Introduction
Ovarian cancer is the fifth most lethal cancer
affecting women, and accounts for the majority of mortalitites from
gynaecological cancer (1). Epithelial
ovarian cancer (EOC) accounts for 90% of all cases of ovarian
cancer and is typically a ‘silent killer’ because patients do not
present with symptoms until advanced disease is present. As a
result, 70% of patients have stage III/IV (International Federation
of Gynaecology and Obstetrics-FIGO staging) advanced EOC at the
time of diagnosis, resulting in a 5-year survival <30% (2). By contrast, if EOC is diagnosed early, at
stage I, the 5-year survival rate is 90%. This high survival rate
with early disease diagnosis highlights the need for an accurate
test to detect ovarian cancer. Screening with CA 125 and/or pelvic
ultrasound has been extensively studied but has not been shown to
improve mortality, due to poor sensitivity and specificity for
ovarian cancer (3). Novel biomarkers
are therefore required to enhance screening strategies and improve
survival rates.
MicroRNAs (miRs) are small non-coding RNAs that are
essential for regulating gene expression. They play important roles
in the regulation of a wide variety of genes by binding to the
3′-untranslated regions of the target mRNA and are involved in the
pathogenesis of various types of cancer (4). The fact that miRs circulate in the blood
where they may reflect gene expression in a distant tumour, has led
to enthusiasm for the role of circulating miRs as biomarkers for
cancer detection and prognosis. Multiple miRs appear to be
dysregulated in EOC tissue but it is unclear whether circulating
miRs can be used to identify women with EOC compared to
controls.
We selected a panel of five miRs that have been
shown to be dysregulated in ovarian cancer tissue. The miR-200
family (miR-200a, miR-200b and miR-200c) plays a key role in
initiating cancer development and invasion by regulating the
transformation of epithelial to mesenchymal cells (5). Hypoxia-regulated miRs are also involved
in cancer progression, modulating the cell response to increasing
hypoxia as the tumour outgrows its vascular supply (6). Both miR-21 and miR-210 are consistently
upregulated in response to hypoxia and therefore may be upregulated
in EOC compared to controls. In the present study, we examined the
expression of circulating miRs in blood and tissue in EOC and
benign ovarian masses to determine whether they are potential
biomarkers for EOC.
Materials and methods
Study subjects
All the subjects were recruited from the Mercy
Hospital for Women, a tertiary referral hospital in Australia in
2013–2014. Written informed consent was obtained prior to inclusion
and the present study was approved by the Mercy Health Human
Research and Ethics Committee, Melbourne, Australia no. R11/34.
Whole blood was collected prior to surgery in women
suspected of EOC. Biopsies of ovarian tissue were collected
intraoperatively and processed immediately. Cases (n=8) were
confirmed after a multidisciplinary meeting and full
histopathological diagnosis of type, grade and FIGO stage of
ovarian cancer was confirmed. Controls (n=5) were aged-matched but
found to have benign ovarian masses surgically and
histopathologically. Patients with a history of previous or
recurrent cancer, as well as non-EOC cases were excluded.
Sample collection
Peripheral whole blood samples (2.5 ml) were
collected in PAXgene blood RNA tubes (PreAnalytix, Hombrechtikon,
Switzerland). As per the manufacturer's instructions, the samples
were stored at room temperature for 24–72 h, transferred to −20°C
for 24 h, and then stored at −80°C until processing.
Ovarian tissue biopsies were collected
intraoperatively and immediately transferred into RNALater (Qiagen,
Hilden, Germany) and snap frozen in liquid nitrogen. They were then
stored at −80°C until processing.
miRNA preparation
miRNA was extracted from peripheral whole blood
using the PAXgene Blood miRNA kit (PreAnalytix) and automated
QiaCube (Qiagen) protocol according to the manufacturer's
instructions. Genomic DNA was removed using DNase treatment, and
total RNA was eluted and stored at −80°C if not used immediately.
The concentration of total RNA was quantified by NanoDrop 1000
(Nanodrop Technologies; Thermo Fisher Scientific, Waltham, MA,
USA).
miRNA was extracted from ovarian tissue using the
RNeasy Kit (Qiagen). In brief, ovarian tissue was homogenized in
buffer. The lysate was centrifuged at 8,000 × g for 3 min, and the
supernatant added to 350 µl of 70% ethanol. The sample was then run
through the RNeasy spin column and centrifuged with buffers as per
protocol. The final RNA was eluted into 30 µl of RNase free water
and stored at −80°C, if not used immediately.
RT-PCR
Specific miRNA Taqman assays (Applied Biosystems
Life Technologies, Foster City, CA, USA) were used for the
quantification of the miRNAs according to the manufacturer's
instructions. To assess miR-200a, miR-200b and miR-200c the
High-Capacity cDNA Reverse Transcription Kit was used. Total RNA
(150 ng) was reverse transcribed for each target miRNA and the
housekeeping miRNA miR-191 was used for normalization. To assess
miR-21 and miR-210 the TaqMan MicroRNA Reverse Transcription kit
was used (with a cDNA conversion and preamplification step) for the
two miRs (miR-21 and miR-210) and the housekeeping miRNA RNU-48.
Following cDNA generation qPCR was performed in duplicate, with
multiple negative controls, using TaqMan Universal PCR Master Mix
II on a CFX 384 (Bio-Rad, Berkeley, CA, USA) with the following
cycling conditions: 95°C for 10 min, and 40 cycles of 95°C for 15
sec, and 60°C for 1 min. Fold-changes in expression were determined
by the comparative CT method normalized against the mean expression
of the housekeeping gene.
Statistical analysis
Data were statistically analyzed using R version
2.12.0 (http://www.r-project.org) or GraphPad
Prism v 5 (GraphPad Software, Inc., La Jolla, CA, USA). Differences
in RT-PCR gene expression were assessed using the Mann-Whitney U or
Kruskal-Wallis test, or where the data were considered normally
distributed, the t-test or ANOVA. Patient characteristics were
compared using χ2 where appropriate. Data were presented
as mean ± standard deviation. P<0.05 was considered to indicate
a statistically significant difference.
Results
Clinical characteristics
Clinical characteristics for the EOC cases and
controls are presented in Table I. The
EOC patients had predominately (>80%) high-grade serous
carcinoma of the ovary, 50% of whom had stage I/II and 50% had
stage III/IV at the time of diagnosis.
 | Table I.Clinical characteristics of EOC cases
and benign controls. |
Table I.
Clinical characteristics of EOC cases
and benign controls.
| Characteristics | Case | Control |
|---|
| Age (years) | 56.3 | 63.5 |
| Benign cystadenoma
(n, %) | 0 | 5 (100) |
| FIGO stage (n,
%) |
| 0 |
| I–II | 4 (50) |
|
|
III–IV | 4 (50) |
|
| Grade (n, %) |
| 0 |
| Low | 0 |
|
| High | 8 (100) |
|
Circulating miRNA expression in
EOC
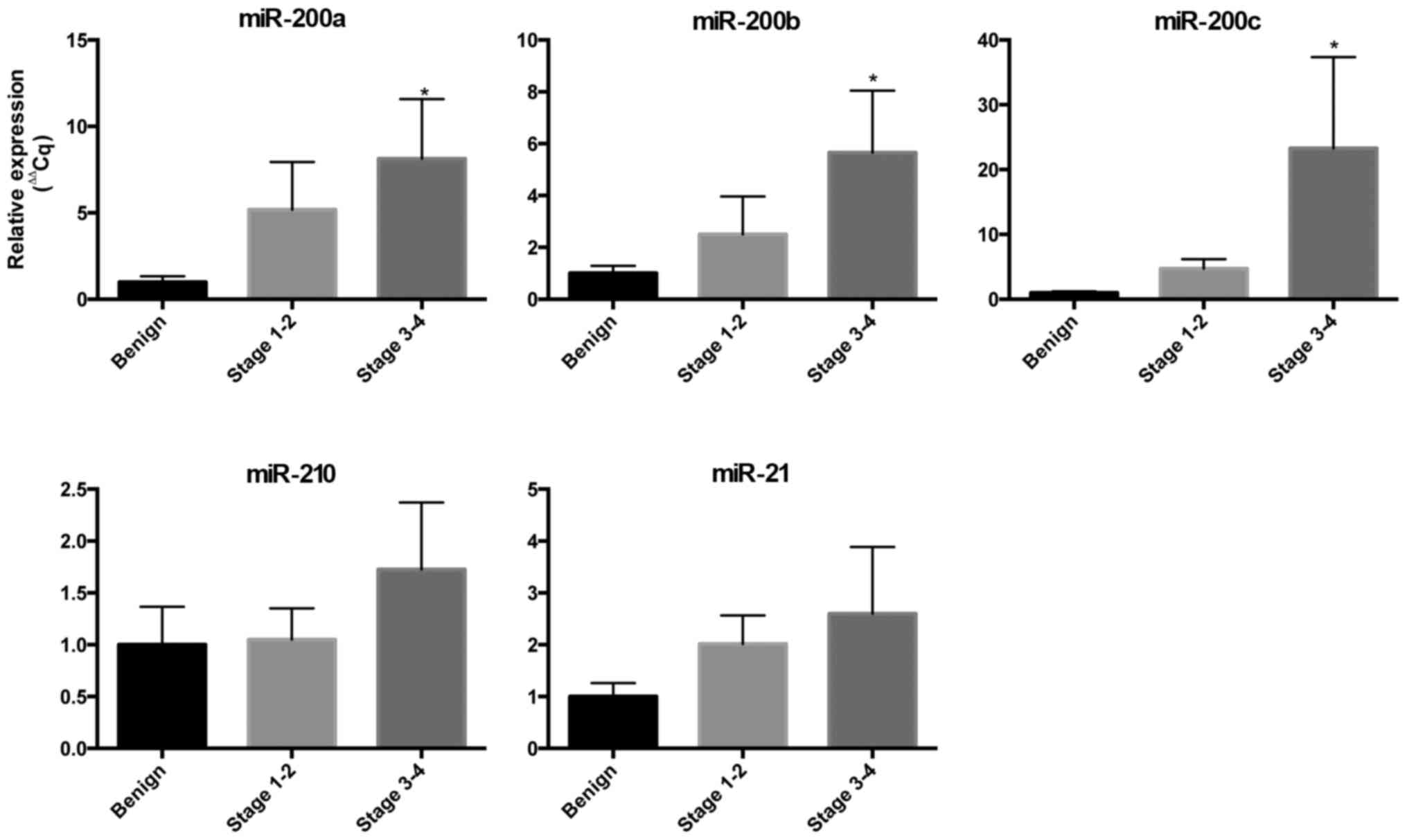
We examined the expression of miRs in the
circulation to determine their potential to be blood-based
biomarkers for EOC (Fig. 1). We found
an increased expression of miR-200a, miR-200b and miR-200c in the
blood in cases with EOC compared to controls and the expression
increased as the stage of EOC increased. The circulating expression
of miR-200a in stage I/II EOC was 5-fold greater than the controls
and 8-fold greater for stage III/IV (P<0.05). The circulating
expression of miR-200b in stage I/II EOC was 2.5-fold greater than
controls and 5.6-fold greater for stage II/IV (P<0.05). The
circulating expression of miR-200c in stage I/II EOC was 4.7-fold
greater than controls and 23.3-fold greater for stage II/IV
(P<0.05). The expression of circulating miR-21 and miR-210 was
increased in EOC but did not meet statistical significance in this
small cohort.
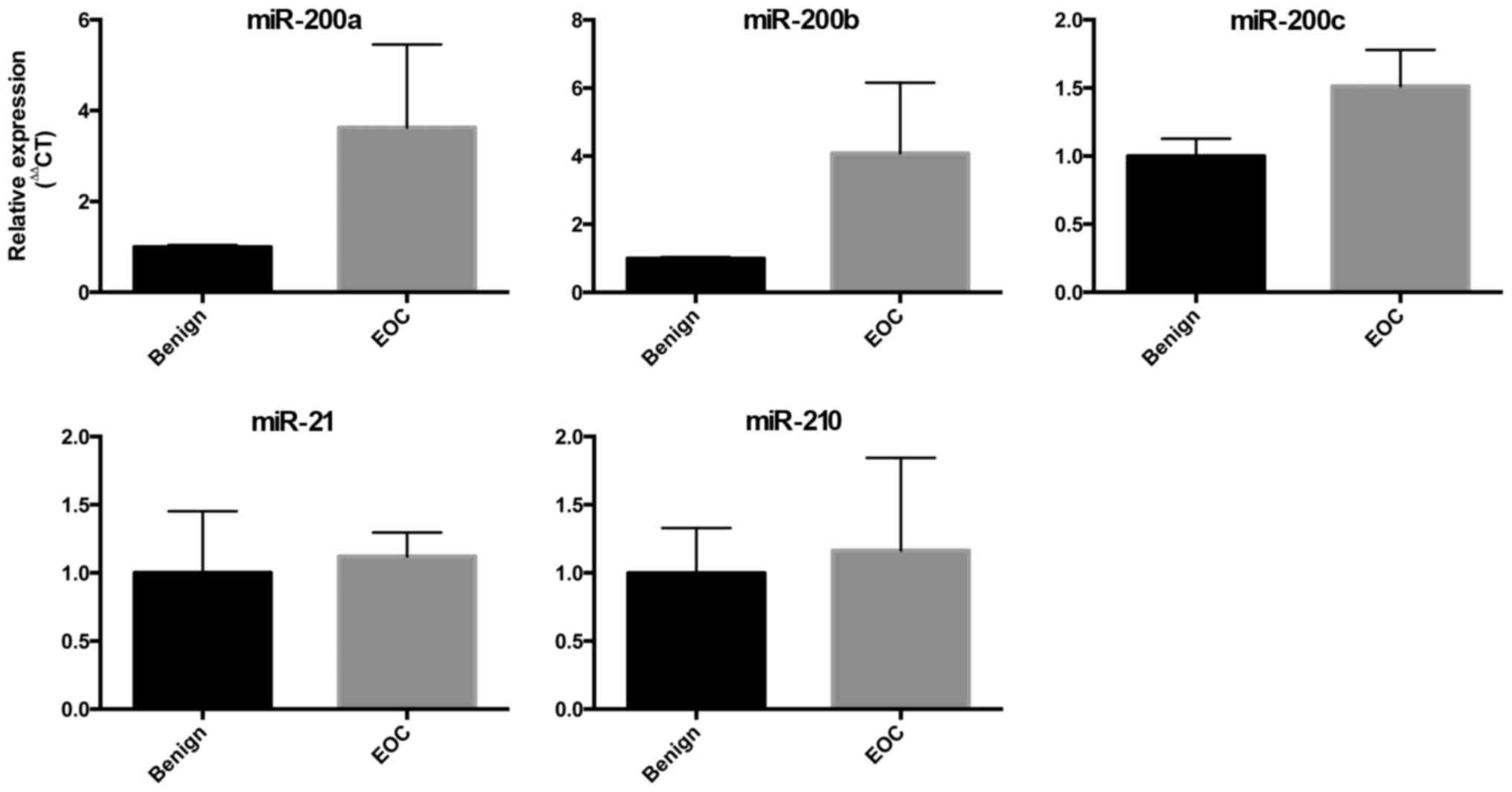
miRNA expression in EOC tissue
We also compared the expression of the miRs in
ovarian tissue collected intraoperatively from women with benign
ovarian masses and women with EOC. The expression of the miR-200
family showed a trend towards increased expression in ovarian
tissue from women with EOC, but this did not meet statistical
significance. There were no differences in the expression of
miR-210 or miR-21 in this small cohort (Fig. 2).
Discussion
Early diagnosis of EOC is dependent on the
development of a reliable biomarker that can discriminate between
benign and malignant pelvic tumours. To the best of our knowledge,
currently, no screening modality for EOC exists. miRs are
aberrantly expressed in a number of cancers, including EOC where
they regulate tumour growth (7).
Circulating miRs are stable in the blood and reflect changes in the
distant tumour allowing a ‘liquid biopsy’ of the tumour to be
non-invasively obtained.
In the present study, we examined the expression of
five miRs that are involved in the pathogenesis of EOC, although,
to the best of our knowledge, their expression in blood has not
been investigated or consistently reproduced in previous studies.
We found significant upregulation of the miR-200 family in EOC
compared to benign ovarian tumours. The miR-200 family likely plays
a key role in carcinogenesis. They are critical regulators of the
epithelial-mesenchymal transition (7).
This process describes the transformation of non-motile epithelial
to motile mesenchymal cells, which therefore have the potential to
invade surrounding tissue and metastasize to distant sites. The
miR-200s target two E-box binding transcription factors, ZEB1 and
ZEB2, which regulate E-cadherin expression and cellular polarity
(7). Increased ovarian tissue
expression of miR-200a and miR-200c have been associated with a
reduced overall survival in EOC patients, which may be attributed
to alterations in response to chemotherapeutics with high levels of
miR-200s (8,9). As a result, the miR-200 family may not
only be used to detect and stage EOC but may provide information
regarding prognosis.
miR-210 has been shown to be increased in EOC tissue
and cell lines in response to tissue hypoxia; however, little is
known with regard to circulating miR-210 in EOC (10). miR-21 has been identified as an
important oncogene in multiple types of cancer and has been shown
to be increased in EOC tissue; nevertheless, little is known
regarding circulating miR-21 in EOC (5,11). In this
pilot study of a limited number of patients, we saw a trend towards
increased miR-210 with advanced EOC, but which did not meet
statistical significance. The potential of miR-210 and miR-21 as
useful EOC biomarkers requires further investigation in a larger
cohort study.
Circulating miRs may be detected in whole blood,
plasma or serum. To the best of our knowledge, this is the first
study to describe their expression in EOC in whole blood collected
prior to initial surgery (12). Whole
blood collected directly into tubes containing RNA stabilizer
(PAXgene tubes) ensures that the cells are lysed and RNA stabilized
immediately to inhibit ex vivo transcription. By contrast,
the expression of miRs in plasma and serum have consistently been
shown to be affected by ex vivo sample handling (13). This may account for the differing
results obtained in studies with different collection methods.
Whole blood overcomes these differences, and may be an ideal medium
for sample collection in large multi-centre studies.
A strength of this study is that it was prospective
in design and we only included high-grade serous EOC patients. Many
biomarker studies include serous, endometriod, clear cell and
non-EOC altogether. Although it would be ideal to develop a
biomarker that can detect all forms of ovarian cancer, there is a
difference in the pathophysiology and genomics of each histological
subtype and therefore EOC should be considered separately in
biomarker discovery experiments (14).
Nevertheless, although our results are promising, there are
important limitations. These include the small sample size in this
pilot study, which restricts the ability to determine the accuracy
of these miRs to diagnose EOC. Therefore, these preliminary
findings require further validation to assess the diagnostic
potential of circulating miRs in ovarian cancer.
In conclusion, circulating miRs, specifically from
the miR-200 family, are increased in high-grade serous EOC, reflect
the extent of disease and constitute potential candidate biomarkers
for non-invasive screening for EOC. Large validation studies are
required to develop biomarkers to ensure early diagnosis and timely
treatment to improve outcomes for women with EOC.
Acknowledgements
We would like to thank Gabrielle Fleming, research
nurse for assisting in subject recruitment and sampling.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Scholz HS, Tasdemir H, Hunlich T, Turnwald
W, Both A and Egger H: Multivisceral cytoreductive surgery in FIGO
stages IIIC and IV epithelial ovarian cancer: Results and 5-year
follow-up. Gynecol Oncol. 106:591–595. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Buys SS, Partridge E, Black A, Johnson CC,
Lamerato L, Isaacs C, Reding DJ, Greenlee RT, Yokochi LA, Kessel B,
et al: PLCO Project Team: Effect of screening on ovarian cancer
mortality: The Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer
Screening Randomized Controlled Trial. JAMA. 305:2295–2303. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shen J, Stass SA and Jiang F: MicroRNAs as
potential biomarkers in human solid tumors. Cancer Lett.
329:125–136. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pal MK, Jaiswar SP, Dwivedi VN, Tripathi
AK, Dwivedi A and Sankhwar P: MicroRNA: A new and promising
potential biomarker for diagnosis and prognosis of ovarian cancer.
Cancer Biol Med. 12:328–341. 2015.PubMed/NCBI
|
|
6
|
Harris AL: Hypoxia - a key regulatory
factor in tumour growth. Nat Rev Cancer. 2:38–47. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bendoraite A, Knouf EC, Garg KS, Parkin
RK, Kroh EM, O'Briant KC, Ventura AP, Godwin AK, Karlan BY,
Drescher CW, et al: Regulation of miR-200 family microRNAs and ZEB
transcription factors in ovarian cancer: Evidence supporting a
mesothelial-to-epithelial transition. Gynecol Oncol. 116:117–125.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim SW, Kim JW, Kim YT, Kim JH, Kim S,
Yoon BS, Nam EJ and Kim HY: Analysis of chromosomal changes in
serous ovarian carcinoma using high-resolution array comparative
genomic hybridization: Potential predictive markers of
chemoresistant disease. Genes Chromosomes Cancer. 46:1–9. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zuberi M, Mir R, Das J, Ahmad I, Javid J,
Yadav P, Masroor M, Ahmad S, Ray PC and Saxena A: Expression of
serum miR-200a, miR-200b, and miR-200c as candidate biomarkers in
epithelial ovarian cancer and their association with
clinicopathological features. Clin Transl Oncol. 17:779–787. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li L, Huang K, You Y, Fu X, Hu L, Song L
and Meng Y: Hypoxia-induced miR-210 in epithelial ovarian cancer
enhances cancer cell viability via promoting proliferation and
inhibiting apoptosis. Int J Oncol. 44:2111–2120. 2014.PubMed/NCBI
|
|
11
|
Chen Y, Chen Q, Liu Q and Gao F: Human
epididymis protein 4 expression positively correlated with miR-21
and served as a prognostic indicator in ovarian cancer. Tumour
Biol. 37:8359–8365. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Häusler SF, Keller A, Chandran PA, Ziegler
K, Zipp K, Heuer S, Krockenberger M, Engel JB, Hönig A, Scheffler
M, et al: Whole blood-derived miRNA profiles as potential new tools
for ovarian cancer screening. Br J Cancer. 103:693–700. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Malentacchi F, Pizzamiglio S, Wyrich R,
Verderio P, Ciniselli C, Pazzagli M and Gelmini S: Effects of
Transport and Storage Conditions on Gene Expression in Blood
Samples. Biopreserv Biobank. 14:122–128. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bell D, Berchuck A, Birrer M, Chien J,
Cramer DW, Dao F, Dhir R, DiSaia P, Gabra H, Glenn P, et al: Cancer
genome atlas research network: Integrated genomic analyses of
ovarian carcinoma. Nature. 474:609–615. 2011. View Article : Google Scholar : PubMed/NCBI
|