Introduction
Asthma is a chronic inflammatory disease of the
airway. More than 100 million people across the globe suffer from
this disorder (Global strategy for asthma management and
prevention; http://www.ginasthma.org). Airway
inflammation, bronchial hyperresponsiveness (BHR), and reversible
airflow obstruction are the primary characteristics of asthma.
Among these, airway inflammation is most closely associated with
the clinical symptoms. Currently, genetic and environmental factors
have been found to be involved in the generation of asthma
pathologies. Thus, the potential contribution of genetic factors is
considered to be an interesting area of clinical and basic
research.
The A disintegrin and metalloprotease 33
(ADAM33) gene is an asthma susceptibility gene, which was
first reported by Van Eerdewegh et al (1). The ADAM33 gene is mapped to the
short arm of chromosome 20p13 in the human genome, and is
predominantly expressed in airway smooth-muscle cells and lung
fibroblast cells, but not in epithelial cells, T cells or
inflammatory leukocytes (2–6). ADAM proteins are involved in cell-cell
and cell-matrix interactions (5), cell
migration (2,3), cell-cell adhesion and signal transduction
(4). The following are characteristic
of ADAM33: Protease activity, a domain structure composed of
a signal sequence, a prodomain, a metalloprotease domain, a
transmembrane domain, a cysteine-rich domain, a disintegrin domain
and a cytoplasmic domain (6). At
present, >100 single-nucleotide polymorphisms (SNPs) of the
ADAM33 gene have been reported to be associated with asthma
or BHR (7–18). However, the data that demonstrate the
association of ADAM33 SNPs with asthma were obtained from
individuals living in mainland areas, who were predominantly
Caucasian and Asian (10,11,19–21), while there is little data on
ADAM33 SNPs and asthma for ethnic minorities, particularly
those living in isolated islands who seldom intermix with other
ethnic groups. The Li population is a unique ethnic minority whose
members live only on the Hainan Island in southern China. Various
lines of evidence have shown there to be various specific genetic
variations in the Li population, differentiating it from the Han
population, even from the Han population from the same island
(22–25). Therefore, in the present study, the
prevalence and types of ADAM33 polymorphisms in the
asthmatic individuals of the Li population were determined and
compared with those in the control subjects.
Materials and methods
Study participants
The present study was conducted in accordance with
the Declaration of Helsinki. Informed consent was obtained from all
individuals and the protocols in the study were approved by the
College's Ethics Committee (approval ID. HNMCE10011-9). Asthma was
diagnosed according to the Global Initiative for Asthma (GINA)
criteria (Global strategy for asthma management and prevention;
http://www.ginasthma.org). The Li population
study subjects were recruited between March 2008 and October 2013
from The People's Hospital of Sanya City (Sanya, China). A total of
150 patients with asthma (92 men and 58 women; mean age, 22.37
years) and 100 healthy controls (59 men and 41 women; mean age,
21.71 years) were enrolled. Subjects with asthma exhibited a mean
forced expiratory volume in 1 sec (FEV1) of 1.79 l (64.58% of
predicted), a mean forced vital capacity (FVC) of 2.37 l (79.82% of
predicted), and a mean FEV1/FVC of 79.64%. The mean duration of
asthma was 9.27 years. No significant difference between asthma
patients and the normal controls was noted, except for lung
function parameters. Pregnant and lactating women were excluded
from the current study.
Extraction of the DNA samples
Clinical data and peripheral blood samples (5 ml per
subject) were obtained from 150 unrelated asthma patients from the
Li population and 100 unrelated healthy individuals who had no
history of asthma. The genomic DNA samples were extracted from the
peripheral blood of the subjects using a Mammalian Genomic DNA
Miniprep kit (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and
the DNA samples were stored at −80°C.
Polymerase chain reaction (PCR) and
sequencing analysis
PCR-restriction fragment length polymorphism was
performed to analyze the ADAM33 SNPs using eight pairs of
primers (Table I) as reported
previously (26,27). PCR reactions were performed in a 25 µl
system, as described previously (27).
The PCR was run for 40 cycles of denaturation at 94°C for 30 sec,
annealing at the optimal melting temperature (60–65°C) for 30 sec
and extension at 72°C for 1 min on a T-Gradient ThermoBlock
(Biometra GmbH, Göttingen, Germany). In addition, sequencing of the
resultant PCR products was performed by a commercial biotech
company (Takara Biotechnology Co., Ltd., Dalian, China) to confirm
the exact alleles.
 | Table I.General information and
oligonucleotide primers used for the amplification of ADAM33
SNPs. |
Table I.
General information and
oligonucleotide primers used for the amplification of ADAM33
SNPs.
| rs number | SNP | Alleles | Primers (5′-3′) | Length of enzyme and
digested area (bp) |
|---|
| rs44707 | T2 | G >A | F:
TTCTCAGGGTCTGGGAGAAA |
HpyCH4III |
|
|
|
| R:
GCCAACCTCCTGGACTCTTA | A:198+112,
G:310 |
| rs511898 | F+1 | C >T | F:
GTATCTATAGCCCTCCAAATCAGAAGAGCC | BsmBI |
|
|
|
| R:
GGACCCTGAGTGGAAGCTG | C:208+192,
T:400 |
| rs528557 | S2 | G >C | F:
AGAGCTCTGAGGAGGGGAACCG | FseI,
C:148+156 |
|
|
|
| R:
GCAGACCATGACACCTTCCTGCTG | G:304 |
| rs612709 | Q-1 | G >A | F:
GGATTCAAACGGCAAGGAG | BtsCI,
A:20+138 |
|
|
|
| R:
GTTCACCTAGATGGCCAGGA | G:158 |
| rs2280089 | T+1 | G >A | F:
CTGAGCCCAGAAACCTGATT | HpyAV,
A:284+28, |
|
|
|
| R:
AGAAGGGAAGGGCTCATGC | G:312 |
| rs2280091 | T1 | A >G | F:
ACTCAAGGTGACTGGGTGCT | NcoI,
A:140+260, |
|
|
|
| R:
GAGGGCATGAGGCTCACTTG | G:400 |
| rs2787094 | V4 | C >G | F:
CTCAGGAACCACCTAGGGGAGAAG | PstI,
G:168+206, |
|
|
|
| R:
CAAAGGTCACACAGCCCCTGACCT | C:374 |
| rs3918396 | S1 | G >A | F:
TGTGCAGGCTGAAAGTATGC | HinfI,
G:132+172, |
|
|
|
| R:
AGAGCTCTGAGGAGGGGAAC | A:304 |
Statistical analysis
The software SNPAnalyzer (version 2.0; Istech,
Kyungkido, Korea) was used to calculate Lewontin's D' value, and
analyze the linkage disequilibrium (LD) and eight ADAM33
SNPs. Hardy-Weinberg equilibrium was confirmed via the exact
distribution of allele frequencies using the χ2 test.
P<0.05 was considered to indicate a statistically significant
difference. The online software Haploview (http://www.broad.mit.edu/mpg/haploview) was used to
construct the haplotype block. In addition, haplotype association
analysis was conducted in two steps, as previously described
(7,27).
In order to determine whether ADAM33 polymorphisms are
associated with the severity of asthma, the asthma patients were
further sub-categorized into high and low severity groups. The high
severity group included subjects suffering from moderate to severe
persistent asthma, whereas the low severity group consisted of
subjects with intermittent and mild persistent asthma. The
participants were diagnosed according to the GINA criteria
(http://www.ginasthma.org).
Results
Demographic characteristics of study
subjects
A total of 150 patients suffering from asthma (92
men and 58 women; mean age, 22.37 years) and 100 healthy control
subjects (59 men and 41 women; mean age, 21.71 years) were enrolled
into the current study and the SNPs of their ADAM33 genes
were analyzed. The major demographic characteristics are presented
in Table II. The mean duration of
asthma was 9.27 years. The demographic and lung function data
indicated that the asthma patients had significantly lower lung
function parameters than the healthy control subjects. The mean
FEV1 and mean FVC in asthma patients were 1.79 l (64.58% of
predicted) and 2.37 l (79.82% of predicted), respectively. To
determine whether ADAM33 gene polymorphisms are associated
with the severity of asthma, the asthmatic patients were divided
into a high severity group and low severity group. As shown in
Table III, the age of the asthma
patients in the high severity group was significantly older than
that in the low severity group (P<0.0001), and the periods
during which the patients had suffered from asthma in the high
severity group was longer than in the low severity group (P=0.006).
In addition, the mean FEV1 in the high severity group was
significantly lower than that in the low severity group (P
<0.0001).
 | Table II.Demographic characteristics of the
asthmatic patients (n=150) and control subjects (n=100). |
Table II.
Demographic characteristics of the
asthmatic patients (n=150) and control subjects (n=100).
| Characteristic | Case | Control | P-value |
|---|
| Age (years) | 22.37±19.72 | 21.71±20.64 | 0.482a |
| Gender (M/F) | 92/58 | 59/41 | 0.819b |
| Duration of asthma
(years) | 9.27±3.28 |
|
|
| Pack year of
smoking | 31.53±6.22 | 29.57±5.81 | 0.052a |
| FVC (% of
predicted) | 79.82±15.66 | 94.81±8.42 |
<0.0001a |
| FEV1 (% of
predicted) | 64.58±13.85 | 92.77±7.52 |
<0.0001a |
 | Table III.Demographic characteristics of the
asthmatic patients in high (n=62) and low (n=88) severity
groups. |
Table III.
Demographic characteristics of the
asthmatic patients in high (n=62) and low (n=88) severity
groups.
| Characteristic | High severity | Low severity | P-value |
|---|
| Age (years) | 31. 27±18.83 | 20.61±19.21 |
<0.0001a |
| Gender (M/F) | 38/24 | 50/38 | 0.361b |
| Duration of asthma
(years) | 11.57±9.05 | 7.82±5.99 | 0.006a |
| FVC (% of
predicted) | 78.94±17.22 | 82.41±20.79 | 0.091a |
| FEV1 (% of
predicted) | 57.41±14.91 | 77.35±16.33 |
<0.0001a |
Genotype frequencies
Eight ADAM33 SNPs, including rs44707/T2,
rs511898/F+1, rs528557/S2, rs612709/Q-1, rs2280089/T+1,
rs2280091/T1, rs2787094/V4 and rs3918396/S1 have been reported to
be associated with asthma in many of the world's mainland areas,
and the majority of studies were performed on Caucasians and Asian
individuals (10,11,19–21). In the present study, genotyping of
these eight ADAM33 SNPs was performed to evaluate the
association with asthma in a Chinese Li population. These data
indicated that all eight ADAM33 SNPs were present and
distributed in Hardy-Weinberg equilibrium in the Li population. The
genotype frequencies of each SNP are presented in Table IV.
 | Table IV.Genotype frequencies of ADAM33
SNPs in the Li population. |
Table IV.
Genotype frequencies of ADAM33
SNPs in the Li population.
|
|
| Genotype [Case (n =
150)/control (n=100)] |
|---|
|
|
|
|
|---|
| SNP ID | SNP | Homozygous
(wild-type) | Heterozygous | Homozygous
(variant) |
|---|
| rs44707 | T2 | GG (82/71) | GA (58/21) | AA (10/8) |
| rs511898 | F+1 | CC (51/46) | CT (81/42) | TT (18/12) |
| rs528557 | S2 | GG (48/39) | GC (78/41) | CC (24/20) |
| rs612709 | Q-1 | GG (66/54) | GA (74/38) | AA (10/8) |
| rs2280089 | T+1 | GG (80/75) | GA (59/19) | AA (11/6) |
| rs2280091 | T1 | AA (121/87) | AG (28/13) | GG (1/0) |
| rs2787094 | V4 | CC (83/73) | CG (60/19) | GG (8/7) |
| rs3918396 | S1 | GG (148/98) | GA (2/1) | AA (0/1) |
ADAM33 polymorphisms and association with asthma.
Among the eight SNPs observed in the current study, the data shown
in Table V demonstrated that only
three SNPs, including rs44707/T2 (P=0.008), rs2280089/T+1 (P=0.021)
and rs2787094/V4 (P=0.028) were significantly different in the Li
asthma patients when compared with the control subjects. All of
these three ADAM33 SNPs were found to be associated with
asthma in the dominant model. Notably, the association of
rs2280089/T+1 with asthma was only observed in European and Latin
American individuals (28). To the
best of our knowledge, no previous study has demonstrated such an
association in Asian individuals (28)
and this is the first time that rs2280089/T+1 has been reported to
be associated with asthma in an Asian population.
 | Table V.ADAM33 polymorphisms with
asthma susceptibility in the Li population. |
Table V.
ADAM33 polymorphisms with
asthma susceptibility in the Li population.
|
|
|
|
|
χ2 test |
|---|
|
|
|
|
|
|
|---|
| SNP ID | SNP | Genetic model | Genotype | Odds ratio (95%
confidence interval) | P-value |
|---|
| rs44707 | T2 | Dominant | GA + AA | 1.97
(1.12–3.29) | 0.008 |
|
|
|
| GG |
|
|
| rs2280089 | T+1 | Dominant | GA + AA | 1.84
(1.14–3.21) | 0.021 |
|
|
|
| GG |
|
|
| rs2787094 | V4 | Dominant | CG + GG | 1.78
(1.09–2.97) | 0.028 |
|
|
|
| CC |
|
|
Haplotype association with asthma
Three ADAM33 SNPs (rs44707/T2, rs2280089/T+1
and rs2787094/V4) that were statistically significant in the
present study were selected to further analyze their haplotypes,
which was performed using Haploview 4.2 software, as previously
reported (26). The eight haplotypes
with frequencies >2.0% were identified to be suitable for
further analysis. Table VI
demonstrates the frequency of GGAGAGT and GAAGGGT haplotypes to be
significantly higher in the asthmatic patients than in the control
subjects (P=0.003 and P=0.008, respectively). Furthermore, the SNPs
rs44707/T2, rs2280089/T+1 and rs2787094/V4 demonstrated strong
D' values in pair-wise LD analysis (D'=0.9012 between
rs44707/T2 and rs2280089/T+1; D'=0.8931 between rs2280089/T+1 and
rs2787094/V4; Fig. 1), confirming the
validity of the present haplotype analysis.
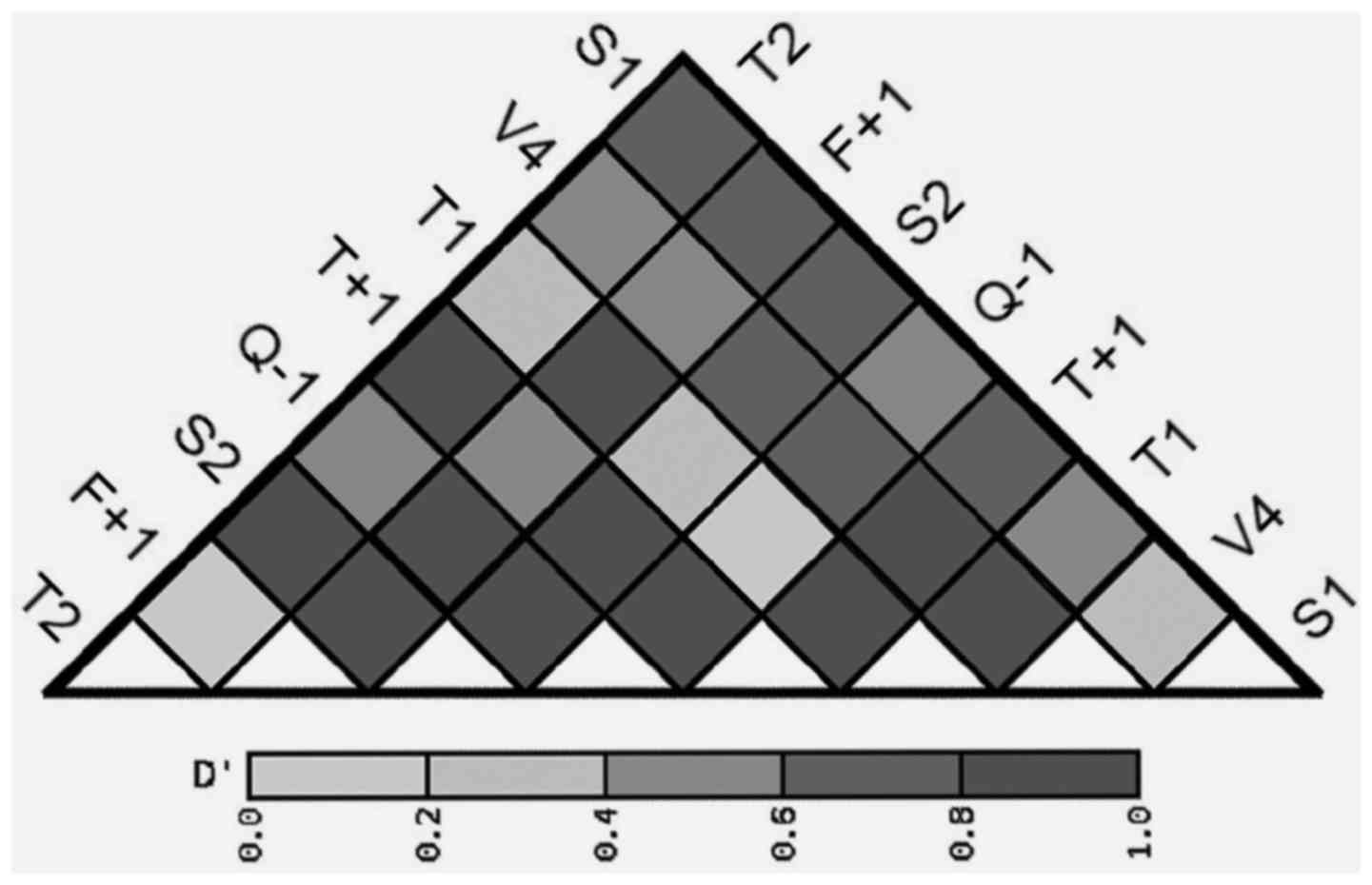 | Figure 1.Pair-wise linkage disequilibrium
analysis of the ADAM33 gene in the Li population. S1,
rs3918396; V4, rs2787094; T1, rs2280091; T+1, rs2280089; Q-1,
rs612709; S2, rs528557; F+1, rs511898; T2, rs44707; ADAM33, A
disintegrin and metalloprotease 33; D', Lewontin's D' value. |
 | Table VI.Frequency of each haplotype from the
five-marker model in the Li population. |
Table VI.
Frequency of each haplotype from the
five-marker model in the Li population.
|
| Haplotype
frequency |
|
|---|
|
|
|
|
|---|
| Haplotype | Case (n=150) | Control
(n=100) | Combined
(n=250) | P-value |
|---|
| GGAGAGT | 0.28 | 0.11 | 0.25 | 0.003 |
| GAAGGGC | 0.04 | 0.03 | 0.03 | NS |
| GGAGGGC | 0.03 | 0.01 | 0.02 | NS |
| GGACGGT | 0.12 | 0.09 | 0.10 | NS |
| GGACGGC | 0.04 | 0.05 | 0.04 | NS |
| GAAGGGT | 0.22 | 0.10 | 0.19 |
0.008 |
| GAAGGAC | 0.09 | 0.08 | 0.09 | NS |
| GGGGAGT | 0.05 | 0.04 | 0.04 | NS |
Association of SNPs in high and low
severity patients
In the present study, the asthma patients were
sub-grouped into high and low severity groups, and the allele and
genotype differences from healthy control subjects were analyzed.
The clinical characteristics of the high and low severity groups
are demonstrated in Table III. The
data concerning allelic distribution analysis showed that allele
frequencies differed significantly between the high severity group
and the control group with respect to SNP rs44707/T2 (P=0.024;
Table VII). In addition, the allele
frequencies demonstrated significant differences between the low
severity group and the control subjects for the SNP rs2787094/V4
(P=0.019; Table VII). However, no
significant differences were noted between the high and low
severity groups in the allele frequencies.
 | Table VII.ADAM33 polymorphisms with
asthma susceptibility in high and low severity patients in the Li
population. |
Table VII.
ADAM33 polymorphisms with
asthma susceptibility in high and low severity patients in the Li
population.
| SNP ID | SNP | Genotype | High
severity/control P-value | Low
severity/control P-value |
|---|
| rs44707 | T2 | GA + AA | 0.024 | NS |
|
|
| GG |
|
|
| rs2280089 | T+1 | GA + AA | NS | NS |
|
|
| GG |
|
|
| rs2787094 | V4 | CG + GG | NS | 0.019 |
|
|
| CC |
|
|
Discussion
Various previous studies have reported that
ADAM33 is a positively cloned gene for asthma (7–18). Thus, it
is necessary to study the association of ADAM33 and
prevalence of asthma in different populations, particularly in
those living in special geographical environments, such as China's
Li population. There are 55 minority ethnic groups in China,
including the Li population (~1.3 million), who reside primarily in
the central and southwestern regions of Hainan Island in the South
China Sea, of which they are the aboriginal inhabitants. The Li
people have maintained much of their traditional lifestyle into the
present day. Although certain members of the Li community have
intermarried with other communities, such as the Han and Miao
populations, most of the Li people have remained relatively
isolated from other populations, meaning that they are likely to
have retained any unique genetic traits. Thus, any genetic study of
the Li people may help to promote personalized therapeutic
strategies in Li patients. Currently, there is a dearth of data
concerning the ADAM33 polymorphisms in the Li population.
Thus, the aim of the present study was to evaluate the variations
in the ADAM33 gene in the Li population. The present study
focused on eight SNPs of the ADAM33 gene that have been
reported to be significantly closely associated with asthma or
airway hyperresponsiveness in several populations (28). Gene segments covering these eight
ADAM33 gene SNPs were successfully amplified and the results
indicated that three SNPs, rs44707/T2, rs2280089/T+1 and
rs2787094/V4, were associated with asthma in a dominant model in a
Li population.
ADAM33 is a polymorphic gene with >100
known SNPs (28). However, not all
these SNPs are associated with asthma. A meta-analysis analyzed 14
SNPs of the ADAM33 gene in 29 case-control studies in
different populations, and the results showed significant
associations with the rs2280091/T1, rs2787094/V4, rs511898/F+1 and
rs2280089/T+1 polymorphisms in the overall population (28). However, positive results were only
found for the rs2280091/T1, rs2787094/V4, rs511898/F+1 and
rs511898/T2 polymorphisms, and only in Asians, not European or
Latin American individuals (28). In
the current study, a specific phenomenon regarding ADAM33
SNPs involving rs2280089/T+1 was observed, which was also observed
in European and Latin American individuals, but never in Asian
individuals (28). To the best of our
knowledge, this is the first study to report that rs2280089/T+1 is
associated with asthma in an Asian population. Although the
underlying mechanism behind the ADAM33 SNP remains unknown,
this finding indicates that the Li population is a particularly
unique minority population in China and the world.
In the present study, the data also demonstrated
ADAM33 SNPs to be significantly closely associated with the
severity of asthma in the Li population. When compared with the
control subjects, significant associations with high severity
asthma were found in SNP rs44707/T2 (P=0.024), whereas a
significant association with low severity asthma patients was
observed in SNP rs2787094/V4 (P=0.019). Therefore, the results are
consistent with previous results where asthma patients demonstrated
significant decreases in lung function over time (29).
Haplotypes contain specific information regarding
potentially unique, unobserved predisposing variants in specific
regions (28,30). In the present study, two common
haplotypes, including GGAGAGT and GAAGGGT, were found to be
asthma-predisposing variants in the Li population. Haplotype data
obtained from other unrelated subjects showed there to be a
significant association between specific ADAM33 haplotypes
and asthma in Chinese Han, Chinese Uygur, Japanese and German
populations, and with BHR in a Korean population (28). Furthermore, family-based studies of
German asthmatic patients confirmed the association of the
ADAM33 haplotype with asthma (17). However, a variety of haplotype analyses
in asthma subjects have already been identified in different
populations. The previous results suggested that a risk haplotype
in one population may be a protective haplotype in another
(28). However, in the present study,
no significant differences were identified in the distribution of
haplotypes between the high and low severity asthma groups. This
may be due to the relatively small sizes of the asthmatic patient
populations. In a previous study, >200 asthmatic subjects were
observed over a period of >20 years, and the resultant data
showed the ADAM33 gene SNPs to be associated with a decline
in FEV1, indicating that the function of the ADAM33 gene may
be involved in all pathologic steps, including asthma prevalence,
severity, airway hyperresponsiveness, progression and prognosis
(9). Various lines of evidence in
previous studies indicated that overexpression of the ADAM33
gene (mRNA and protein) was seen in various types of cells in
asthma patients, including airway smooth muscle cells, fibroblasts,
mesenchymal cells and endothelial cells (2–6).
Furthermore, data from previous studies have demonstrated that the
expression of the ADAM33 gene may regulate the expression of
transforming growth factor-β, which is a major cytokine expressed
and excreted by bronchial macrophages, epithelial cells and
mesenchymal cells for airway remodeling (31). A potential mechanism by which
ADAM33 affects asthma may be associated with the regulation
of airway inflammation, hyperresponsiveness and remodeling
processes; however, the exact molecular mechanism requires further
investigation.
In conclusion, these data demonstrated a positive
association between ADAM33 gene SNPs and asthma prevalence
in a Li minority population in southern China. These data
strengthen the evidence from previous investigations and studies of
other populations elsewhere in the world. Currently, the exact
functions of the ADAM33 gene in the progress of asthma
pathogenesis remain unknown. Further investigations are required to
determine the exact mechanism and the exact role of the
ADAM33 gene in asthma.
Acknowledgements
The present study was funded by the National Natural
Science Foundation of China (grant no. 81460020). The authors would
like to thank LetPub (www.letpub.com) for its linguistic assistance during
the preparation of this manuscript.
References
|
1
|
Van Eerdewegh P, Little RD, Dupuis J, Del
Mastro RG, Falls K, Simon J, Torrey D, Pandit S, McKenny J,
Braunschweiger K, et al: Association of the ADAM33 gene with asthma
and bronchial hyperresponsiveness. Nature. 418:426–430. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Martin J, Eynstone LV, Davies M, Williams
JD and Steadman R: The role of ADAM 15 in glomerular mesangial cell
migration. J Biol Chem. 277:33683–33689. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
McFarlane S: Metalloproteases: Carving out
a role in axon guidance. Neuron. 37:559–562. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Seals DF and Courtneidge SA: The ADAMs
family of metalloproteases: Multidomain proteins with multiple
functions. Genes Dev. 17:7–30. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
White JM: ADAMs: Modulators of cell-cell
and cell-matrix interactions. Curr Opin Cell Biol. 15:598–606.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yoshinaka T, Nishii K, Yamada K, Sawada H,
Nishiwaki E, Smith K, Yoshino K, Ishiguro H and Higashiyama S:
Identification and characterization of novel mouse and human
ADAM33s with potential metalloprotease activity. Gene. 282:227–236.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Awasthi S, Tripathi P, Ganesh S and Husain
N: Association of ADAM33 gene polymorphisms with asthma in Indian
children. J Hum Genet. 56:188–195. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Howard TD, Postma DS, Jongepier H, Moore
WC, Koppelman GH, Zheng SL, Xu J, Bleecker ER and Meyers DA:
Association of a disintegrin and metalloprotease 33 (ADAM33) gene
with asthma in ethnically diverse populations. J Allergy Clin
Immunol. 112:717–722. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jongepier H, Boezen HM, Dijkstra A, Howard
TD, Vonk JM, Koppelman GH, Zheng SL, Meyers DA, Bleecker ER and
Postma DS: Polymorphisms of the ADAM33 gene are associated with
accelerated lung function decline in asthma. Clinical and
experimental allergy. Clin Exp Allergy. 34:757–760. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee JH, Park HS, Park SW, Jang AS, Uh ST,
Rhim T, Park CS, Hong SJ, Holgate ST, Holloway JW and Shin HD:
ADAM33 polymorphism: association with bronchial
hyper-responsiveness in Korean asthmatics. Clin Exp Allergy.
34:860–865. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee YH and Song GG: Association between
ADAM33 T1 polymorphism and susceptibility to asthma in Asians.
Inflamm Res. 61:1355–1362. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Noguchi E, Ohtsuki Y, Tokunaga K,
Yamaoka-Sageshima M, Ichikawa K, Aoki T, Shibasaki M and Arinami T:
ADAM33 polymorphisms are associated with asthma susceptibility in a
Japanese population. Clin Exp Allergy. 36:602–608. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sadeghnejad A, Ohar JA, Zheng SL, Sterling
DA, Hawkins GA, Meyers DA and Bleecker ER: Adam33 polymorphisms are
associated with COPD and lung function in long-term tobacco
smokers. Respir Res. 10:212009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sakagami T, Jinnai N, Nakajima T, Sekigawa
T, Hasegawa T, Suzuki E, Inoue I and Gejyo F: ADAM33 polymorphisms
are associated with aspirin-intolerant asthma in the Japanese
population. J Hum Genet. 52:66–72. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Su D, Zhang X, Sui H, Lü F, Jin L and
Zhang J: Association of ADAM33 gene polymorphisms with adult
allergic asthma and rhinitis in a Chinese Han population. BMC Med
Genet. 9:822008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Uh ST, Jang AS, Park SW, Park JS, Min CG,
Kim YH, Park BL, Shin HD, Kim DS and Park CS: ADAM33 gene
polymorphisms are associated with the risk of idiopathic pulmonary
fibrosis. Lung. 192:525–532. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Werner M, Herbon N, Gohlke H, Altmüller J,
Knapp M, Heinrich J and Wjst M: Asthma is associated with
single-nucleotide polymorphisms in ADAM33. Clin Exp Allergy.
34:26–31. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xue W, Han W and Zhou ZS: ADAM33
polymorphisms are associated with asthma and a distinctive palm
dermatoglyphic pattern. Mol Med Rep. 8:1795–1800. 2013.PubMed/NCBI
|
|
19
|
Fan JG, Wang ZA and Zhao HX: The ADAM33 S2
polymorphism is associated with susceptibility to pediatric asthma
in the Chinese Han population. Genet Test Mol Biomarkers.
19:573–578. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Klaassen EM, Penders J, Jöbsis Q, van de
Kant KD, Thijs C, Mommers M, van Schayck CP, van Eys G, Koppelman
GH and Dompeling E: An ADAM33 polymorphism associates with
progression of preschool wheeze into childhood asthma: A
prospective case-control study with replication in a birth cohort
study. PLoS One. 10:e01193492015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zheng W, Wang L, Su X and Hu XF:
Association between V4 polymorphism in the ADAM33 gene and asthma
risk: A meta-analysis. Genet Mol Res. 14:989–999. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ding Y, Yang D, Zhou L, He P, Yao J, Xie
P, Lin D, Sun D, Sun P, Li Q, et al: Cytochrome P450 2C9 (CYP2C9)
polymorphisms in Chinese Li population. Int J Clin Exp Med.
8:21024–21033. 2015.PubMed/NCBI
|
|
23
|
Ding Y, Yang D, Zhou L, Xu J, Chen Y, He
P, Yao J, Chen J, Niu H, Sun P and Jin T: Variants in multiple
genes polymorphism association analysis of COPD in the Chinese Li
population. Int J Chron Obstruct Pulmon Dis. 10:1455–1463. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ding Y, Yang H, He H, Shi H, He P, Yan S
and Jin T: Plasma adiponectin concentrations and adiponectin gene
polymorphisms are associated with bronchial asthma in the Chinese
Li Population. Iran J Allergy Asthma Immunol. 14:292–297.
2015.PubMed/NCBI
|
|
25
|
Wang J, Wen J, Si-Ma-Yi MH, He YB,
Tu-Er-Xun KL, Xia Y, Zhang JL and Wu-Shou-Er QM: Association of
ADAM33 gene polymorphisms with asthma in the Uygur population of
China. Biomed Rep. 1:447–453. 2013.PubMed/NCBI
|
|
26
|
Tan J, Liu AP, Sun C, Bai YF and Lv F:
Association of ADAM33 gene polymorphisms with COPD in the Mongolian
population of China. Ann Hum Biol. 41:9–14. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Thongngarm T, Jameekornrak A, Limwongse C,
Sangasapaviliya A, Jirapongsananuruk O, Assawamakin A, Chaiyaratana
N, Luangwedchakarn V and Thongnoppakhun W: Association between
ADAM33 polymorphisms and asthma in a Thai population. Asian Pac J
Allergy Immunol. 26:205–211. 2008.PubMed/NCBI
|
|
28
|
Liang S, Wei X, Gong C, Wei J, Chen Z and
Deng J: A disintegrin and metalloprotease 33 (ADAM33) gene
polymorphisms and the risk of asthma: A meta-analysis. Hum Immunol.
74:648–657. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pino-Yanes M, Thakur N, Gignoux CR,
Galanter JM, Roth LA, Eng C, Nishimura KK, Oh SS, Vora H, Huntsman
S, et al: Genetic ancestry influences asthma susceptibility and
lung function among Latinos. J Allergy Clin Immunol. 135:228–235.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cordell HJ: Estimation and testing of
genotype and haplotype effects in case-control studies: Comparison
of weighted regression and multiple imputation procedures. Genet
Epidemiol. 30:259–275. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Girodet PO, Dournes G, Thumerel M,
Begueret H, Dos Santos P, Ozier A, Dupin I, Trian T, Montaudon M,
Laurent F, et al: Calcium channel blocker reduces airway remodeling
in severe asthma. A proof-of-concept study. Am J Respir Crit Care
Med. 191:876–883. 2015. View Article : Google Scholar : PubMed/NCBI
|















