Introduction
Internal control genes are necessary for the
accurate determination of gene expression using techniques such as
quantitative real-time polymerase chain reaction (PCR).
High-throughput real-time reverse transcriptase (RT)-PCR, with its
outstanding sensitivity and accuracy, has rendered the selection of
a housekeeping (HK) gene used as the internal standard for the
estimation and comparison of mRNA levels even more important
(1–3). Previous studies showed that the
expression levels of commonly used HK genes are different in
various tissues or between normal and diseased tissues (1,4–9).
It has been argued that since the expression levels
of various HK genes differ depending on the tissue type or
experimental conditions, more than one or even several various
control genes should be assessed in parallel for certain tissues
(1,3,10–12).
In colorectal cancer (CRC), downregulation of the HK gene β-2
microglobulin (B2M) has been confirmed using real-time RT-PCR
(13,14).
In a previous study, we examined the expression of
toll-like receptors 2 (TLR2) and 4 (TLR4) in sporadic human CRC
tissue (15). TLRs are known to be
involved in innate immunity and to play an important role in immune
surveillance (16–20). In this study, we used a commercially
available kit for analysis of the preferred internal control genes
and selected three reference genes: β-glucuronidase (GUS), β-actin
(BA) and B2M. Using these three internal control genes, the mRNA
expression levels of TLR2 and TLR4 in cancerous and non-cancerous
colorectal tissue specimens were quantified using TaqMan real-time
PCR and compared. Based on the results, we discuss the validity of
each of the internal control genes in terms of accurate analysis of
gene expression in CRC tissue under the present experimental
conditions.
Materials and methods
Tissue specimens and internal control
genes
Surgical specimens of colorectal tissue were
obtained from 50 CRC patients at the Toho University Sakura Medical
Center (Sakura, Japan) as previously described (15). Written informed consent was obtained
from the subjects according to the terms of the Declaration of
Helsinki. Non-cancerous tissue, located proximal to the tumor and
macroscopically free of disease, was obtained immediately after
surgery. The characteristics of the 50 subjects were previously
described (15) and there was no
bias in the sample population.
The 50 cancerous specimens were grouped according to
the histopathological stage (pStage I, II, III and IV), based on
the tumor-node-metastasis (TMN) classification of the International
Union against Cancer (UICC). Differences in TLR2 and TLR4
expression in each of the pStages and non-cancerous tissues were
examined.
To select the optimal internal control gene, the
TaqMan® Human Endogenous Control Plate (Applied
Biosystems, Inc., Foster City, CA, USA) was used, which contained
TaqMan primers and probes for 11 commonly used HK genes and one
internal positive control sequence. Non-cancerous and cancerous
tissue specimens obtained from CRC patients were used as the test
specimens (8 samples from 4 patients) to determine the most
suitable internal reference genes. One of the non-cancerous tissue
specimens was used to calibrate the plate.
This study was approved by the Ethics Committee of
the Toho University Sakura Medical Center.
Real-time PCR
RNA was extracted using an RNeasy® Plus
Mini kit (Qiagen, Hilden, Germany) and cDNA was synthesized using
an AffinityScript® QPCR cDNA synthesis kit (Stratagene,
La Jolla, CA, USA), according to the manufacturer's instructions,
as previously described (15).
Quantitative real-time PCR was performed using a
Stratagene Mx3000P® QPCR System with TaqMan® Gene
Expression Master mix. The thermal profile consisted of precycle
heat activation at 95°C for 10 min, followed by 95°C for 15 sec and
then 60°C for 60 sec for a total of 40 cycles. Data were expressed
as fold-induction relative to the RNA amplified at the lowest
level. Relative quantification of total gene product in each sample
was performed using the comparative CT (ΔΔCT) method.
The primers and probes used in this study can be
found using the assay IDs: Hs00610101_ml for TLR2, Hs00152939_ml
for TLR4, and Hs99999903_ml for BA (http://products.appliedbiosystems.com/ab/en/US/adirect/ab).
Statistical analysis
Data were expressed as the means ± standard
deviation (SD). Mean values were compared in groups using the
unpaired Student's t-test with two-tailed P-values using StatMate
III (ATMS Co., Ltd., Tokyo, Japan). P<0.05 was considered to
indicate statistically significant difference.
Results
Detection of the indicated target genes
to compare gene expression between sample
The effect of several potential internal control
genes on relative expression levels was assessed using the
TaqMan® Human Endogenous Control Plate, according to the
manufacturer's instructions. The three HK genes selected as
internal control genes, GUS, BA and B2M, exhibited minimal
variability at expression levels, based on the total threshold
cycle, in samples under the experimental conditions used (Fig. 1). Relative quantification of gene
expression was performed using each gene individually and
together.
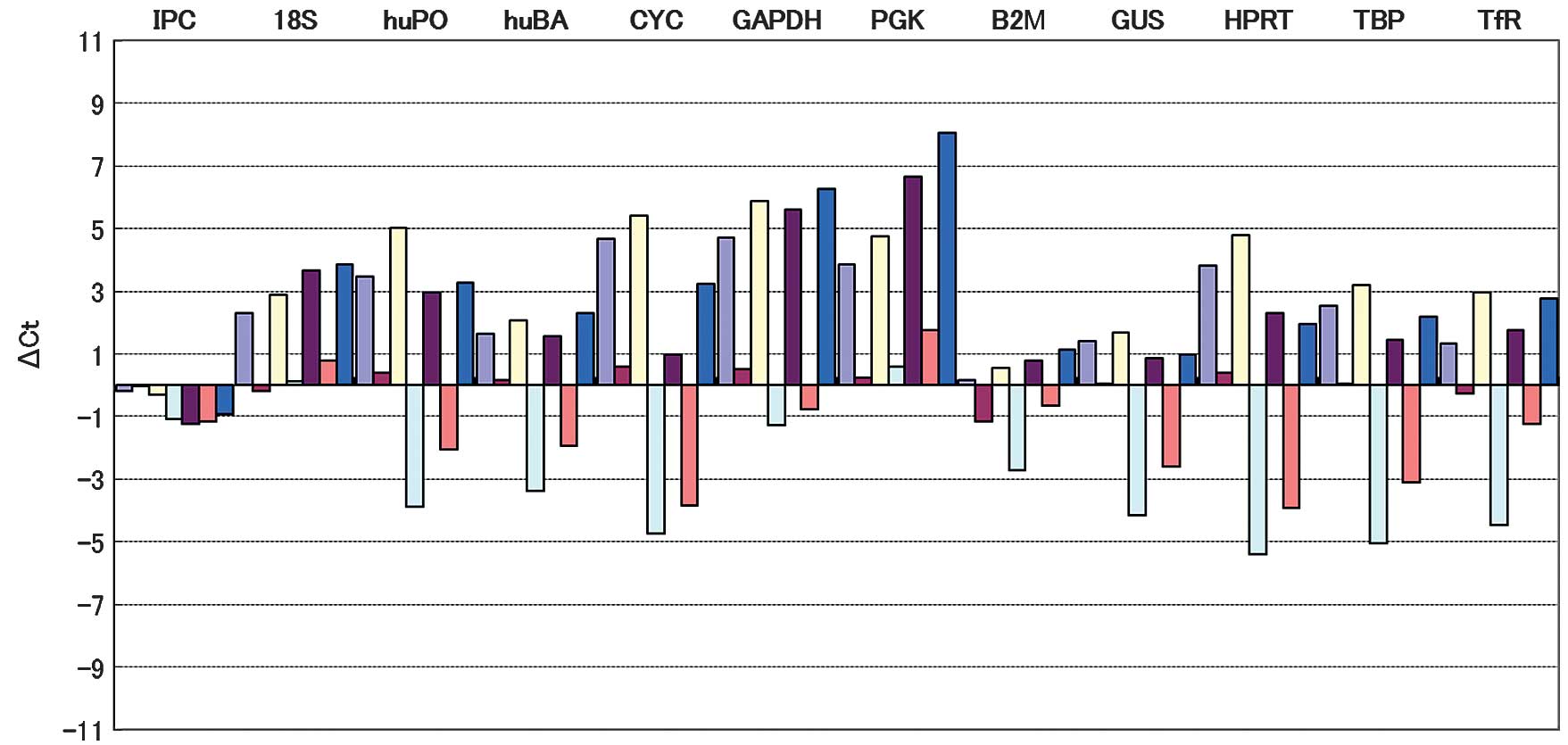 | Figure 1Columns represent individual samples
analyzed using TaqMan primers and probes for the detection of the
indicated target gene in order to compare gene expression between
samples. Less fluctuation in amplification indicated less variable
gene expression. The three HK genes used as internal reference
genes (GUS, BA and B2M) were selected based on these results. IPC,
internal positive control; 18S, 18S rRNA; huPO, acidic ribosomal
protein; huBA, β-actin; CYC, cyclophilin; GAPDH,
glyceraldehyde-3-phosphate dehydrogenase; PGK,
phosphoglycerokinase; B2M, β-2 microglobulin; GUS, β-glucronidase;
HPRT, hypoxanthine ribosyl transferase; TBP, transcription factor
IID, TATA binding protein; TfR, transferrin receptor. |
TLR2 and TLR4 expression in cancerous and
non-cancerous tissues
TLR4 expression in non-cancerous and cancerous
tissues was confirmed in all the 50 subjects using real-time PCR.
TLR4 mRNA expression was significantly higher in cancerous compared
to non-cancerous tissues when B2M was used as the internal control
gene (P<0.001). No significant difference was observed in TLR4
expression between non-cancerous and cancerous tissues when GUS and
BA were used as the control genes (Table I). TLR2 mRNA expression was also
confirmed in the 50 cancerous and in 49 of the non-cancerous tissue
specimens using real-time PCR. TLR2 expression was significantly
higher in cancerous compared to non-cancerous tissues when analyzed
using each of the three internal control genes (P<0.001)
(Table I).
 | Table ITLR2 and TLR4 expression in cancerous
and non-cancerous tissues (mean ± SD). |
Table I
TLR2 and TLR4 expression in cancerous
and non-cancerous tissues (mean ± SD).
| BA | GUS | B2M |
|---|
|
|
|
|---|
| Receptor | NCT | CT | NCT | CT | NCT | CT |
|---|
| TLR2 | 2.61±1.82 | 6.60±4.15b | 5.08±4.44 | 13.61±12.26a | 5.14±3.01 | 30.84±22.61b |
| TLR4 | 1.61±1.04 | 2.17±2.22 | 3.24±3.40 | 2.72±2.37 | 2.92±21.13 | 5.22±3.23b |
TLR2 and TLR4 expression according to
histological stage
TLR2 and TLR4 expression was compared between
cancerous and non-cancerous tissues at various pStages. TLR4
expression was significantly higher in pStage I and II cancerous
compared to non-cancerous tissues when B2M was used as an internal
control gene, while no statistically significant differences were
detected in pStage II and IV cancerous tissues (Table II). TLR2 expression was
significantly higher in pStage I, II and III cancerous compared to
non-cancerous tissues with the three control genes (pStage I,
P<0.05 for the control genes; pStage II, P<0.05 with GUS and
P<0.01 with B2M and BA; pStage III, P<0.05 with GUS and BA
and P<0.001 with B2M). In pStage IV tissues, significant
differences in TLR2 expression were observed only with GUS and BA
(P<0.01) (Table II). TLR2
expression did not differ significantly in non-cancerous and
cancerous tissues in pStage IV with B2M as the control gene.
 | Table IITLR2 and TLR4 expression at various
histological stages (mean ± SD). |
Table II
TLR2 and TLR4 expression at various
histological stages (mean ± SD).
| BA | GUS | B2M |
|---|
|
|
|
|---|
| Receptor
(pStage) | NCT | CT | NCT | CT | NCT | CT |
|---|
| TLR2 (I) | 1.76±0.96 | 7.02±4.55b | 2.70±1.20 | 8.84±6.13a | 4.02±0.15 | 11.21±5.64a |
| TLR4 (I) | 1.14±0.43 | 2.10±1.41 | 1.15±0.11 | 1.31±0.13 | 1.30±0.64 | 5.43±3.17a |
| TLR2 (II) | 2.89±2.05 | 10.90±8.61b | 5.10±4.35 | 18.02±14.85a | 5.32±3.40 | 52.84±58.88b |
| TLR4 (II) | 1.11±0.55 | 1.82±1.74 | 2.47±2.63 | 2.03±1.37 | 4.16±4.37 | 4.69±3.37 |
| TLR2 (III) | 4.02±3.19 | 7.73±5.55b | 5.07±5.40 | 12.44±9.71a | 6.11±2.83 | 34.17±19.96c |
| TLR4 (III) | 1.72±0.78 | 1.33±1.09 | 4.09±3.08 | 2.86±2.50 | 3.79±2.00 | 6.80±4.00a |
| TLR2 (IV) | 1.77±1.06 | 5.59±1.94b | 1.50±0.72 | 5.94±2.18b | 3.94±1.83 | 16.64±10.22 |
| TLR4 (IV) | 1.67±1.04 | 3.50±3.33 | 2.65±1.63 | 3.92±3.17 | 2.86±1.81 | 4.63±0.03 |
Discussion
Common HK genes, such as GAPDH and BA are
traditionally used as internal controls for the assessment of gene
expression using techniques such as quantitative real-time PCR
(12). However, previous studies
have indicated that the expression levels of HK genes may be
different in various tissues or between normal and diseased tissues
(2,4). Shrout et al (21) and Bianchini et al (13) reported that low levels of B2M in CRC
tissue might serve as a prognostic indicator in CRC patients with
lymph node metastasis. Vandesompele et al (1) have recommended the use of at least
three appropriate control genes for the calculation of a
normalization factor in view of the inherent variation in the
expression of HK genes.
Bustin (22)
proposed the use of the TaqMan® Human Endogenous Control
Plate for normalizing mRNA levels in tissue culture cells using HK
genes. Other groups have similarly emphasized the convenience of
this system for initial screening to select an appropriate internal
reference gene (11,23). In the present study, three HK genes
(GUS, B2M and BA) were selected as the internal control genes.
These genes exhibited a relatively stable expression under the
present conditions.
In the majority of cases, consistent results were
obtained when GUS and BA were used as the internal control genes.
However, TLR4 expression was higher in cancerous compared to
non-cancerous tissues, when B2M was used as the internal control
gene. These results may be associated with the low level of B2M
expression in cancerous tissues and are consistent with the
findings of Shrout et al (21) and Bianchini et al (13).
Acknowledgements
This study was supported in part by a
grant from the Cancer Research Funds for Patients and Family of the
Medical treatment and Welfare Network Chiba, Japan.
References
|
1
|
Vandesompele J, De Preter K, Pattyn F,
Poppe B, Van Roy N, De Paepe A and Speleman F: Accurate
normalization of real-time quantitative RT-PCR data by genometric
averaging of multiple internal control genes. Genome Biol.
3:RESEARCH0034. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bas A, Forsberg G, Hammarström S and
Hammarström ML: Utility of the housekeeping genes 18S rRNA, β-actin
and glyceraldehyde-3-phosphate-dehydrogenase for normalization in
real-time quantitative reverse transcriptase-polymerase chain
reaction analysis of gene expression in human T lymphocytes. Scand
J Immunol. 59:566–573. 2004.
|
|
3
|
Radonić A, Thulke S, Mackay IM, Landt O,
Siegert W and Nitsche A: Guideline to reference gene selection for
quantitative real-time PCR. Biochem Biophys Res Commun.
313:856–862. 2004.PubMed/NCBI
|
|
4
|
Barber RD, Harmer DW, Coleman RA and Clark
BJ: GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression
in a panel of 72 human tissues. Physiol Genomics. 21:389–395. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
van der Woude CJ, Moshage H, Homan M,
Klebeuker JH, Jansen PLM and van Dekken H: Expression of apotosis
related proteins during malignant progression in chronic ulcerative
colitis. J Clin Pathol. 58:811–814. 2005.PubMed/NCBI
|
|
6
|
Ernst PB, Takaishi H and Crowe SE:
Helicobacter pylori infection as a model for
gastrointestinal immunity and chronic inflammatory disease. Dig
Dis. 19:104–111. 2001. View Article : Google Scholar
|
|
7
|
Itzkowitz SH and Yio X: Inflammation and
cancer IV. Colorectal cancer in inflammatory bowel disease: the
role of inflammation. Am J Physiol Gastrointest Liver Physiol.
287:G7–G17. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rutter M, Saunders B, Wilkinson K, Rumbles
S, Schofield G, Kamm M, Williams C, Price A, Talbot I and Forbes A:
Severity of inflammation is a risk factor for colorectal neoplasma
in ulcerative colitis. Gastroenterology. 126:451–459. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cario E and Podolsky DK: Differential
alternation in intestinal epithelial cell expression of Toll-like
receptor 3 (TLR3) and TLR4 in inflammatory bowel disease. Infect
Immun. 68:7010–7017. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schmid H, Cohen CD, Henger A, Irrgang S,
Schlondorff D and Kretzler M: Validation of endogenous controls for
expression analysis in microdissected human renal biopsies. Kidney
Int. 64:356–360. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
de Kok JB, Roelofs RW, Giesendorf BA,
Pennings JL, Waas ET, Feuth T and Swinkels DW: Normalization of
gene expression measurements in tumor tissues: comparison of 13
endogenous control genes. Lab Invest. 85:154–159. 2005.PubMed/NCBI
|
|
12
|
Ohl F, Jung M, Xu C, Stephan C, Rabien A,
Burkhardt M, Nitsche A, Kristiansen G, Loening SA, Radonić A and
Jung K: Gene expression studies in prostate cancer tissue: which
reference gene should be selected for normalization? J Mol Med
(Berl). 83:1014–1024. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bianchini M, Levy E, Zucchini C, Pinski V,
Macagno C, De Sanctis P, Valvassori L, Carinci P and Mordoh J:
Comparative study of gene expression by cDNA microarray in human
colorectal cancer tissues and normal mucosa. Int J Oncol. 29:83–94.
2006.PubMed/NCBI
|
|
14
|
Takeuchi O, Hoshino K, Kawai T, Sanjo H,
Takada H, Ogawa T, Takeda K and Akira S: Differential roles of TLR2
and TLR4 in recognition of Gram-negative and Gram-positive
bacterial cell wall components. Immunity. 11:443–451. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nihon-Yanagi Y, Terai K, Murano T,
Matsumoto T and Okazumi S: Tissue expression of Toll-like receptors
2 and 4 in sporadic human colorectal cancer. Cancer Immunol
Immunother. 61:71–77. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ortega-Cava CF, Ishihara S, Rumi MAK,
Kawashima K, Ishimura N, Kazumori H, Udagawa J, Kadowaki Y and
Kinoshita Y: Strategic compartmentalization of Toll-like receptor 4
in the mouse gut. J Immunol. 170:3977–3985. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Aderem A and Ulevitch RJ: Toll-like
receptors in the induction of the innate immune response. Nature.
406:782–787. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Janeway CA Jr and Medzhitov R: Innate
immune recognition. Annu Rev Immunol. 20:197–216. 2002. View Article : Google Scholar
|
|
19
|
Akira S and Hemmi H: Recognition of
pathogen-associated molecular patterns by TLR family. Immunol Lett.
85:85–95. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Miyake K: Innate immune sensing of
pathogens and danger signals by cell surface Toll-like receptors.
Semin Immunol. 19:3–10. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shrout J, Yousefzadeh M, Dodd A, Kirven K,
Blum C, Graham A, Benjamin K, Hoda R, Krishana Romano M, Wallace M,
Garrett-Mayer E and Mitas M: β-2 microglobulin mRNA expression
levels are prognostic for lymph node metastasis in colorectal
cancer patients. Br J Cancer. 98:1999–2005. 2008.
|
|
22
|
Bustin SA: Quantification of mRNA using
real-time reverse transcription PCR (RT-PCR): trends and problems.
J Mol Endocrinol. 29:23–39. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dheda K, Huggett JF, Bustin SA, Johnson
MA, Rook G and Zumla A: Validation of housekeeping genes for
normalizing RNA expression in real-time PCR. Biotechniques.
37:112–119. 2004.PubMed/NCBI
|















