speirs V: Choosing the
right cell line for breast cancer research. Breast Cancer Res.
13(215)2011.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Fedele P, Orlando L and Cinieri S:
Targeting triple negative breast cancer with histone deacetylase
inhibitors. Expert Opin Investig Drugs. 26:1199–1206.
2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Fathi SM, Alhammer AH and Ali IA: Testing
the cytotoxic potential of biosynthesized nanoparticles using
Conocarpus erectus Leaves against human breast cancer cells. AIP
Conf Proc. 2922(040008)2024.
|
|
26
|
Zaki NH, Ali AM, Al-Rubaiee GH and
Alhammer AH: Malaysian Anti-bacterial and Anti-tumoral activities
of spirulina platensis extracellular extract. J Med Health Sci.
18:11–16. 2022.
|
|
27
|
Ali SM, Al-Karam LQ and Alhammer AJN: In
Vitro cancer cells therapy with nano-gold depending on its optical
properties. NeuroQuantology. 20:57–61. 2022.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Lafta FM, Mohammed RK, Alhammer AH and
Ahmed ME: Cytotoxic potential of Neem (Azadirachta indica A. Juss)
Oil. Tropical J Natural Product Res. 7:5436–5440. 2023.
|
|
29
|
Alhammer AH, Al-juboori SI and Mudhafar
SA: APR-246 enhances the anticancer effect of doxorubicin against
p53-mutant AsPC-1 pancreatic cancer cells. Baghdad Sci J.
21:2551–2560. 2024.
|
|
30
|
Nashaan FA, Al-Rawi MS, Alhammer AH, Rabie
AM and Tomma JH: Synthesis, characterization, and cytotoxic
activity of some imides from galloyl hydrazide. Eurasian Chem
Commun. 4:966–975. 2022.
|
|
31
|
Mohammed I, Alhammer AH and Arif IS: The
p53 reactivator PRIMA-1MET synergises with
5-fluorouracil to induce apoptosis in pancreatic cancer cells.
Invest New Drugs. 41:587–595. 2023.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Ahmadiankia N, Bagheri M and Fazli MJAB:
Differential migration-related gene expression and altered cytokine
secretion in response to serum starvation in cultured MDA-MB-231
cells. Asian Biomed. 13:123–129. 2019.
|
|
33
|
Chen M, Huang J, Yang X, Liu B, Zhang W,
Huang L, Deng F, Ma J, Bai Y, Lu R, et al: Serum starvation induced
cell cycle synchronization facilitates human somatic cells
Reprogramming. PLoS One. 7(e28203)2012.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Aghababazadeh M and Kerachian MA: Cell
fasting: Cellular response and application of serum starvation. J
Fasting Health. 2:147–150. 2014.
|
|
35
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Lei S, Zheng R, Zhang S, Wang S, Chen R,
Sun K, Zeng H, Zhou J and Wei W: Global patterns of breast cancer
incidence and mortality: A population-based cancer registry data
analysis from 2000 to 2020. Cancer Commun (Lond). 41:1183–1194.
2021.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Qiao Y, Chen J, Ma C, Liu Y, Li P, Wang Y,
Hou L and Liu Z: Increased KIF15 expression predicts a poor
prognosis in patients with lung adenocarcinoma. Cell Physiol
Biochem. 51:1–10. 2018.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Alfarsi LH, Elansari R, Toss MS,
Diez-Rodriguez M, Nolan CC, Ellis IO, Rakha EA and Green AR:
Kinesin family member-18A (KIF18A) is a predictive biomarker of
poor benefit from endocrine therapy in early ER+ breast cancer.
Breast Cancer Res Treat. 173:93–102. 2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Kang H, Ga YJ, Kim SH, Cho YH, Kim JW, Kim
C and Yeh JY: Small interfering RNA (siRNA)-based therapeutic
applications against viruses: Principles, potential, and
challenges. J Biomed Sci. 30(88)2023.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Stapleton JA, Endo K, Fujita Y, Hayashi K,
Takinoue M, Saito H and Inoue T: Feedback control of protein
expression in mammalian cells by tunable synthetic translational
inhibition. ACS Synth Biol. 1:83–88. 2012.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Wang Z, Chen M, Fang X, Hong H, Yao Y and
Huang H: KIF15 is involved in development and progression of
Burkitt lymphoma. Cancer Cell Int. 21(261)2021.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Sun X, Chen M, Liao B and Liang Z:
Knockdown of KIF15 promotes cell apoptosis by activating crosstalk
of multiple pathways in ovarian cancer: Bioinformatic and
experimental analysis. Int J Clin Exp Pathol. 14:267–291.
2021.PubMed/NCBI
|
|
43
|
Bi H, Hou X, Shen Q, Liu Z, Zhu X, Ma L
and Lu J: Knockdown of KIF15 suppresses proliferation of prostate
cancer cells and induces apoptosis through PI3K/Akt signaling
pathway. Cell Death Discov. 9(326)2023.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Mann BJ, Balchand SK and Wadsworth P:
Regulation of Kif15 localization and motility by the C-terminus of
TPX2 and microtubule dynamics. Mol Biol Cell. 28:65–75.
2017.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Xue G and Hemmings BA: PKB/Akt-dependent
regulation of cell motility. J Natl Cancer Inst. 105:393–404.
2013.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Malaby HLH, Dumas ME, Ohi R and Stumpff J:
Kinesin-binding protein ensures accurate chromosome segregation by
buffering KIF18A and KIF15. J Cell Biol. 218:1218–1234.
2019.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Klejnot M, Falnikar A, Ulaganathan V,
Cross RA, Baas PW and Kozielski F: The crystal structure and
biochemical characterization of Kif15: A bifunctional molecular
motor involved in bipolar spindle formation and neuronal
development. Acta Crystallogr D Biol Crystallogr. 70:123–133.
2014.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Dumas ME, Chen GY, Kendrick ND, Xu G,
Larsen SD, Jana S, Waterson AG, Bauer JA, Hancock W, Sulikowski GA
and Ohi R: Dual inhibition of Kif15 by oxindole and
quinazolinedione chemical probes. Bioorg Med Chem Lett. 29:148–154.
2019.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Solon AL, Zaniewski TM, O'Brien P, Clasby
M, Hancock WO and Ohi R: Synergy between inhibitors of two mitotic
spindle assembly motors undermines an adaptive response. Mol Biol
Cell. 33(ar132)2022.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Milic B, Chakraborty A, Han K, Bassik MC
and Block SM: KIF15 nanomechanics and kinesin inhibitors, with
implications for cancer chemotherapeutics. Proc Natl Acad Sci USA.
115:E4613–E4622. 2018.PubMed/NCBI View Article : Google Scholar
|