Introduction
Insulin resistance is a condition of the body not
appropriately responding to circulating insulin; furthermore,
insulin resistance in adipose tissue is an important characteristic
of gestational diabetes mellitus (GDM) (1). To compensate for the insulin resistance
in the adipose tissue, pancreatic β-cells initially produce more
insulin; however, beyond a certain limit, β-cell failure and DM
occur (2). At the cellular level, the
insulin signaling cascade has an important metabolic role, while
its disruption may induce insulin resistance and is closely
associated with GDM (3). However, to
date, the causes of insulin resistance in adipose tissue at the
cellular and molecular level have remained elusive. As aberration
of gene expression and molecular pathways may be involved in GDM,
the present study determined and analyzed the gene expression
profiles of adipose tissues from pregnant women with and without
GDM. To the best of our knowledge, the present study was the first
to show a distinct pattern of differentially expressed genes (DEGs)
in omental visceral adipose tissue (OVAT) from Chinese GDM patients
by using a whole genome microarray.
While adipose tissue has been regarded as a storage
organ, it is likely to have further functions, as it has been
convincingly shown to secrete a number of cytokines, which have a
fundamental role in causing insulin resistance in GDM (4,5). Adipose
tissues in different sites of the body have distinct biochemical
properties (6). It has been reported
that the accumulation of OVAT is mainly correlated with an
increased risk of altered glucose homeostasis and insulin
resistance (7).
Microarrays represent a powerful tool for studying
the mechanisms of complex diseases and enable for comprehensive
analysis of the interaction between multiple genes simultaneously
implicated in pathological processes (8–10).
In the present study, the gene expression profiles
of OVATs from pregnant women with or without GDM were compared with
the aim of identifying DEGs. Functional enrichment analysis was
performed to determine the gene ontology (GO) functions and the key
signaling pathways in the pathogenetic processes of GDM. Several
molecular mechanisms and pathways that may be responsible for the
insulin resistance and progression of GDM were identified.
Materials and methods
Patients and tissues
OVATs were obtained from six patients during
C-section at the Department of Obstetrics and Gynaecology, the
First Affiliated Hospital of Kunming Medical College (Kunming,
China) from January 2012 to September 2013, including three cases
with normal glucose tolerance and three cases with GDM. The
diagnosis of GDM was made by OGTT 75 g, according to the World
Health Organization criteria, during the second trimester (24–28
weeks of gestation). Exclusion criteria for participation included
multiple gestation, infection, pregnancy with complications,
congenital or chromosomal abnormalities of the fetus, a family
history of diabetes, and pregnancy with alcohol or drug abuse.
Samples were obtained and immediately snap-frozen in liquid
nitrogen. Informed consent was obtained prior to caesarean section.
The present study was approved by the Ethics Committee of Kunming
Medical College (Kunming, China).
RNA extraction and isolation
Adipose tissues from each of the six patients were
individually minced with small scissors and total RNA was extracted
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) and purified using the RNeasy Mini kit (Qiagen,
Hilden, Germany). The RNA concentration was detected using a 2100
Bioana bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA,
USA). RNA purity and integrity were assessed by gel electrophoresis
as well as spectrophotometrically using a Nanodrop (Thermo Fisher
Scientific, Inc.). The extracted RNA was temporarily stored in a
−70°C freezer in 95% ethanol for further analysis.
Microarray assay and data
normalization
The whole Human Gene Expression Array (Affymetrix
GeneChip® PrimeView™, Homo sapiens; Affymetrix, Santa
Clara, CA, USA) was used to screen for gene expression in the OVAT
cells of pregnant women according to the manufacturer's
instructions.
First, the ribosomal RNA was removed from the total
RNA, and the purified mRNA was amplified and reversely transcribed
into fluorescent/biotinylated complementary (c)DNA using the ENZO
kit (Affymetrix). The cDNA was fragmented and then hybridized to
the Affymetrix GeneChip® PrimeView™ containing 36,000
probe-sets representing ~20,000 unique genes. Six microarrays were
run for the samples (from the six patients including three cases
with normal glucose tolerance and three cases with GDM mentioned
above) tested and each array was replicated twice. Subsequently,
the arrays were processed on an Affymetrix fluidics station
(Affymetrix), where they were subjected to automated washing and
staining. The arrays were scanned using an Affymetrix GeneChip
scanner 3000 (Affymetrix), raw data were obtained using the Feature
Extraction Software 10.7 (Agilent Technologies, Inc.) and
normalized using the quantile algorithm with Gene Spring 11.0
(Agilent Technologies, Inc.) to remove background bias with the
normalization value set to 1. Systematic bioinformatic analyses of
the microarray data were performed by Novel Bioinformatics Co.,
Ltd. (Shanghai, China).
Differential expression analysis, GO
analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
analysis, gene-gene interaction network (Gene-Act-Net) and
pathway-pathway (Path)-Act-Net construction
Genes with fold-changes of ≥2 or <0.5 and
P<0.05 were subjected to a secondary selection based on the size
of the negative log2 of their P-values, and the thereby
selected genes were considered to be DEGs.
To assess the function of the DEGs, they were
enriched into terms of the GO categories cellular component (CC),
biological process (BP) and molecular function (MF) using the GO
chart feature of the Database for Annotation, Visualization and
Integrated Discovery.
Significantly enriched pathways of these DEGs were
then determined using the KEGG database, and Gene-Act-Net
interaction network was constructed with regard to genes, included
in the database.
Based on the results of the KEGG enrichment
analyses, the Path-Act-Net was constructed. Furthermore, the
Gene-Act-Net was constructed based on the DEGs in the GO terms and
pathways.
Results
Patient characteristics
Between the patients with and without GDM, no
significant differences in age, G1P0 parity, gestational week or
height were present. However, the weight and body mass index were
significantly higher in the GDM group.
DEGs in OVATS of GDM patients
A total of 935 DEGs were identified, including 450
genes which were downregulated and 485 which were upregulated in
the OVATS of pregnant women with GDM compared with those
without.
GO analysis of DEGs
In the GO category MF, terms including receptor
binding, actin binding, extracellular matrix binding and C-X-C
motif chemokine receptor R3 binding were found to be enriched.
Moreover, in the category CC, DEGs were enriched in the terms
extracellular matrix/region or actin cytoskeleton, while in the
category BP, antigen processing and presentation, extracellular
matrix organization, positive regulation of cell-substrate
adhesion, response to nutrients and response to dietary excess were
significantly over-represented.
In the BP category, antigen processing and
presentation, immune response and regulation of immune response
were among the major downregulated GO terms (Fig. 1A), while vascular smooth muscle
contraction, extracellular matrix organization and response to
nutrients were among the major upregulated GO terms (Fig. 1B). Similarly, the top 10 down- and
upregulated GO terms in the CC category are shown in Fig. 1C and D, respectively, and those in the
MF category are shown in Fig. 1E and
F, respectively.
 | Figure 1.Top 10 GO terms in the categories (A
and B) MF and (C and D) CC for down- and upregulated DEGs,
respectively, in omental visceral adipose tissues of women with
gestational diabetes mellitus. The longer the bar, the smaller the
P-value. DEGs were rated according to their
log2(P-value) using the Database for the Annotation,
Visualization and Integrated Discovery (P<0.05). Top 10 GO terms
in the categories (E and F) BP for down- and upregulated DEGs,
respectively, in omental visceral adipose tissues of women with
gestational diabetes mellitus. The longer the bar, the smaller the
P-value. DEGs were rated according to their
log2(P-value) using the Database for the Annotation,
Visualization and Integrated Discovery (P<0.05). GO, gene
ontology; DEG, differentially expressed gene; MHC, major
histocompatibility complex; BP, biological process; CC, cellular
component; MF, molecular function; up, upregulated; down,
downregulated; ER, endoplasmic reticulum, MHC, major
histocompatibility complex; IgG, immunoglobulin G. |
In the CC category, extracellular region, major
histocompatibility complex class II and nuclear RNA export factor
complex were the major donwnregulated GO terms (Fig. 1C), while melanosome, smooth muscle
contractile fiber and extracellular matrix were the major
downregulated GO terms (Fig. 1D).
In the MF category, natural killer (NK) cell
lectin-like receptor binding, serine-type endopeptidase activity
and peptide antigen binding were the top three donwnregulated GO
terms (Fig. 1E), while heparin
binding, phosphatidylserine binding and
phosphatidylinositol-4,5-bisphosphate binding were the top three
upregulated GO terms (Fig. 1F). The
terms likely to be relevant for GDM were antigen processing and
presentation, immune response and response to nutrients.
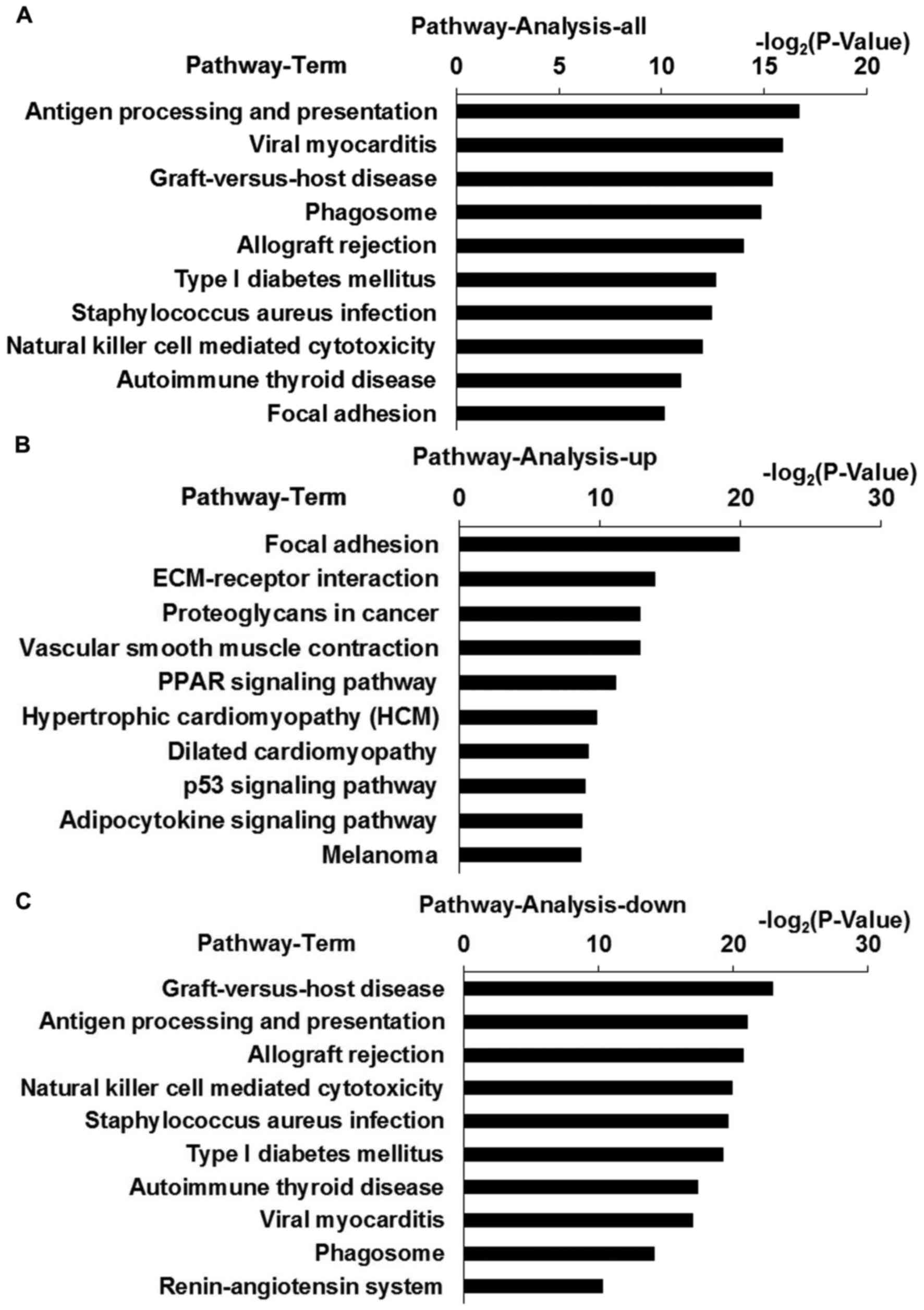
KEGG pathway analysis
To identify the most commonly dysregulated pathways
in OVATs of pregnant women with GDM, KEGG pathway analysis was
performed. The top six enriched pathways of the DEGs were antigen
processing and presentation, viral myocarditis, graft-versus-host
disease, phagosome, allograft rejection and type I diabetes
mellitus (T1D) (Fig. 2A). With regard
to the antigen processing and presentation pathway, 79 genes were
represented on the array chips, of which 14 were differentially
expressed [transporter 2, ATP-Binding Cassette, Sub-Family B
(P=0.0046), LOC100287534 (P=0.0160), major histocompatibility
complex, class I, A (HLA-A; P=0.011), killer cell
immunoglobulin-like receptor (KIR)2DL4 (P=0.0160), KIR2DL5A
(P=0.0161), HLA-DQA1 (P=0.0139), heat shock protein (HSP)A5
(P=0.0189), HLA-E (P=0.0199), HLA-DOA (P=0.0251), HLA-F (P=0.0299),
HLA-DMB (P=0.033), HSPA4 (P=0.0379), HSPA6 (P=0.0390), HLA-DPB1
(P=0.0489)].
The top five upregulated pathways included focal
adhesion, extracellular matrix (ECM)-receptor interaction,
proteoglycans in cancer, vascular smooth muscle contraction and the
peroxisome proliferator-activated receptor signaling pathway
(Fig. 2B). The top four downregulated
pathways included graft-versus-host disease, antigen processing and
presentation, allograft rejection and NK cell-mediated cytotoxicity
(Fig. 2C).
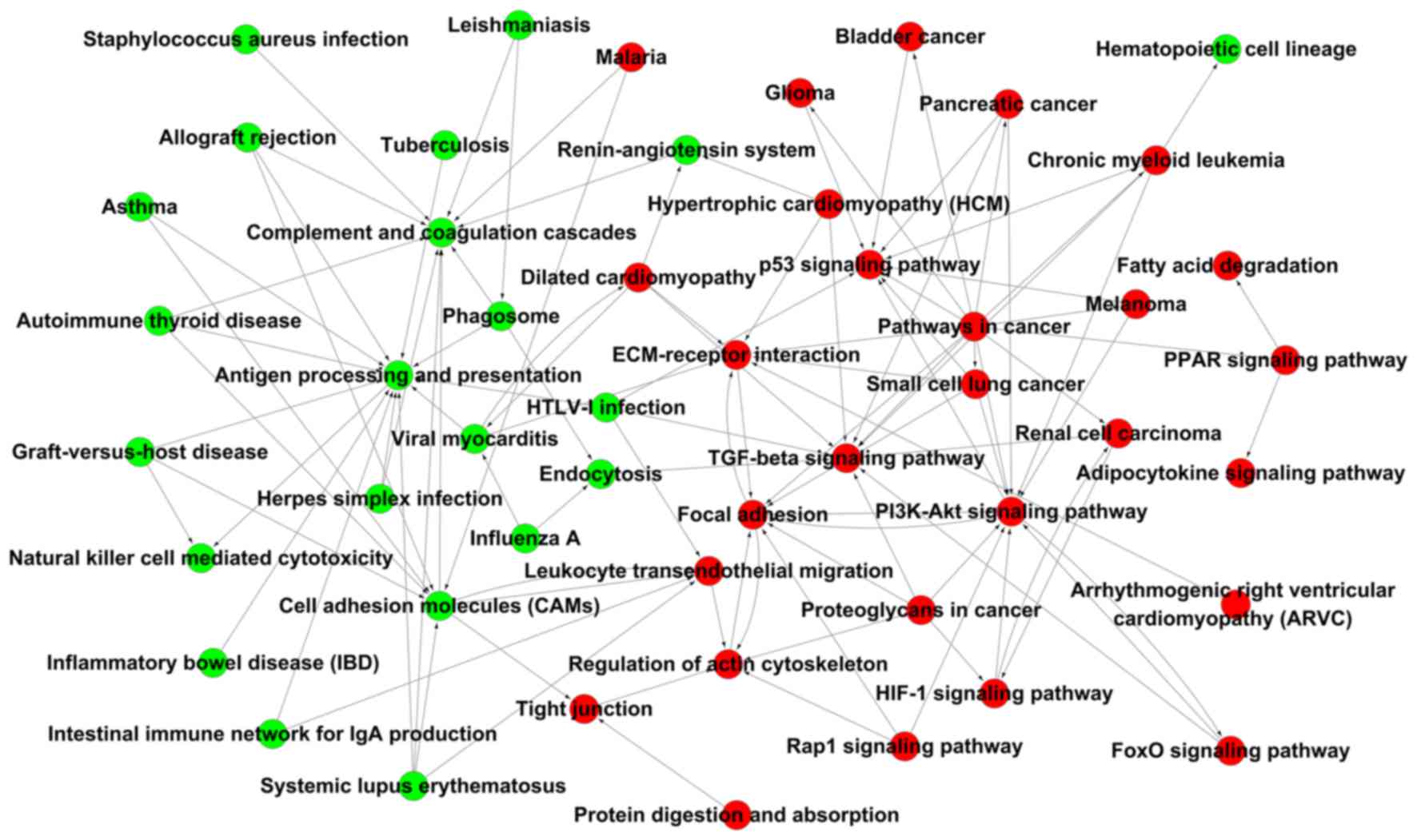
Gene-Act-Net and Pathway-Act-Net
The Gene-Act-Net and the Path-Act-Net were
constructed based on the interaction of the DEGs or their enriched
pathways, respectively (P<0.05). In the Path-Act-Net, antigen
processing and presentation was the top-scoring component
identified. Furthermore, complement and coagulation cascades, cell
adhesion molecules, the transforming growth factor (TGF)-β,
phosphoinositide-3 kinase (PI3K)-Akt and P53 signaling pathways,
and regulation of the actin cytoskeleton were high-scoring
components in the network, suggesting that immune response and
signaling pathways associated with insulin are the major pathways
associated with the pathogenesis of GDM. Within the network,
antigen processing and presentation as well as cell adhesion
molecules were pathways associated with downregulated DEGs with the
highest number of connections, while TGF-β signaling, focal
adhesion, PI3K-Akt signaling, P53 signaling, ECM-receptor
interaction and regulation of the actin cytoskeleton were pathways
associated with upregulated DEGs with the highest number of
connections (Fig. 3).
In the Gene-Act-Net, the four genes with the highest
degree of interaction were integrin subunit β-1 (ITGB1), AKT
serine/threonine kinase 3 (AKT3), integrin subunit α V (ITGAV) and
fibroblast growth factor receptor (FGFR)3 (≥10 connections)
(Table I).
 | Table I.Differentially expressed genes with
the highest degree of interaction in the gene-gene interaction
network. |
Table I.
Differentially expressed genes with
the highest degree of interaction in the gene-gene interaction
network.
| Gene ID | Symbol | Description | InDegree | OutDegree | Degree |
|---|
| 3688a | ITGB1 | Integrin subunit
β-1 | 9 | 7 | 16 |
| 10000a | AKT3 | AKT
serine/threonine kinase 3 | 2 | 10 | 12 |
| 3685a | ITGAV | Integrin subunit α
V | 9 | 1 | 10 |
| 2261b | FGFR3 | Fibroblast growth
factor receptor 3 | 8 | 2 | 10 |
| 3105b | HLA-A | Major
histocompatibility complex, class I, A | 3 | 5 | 8 |
| 3134b | HLA-F | Major
histocompatibility complex, class I, F | 3 | 5 | 8 |
| 3133b | HLA-E | Major
histocompatibility complex, class I, E | 3 | 5 | 8 |
| 10398a | MYL9 | Myosin light chain
9 | 6 | 1 | 7 |
| 6385b | SDC4 | Syndecan-4 | 6 | 0 | 6 |
| 4659a | PPP1R12A | Protein phosphatase
1 regulatory subunit 12A | 3 | 3 | 6 |
| 5500a | PPP1CB | Protein phosphatase
1 catalytic subunit β | 3 | 3 | 6 |
| 3479a | IGF1 | Insulin-like growth
factor 1 | 3 | 3 | 6 |
| 2257a | FGF12 | Fibroblast growth
factor 12 | 3 | 3 | 6 |
| 2259a | FGF14 | Fibroblast growth
factor 14 | 3 | 3 | 6 |
| 7410b | VAV2 | VAV2 guanine
nucleotide exchange factor | 3 | 3 | 6 |
| 2952a | GSTT1 | Glutathione
S-transferase theta-1 | 3 | 3 | 6 |
| 4257a | MGST1 | Microsomal
glutathione S-transferase 1, isoform CRA_a | 3 | 3 | 6 |
| 2949a | GSTM5 | Glutathione
S-transferase Mu 5 | 3 | 3 | 6 |
| 3111b | HLA-DOA | Major
histocompatibility complex, class II, DO α | 3 | 3 | 6 |
| 3109b | HLA-DMB | Major
histocompatibility complex, class II, DM β | 3 | 3 | 6 |
| 3115b | HLA-DPB1 | Major
histocompatibility complex, class II, DP β 1 | 3 | 3 | 6 |
| 3117b | HLA-DQA1 | cDNA FLJ51239,
moderately similar to major histocompatibility complex class II
histocompatibility antigen, DQ(W3) α chain | 3 | 3 | 6 |
| 7057a | THBS1 | Thrombospondin 1,
isoform CRA_a | 1 | 4 | 5 |
| 21826a | THBS2 |
Thrombospondin-2 | 1 | 4 | 5 |
| 998a | CDC42 | Cell division cycle
42, isoform CRA_a | 1 | 4 | 5 |
| 8644a | AKR1C3 | Aldo-keto reductase
family 1 member C3 homolog | 4 | 1 | 5 |
| 7363a | UGT2B4 |
UDP-glucuronosyltransferase 2 member
B4 | 1 | 4 | 5 |
| 7037b | TFRC | Transferrin
receptor (p90, CD71) mRNA | 4 | 1 | 5 |
| 1282a | COL4A1 | Collagen type IV α
1 chain | 1 | 3 | 4 |
| 1281a | COL3A1 | Collagen type III α
1 chain | 1 | 3 | 4 |
| 4638a | MYLK | Myosin light chain
kinase | 3 | 1 | 4 |
| 1277a | COL1A1 | Collagen type I α-1
chain | 1 | 3 | 4 |
| 3371a | TNC | Tenascin C | 1 | 3 | 4 |
| 57292b | KIR2DL5A | Killer cell
immunoglobulin like receptor, two Ig domains and long cytoplasmic
tail 5A | 3 | 1 | 4 |
| 3805b | KIR2DL4 | Killer cell
immunoglobulin like receptor, two Ig domains and long cytoplasmic
tail 4 | 3 | 1 | 4 |
| 6891b | TAP2 | Transporter 2, ATP
binding cassette subfamily B member | 0 | 4 | 4 |
| 4192a | MDM2 | E3
ubiquitin-protein ligase Mdm2 | 2 | 1 | 3 |
| 5781a | PTPN11 | Protein tyrosine
phosphatase, non-receptor type 11 | 2 | 1 | 3 |
| 1962a | EHHADH | Enoyl-CoA hydratase
and 3-hydroxyacyl CoA dehydrogenase | 1 | 2 | 3 |
| 2180a | ACSL1 | Acyl-CoA synthetase
long-chain family member 1 mRNA | 1 | 2 | 3 |
| 4773b | NFATC2 | Nuclear factor of
activated T-cells 2 | 1 | 1 | 2 |
| 948a | CD36 | CD36 antigen
mRNA | 2 | 0 | 2 |
| 4629a | MYH11 | Myosin heavy chain
11 | 1 | 1 | 2 |
| 2316a | FLNA | Filamin-A | 0 | 2 | 2 |
| 94274a | PPP1R14A | Protein phosphatase
1 regulatory subunit 14A | 0 | 2 | 2 |
| 51a | ACOX1 | Acyl-CoA oxidase
1 | 2 | 0 | 2 |
| 7043a | TGFB3 | Transforming growth
factor, β 3 | 2 | 0 | 2 |
| 4734b | NEDD4 | Neural precursor
cell expressed, developmentally downregulated 4, E3 ubiquitin
protein ligase | 1 | 1 | 2 |
| 548596b | CKMT1A | Creatine kinase,
mitochondrial 1A | 1 | 1 | 2 |
| 1159b | CKMT1B | Creatine kinase,
mitochondrial 1B (EC 2.7.3.2) | 1 | 1 | 2 |
| 10725b | NFAT5 | Nuclear factor of
activated T-cells 5, tonicity-responsive, isoform CRA_b | 1 | 0 | 1 |
| 3952a | LEP | Leptin | 0 | 1 | 1 |
| 3563b | IL3RA | Interleukin-3
receptor subunit α | 1 | 0 | 1 |
| 284a | ANGPT1 | Angiopoietin-1 | 0 | 1 | 1 |
| 2034a | EPAS1 | Endothelial PAS
domain-containing protein 1 | 0 | 1 | 1 |
| 10332b | CLEC4M | C-type lectin
domain family 4, member M, transcript variant 6 mRNA | 0 | 1 | 1 |
| 1759b | DNM1 | Dynamin-1 (EC
3.6.5.5) | 0 | 1 | 1 |
Discussion
GDM is defined as the first occurrence of glucose
intolerance during pregnancy. Although it is a common disorder, its
pathophysiology has remained to be fully elucidated. Adipose tissue
has been implicated in the development of insulin resistance and
excess adipose tissue has been shown to be associated with GDM
(11,12).
Adipose tissue can be further classified as
sub-cutaneous and visceral adipose tissue. The human body has six
visceral fat depots: Perirenal, gonadal, epicardial,
retroperitoneal, omental and mesenteric adipose, each of which has
distinct characteristics. Human omental adipose tissue exhibits
specific mRNA expression profiles. Only few studies have compared
the difference in expression profiles in OVATs of pregnant women
with and without GDM (13,14). To the best of our knowledge, the
present study was the first to assess the DEGs in OVATs from
pregnant women with vs. without GDM using the Affymetrix chip due
to the difficulty in obtaining OVAT samples during the same period,
which are matched with regard to age, gestational age, G1P0 parity
and height.
The microarray approach and bioinformatic
technologies were combined to perform a comparative gene expression
profiling analysis of normal and pathological OVATs. Subsets of
DEGs were identified, GO and KEGG pathways were analyzed, and
interaction networks were generated.
The DEGs identified between the two groups may
represent new candidate genes associated with GDM. In the present
study, the top four genes in the Gene-Act-Net with the highest
degree of interaction were ITGB1, AKT3, ITGAV (upregulated) and
FGFR3 (downregulated), suggesting their function as hub genes with
a high regulatory ability. ITGB7 and ITGB1 have been previously
reported to be associated with type 1 diabetes and their proteins
engage in direct mutual receptor-ligand interactions associated
with the homing of T cells from blood to tissues, such as the
intestine and pancreas (15). ITGB1
was also found to be involved in the regulation of cell migration
associated with numerous pathologies (16–18). AKT3 is
activated by growth factors and other extracellular stimuli,
including glucose, as well as key upstream regulatory proteins
including Ras, PI3K subunits and phosphatase and tensin homolog,
which are involved in the regulation of a diversity of biological
roles of activated Akt, including the regulation of cell
metabolism, survival and proliferation (19,20). ITGAV
and FGFR3 are involved in cell migration and proliferation, and
have been reported to be associated with the progression and
dissemination of cancer (21–24).
GO analysis revealed enrichment of the DEGs in the
MF category in terms including receptor binding, extracellular
matrix binding and CXCR3, as well as in the BP category in terms
including antigen processing and presentation, extracellular matrix
organization, positive regulation of cell-substrate adhesion, which
were also closely associated with inflammation response or immune
response. In addition, the DEGs in the GDM group were enriched in
GO terms including vascular smooth muscle contraction (GO:0014829)
and muscle contraction (GO:0006936), possibly due to the impaired
vasodilation in the GDM group. The function of vascular smooth
muscle cells is to regulate blood flow and pressure through
contraction and relaxation. The principle complications of GDM are
cardiovascular disease, the risk of which is potentiated by
obesity, hypertensive disorders of pregnancy. Studies have shown
that women with GDM have higher risk of cardiac dysfunction and
endothelial dysfunction very soon after pregnancy (25), which may explain for the upregulation
of these pathways compared with those in normal women in the same
gestational week. Furthermore, preterm delivery is relatively
common in women with GDM and birth-associated muscle contractions
may be another reason for the enrichment of these pathways in the
GDM group (26).
In the present study, KEGG analysis revealed that
the DEGs in the OVATs from patients with GDM were enriched in
pathways including antigen processing and presentation, cell
adhesion molecules, T1D, NK cell-mediated cytotoxicity and TGF-β
signaling. These pathways were categorized as ‘signaling molecules
and interaction’, ‘immune system’ and ‘inflammatory response’,
suggesting that these processes are involved in GDM. Among them,
four pathways (antigen processing and presentation, cell adhesion
molecules, T1D and NK cell-mediated cytotoxicity) have been
previously reported in the blood of Chinese women with GDM
(27). Considering that OVAT is
composed of several different cell types, including adipocytes,
pre-adipocytes, macrophages, vascular cells and other blood cells,
it is reasonable to assume that certain significant pathways are
similar between OVAT and peripheral blood, and insulin resistance
in peripheral blood and adipose tissue may also share certain
pathways. A study by Yang et al (28) demonstrated that TGF-β signaling
participates in steatohepatitis through the regulation of lipid
metabolism and apoptosis in hepatocytes. Considering that the
present study used adipose tissues, it is expected that pathways
associated with lipid metabolism dysfunction have a marked role in
the insulin resistance of patients with GDM. In the present study,
DEGs were enriched in pathways including peroxisome
proliferator-activated receptor signaling, adipocytokine signaling
and fatty acid degradation, which are associated with lipid
metabolism.
As for pathways associated with T1D, the results of
the present study suggested that GDM is mainly facilitated by
autoimmune destruction of insulin-producing pancreatic β-cells. The
nine genes found to be involved, HLA-A, HLA-DQA1, granzyme B,
HLA-F, HLA-E, perforin 1, HLA-DOA, HLA-DMB and HLA-DPB1, were all
downregulated. These results may indicate similarities between GDM
and T1D. However, it has been reported that pregnant women have an
increased risk of developing T2D. GDM may share common
characteristics with T1D, including insulin resistance and the
immune response; the findings of the present study are therefore in
accordance with those of previous ones, which appeared to have
detected autoimmune phenomena in patients with GDM (29). Evangelista et al (30) reported that gene expression signatures
of GDM patients were closer to those of T1D patients than to those
of T2D patients, which may provide an explanation for the findings
of the present study.
In the present study, KEGG analysis also suggested
that pathways associated with antigen processing and presentation
were dysregulated in the GDM group. The findings highlight a
significant role of HLA genes in the adipose tissue of women with
GDM. A general downregulation of HLA genes has been observed among
placentas and blood samples from women with GDM (31). In general, GDM has been associated with
increased anti-HLA-class II antibodies in the maternal circulation
and reduced tolerance towards alloantigen through inflammatory
activation (32).
In the present study, the relevant terms in which
DEGs were enriched were antigen processing and presentation, immune
response and response to nutrients. Maternal over-nutrition has
been reported to be associated with elevated triglyceride levels,
increased inflammatory markers and fatty livers in offspring
(33). GDM is reconsidered to be a
state of chronic, low-grade inflammation and is, distinct from an
acute pro-inflammatory response, primarily triggered by metabolites
and nutrients, leading to systemic insulin resistance.
As for NK cell-mediated cytotoxicity pathways, their
participation in GDM progression is expected due to the proposed
involvement of the NK pathway in T1D (34). It is thought that the NK pathway causes
the release of cytotoxic granules, penetration of effector proteins
through the cell membrane and subsequent induction of apoptosis.
Considering adipose tissue being the main inflammatory organ and
highly expressing numerous inflammatory mediators, it is expected
that inflammatory pathways are associated with its role in GDM. It
has been speculated that cytokine-mediated inflammation leads to
metabolic abnormalities by increasing insulin resistance in
patients (35). Numerous pre-clinical
and clinical studies support the notion that obesity-induced
inflammation may be a specific type of inflammation resulting from
overnutrition and stress pathways that drive abnormal metabolic
homeostasis and lead to insulin resistance (36). It has been widely evidenced that
obesity is a major cause of impaired insulin signaling (37–39).
However, the precise molecular mechanisms of the obesity-induced
inflammation causing insulin resistance have remained to be
elucidated.
As for focal adhesion, the present study revealed a
hub function in the Path-Act-Net of upregulated DEGs. The focal
adhesion pathway is closely associated with the insulin signaling
pathway. Focal adhesion proteins are integrin-rich microdomains,
which transmit mechanical signals from the ECM to activate
signaling pathways inside the cell and structurally link the
cytoskeleton to the ECM. Cell adhesion and focal adhesion kinases
regulate insulin receptor substrate-1 expression (40). Focal adhesion kinase is a substrate for
the insulin and insulin-like growth factor-1 tyrosine kinase
receptors (41–43). For instance, Bisht and Dey (43) reported that focal adhesion kinase
regulates insulin resistance in skeletal muscle. Zhang et al
(44) also reported that the focal
adhesion pathway was of high significance in the functional
interaction network of DEGs in adipose tissues of patients with
insulin resistance.
The results of the KEGG and GO enrichment analyses
suggested that antigen processing and presentation was
significantly different in OVAT tissues of pregnant women with and
without GDM, suggesting that immune responses are involved in GDM.
This may be due to the fact that the OVAT tissues are under
pathological stress and hypoxia, leading to immune-associated
responses, such as the feto-matenal tolerance, innate immunity and
inflammation response. Based on the previously suggested
involvement of immune responses T1D, it is likely that it is also
involved in the development and progression of GDM.
In conclusion, the results of the present study
suggested that hub genes of the Gene-Act-Net, including ITGB1,
AKT3, ITGAV and FGFR3, in OVAT are associated with GDM.
Furthermore, molecular signaling pathways associated with immune
responses, inflammatory processes, focal adhesion and signal
transduction were identified to be closely associated with GDM in
Chinese women. These candidate genes and pathways provided novel
insight into the pathogenesis of insulin resistance in OVAT of GDM
patents. However, the present study was limited by the low number
of study subjects; furthermore, the differential expression of
certain genes may have been a result, but not necessarily a cause
of GDM. Further experimental studies are therefore required to
confirm or expand the conclusions of the present study.
Acknowledgements
The present study was supported by the Natural
Science Foundation of China (no. 81360103) and the Application of
Basic Research Project of Yunnan Province (no. 2012FB039).
References
|
1
|
Kjos SL and Buchanan TA: Gestational
diabetes mellitus. N Engl J Med. 341:1749–1756. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kasuga M: Insulin resistance and
pancreatic beta cell failure. J Clin Invest. 116:1756–1760. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
White MF: Insulin signaling in health and
disease. Science. 302:1710–1711. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wójcik M, Chmielewska-Kassassir M,
Grzywnowicz K, Woźniak L and Cypryk K: The relationship between
adipose tissue-derived hormones and gestational diabetes mellitus
(GDM). Endokrynol Pol. 65:134–142. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fasshauer M, Blüher M and Stumvoll M:
Adipokines in gestational diabetes. Lancet Diabetes Endocrinol.
2:488–499. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ibrahim MM: Subcutaneous and visceral
adipose tissue: Structural and functional differences. Obes Rev.
11:11–18. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Insenser M, Montes-Nieto R, Vilarrasa N,
Lecube A, Simó R, Vendrell J and Escobar-Morreale HF: A nontargeted
proteomic approach to the study of visceral and subcutaneous
adipose tissue in human obesity. Mol Cell Endocrinol. 363:10–19.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sun G: Application of DNA microarrays in
the study of human obesity and type 2 diabetes. OMICS. 11:25–40.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Draghici S, Khatri P, Eklund AC and
Szallasi Z: Reliability and reproducibility issues in DNA
microarray measurements. Trends Genet. 22:101–109. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Allison DB, Cui X, Page GP and Sabripour
M: Microarray data analysis: From disarray to consolidation and
consensus. Nat Rev Genet. 7:55–65. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guilherme A, Virbasius JV, Puri V and
Czech MP: Adipocyte dysfunctions linking obesity to insulin
resistance and type 2 diabetes. Nat Rev Mol Cell Biol. 9:367–377.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Maassen JA, Romijn JA and Heine RJ: Fatty
acid-induced mitochondrial uncoupling in adipocytes as a key
protective factor against insulin resistance and beta cell
dysfunction: A new concept in the pathogenesis of
obesity-associated type 2 diabetes mellitus. Diabetologia.
50:2036–2041. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ma Y, Gao J, Yin J, Gu L, Liu X, Chen S,
Huang Q, Lu H, Yang Y, Zhou H, et al: Identification of a Novel
Function of Adipocyte Plasma Membrane-Associated Protein (APMAP) in
Gestational Diabetes Mellitus by Proteomic Analysis of Omental
Adipose Tissue. J Proteome Res. 15:628–637. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Oliva K, Barker G, Rice GE, Bailey MJ and
Lappas M: 2D-DIGE to identify proteins associated with gestational
diabetes in omental adipose tissue. J Endocrinol. 218:165–178.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Evangelou M, Smyth DJ, Fortune MD, Burren
OS, Walker NM, Guo H, Onengut-Gumuscu S, Chen WM, Concannon P, Rich
SS, et al: A method for gene-based pathway analysis using
genomewide association study summary statistics reveals nine new
type 1 diabetes associations. Genet Epidemiol. 38:661–670. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou F, Huang X, Zhang Z, Chen Y, Liu X,
Xing J and He X: Functional polymorphisms of ITGB1 are associated
with clinical outcome of Chinese patients with resected colorectal
cancer. Cancer Chemother Pharmacol. 75:1207–1215. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang L, Zhang Y, Lv W, Lu J, Mu J, Liu Y
and Dong P: Long non-coding RNA Linc-ITGB1 knockdown inhibits cell
migration and invasion in GBC-SD/M and GBC-SD gallbladder cancer
cell lines. Chem Biol Drug Des. 86:1064–1071. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fabbri C, Crisafulli C, Gurwitz D, Stingl
J, Calati R, Albani D, Forloni G, Calabrò M, Martines R, Kasper S,
et al: Neuronal cell adhesion genes and antidepressant response in
three independent samples. Pharmacogenomics J. 15:538–548. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Eves R, Oldham R, Jia L and Mak AS: The
roles of akt isoforms in the regulation of podosome formation in
fibroblasts and extracellular matrix invasion. Cancers (Basel).
7:96–111. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Phung TL, Du W, Xue Q, Ayyaswamy S, Gerald
D, Antonello Z, Nhek S, Perruzzi CA, Acevedo I, Ramanna-Valmiki R,
et al: Akt1 and akt3 exert opposing roles in the regulation of
vascular tumor growth. Cancer Res. 75:40–50. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Waisberg J, De Souza Viana L, Affonso
Junior RJ, Silva SR, Denadai MV, Margeotto FB, De Souza CS and
Matos D: Overexpression of the ITGAV gene is associated with
progression and spread of colorectal cancer. Anticancer Res.
34:5599–5607. 2014.PubMed/NCBI
|
|
22
|
Denadai MV, Viana LS, Affonso RJ Jr, Silva
SR, Oliveira ID, Toledo SR and Matos D: Expression of integrin
genes and proteins in progression and dissemination of colorectal
adenocarcinoma. BMC Clin Pathol. 13:162013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hosen I, Rachakonda PS, Heidenreich B, de
Verdier PJ, Ryk C, Steineck G, Hemminki K and Kumar R: Mutations in
TERT promoter and FGFR3 and telomere length in bladder cancer. Int
J Cancer. 137:1621–1629. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guancial EA, Werner L, Bellmunt J, Bamias
A, Choueiri TK, Ross R, Schutz FA, Park RS, O'Brien RJ, Hirsch MS,
et al: FGFR3 expression in primary and metastatic urothelial
carcinoma of the bladder. Cancer Med. 3:835–844. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Noctor E, Crowe C, Carmody LA, Kirwan B,
O'Dea A, Glynn LG, McGuire BE, O'Shea PM and Dunne FP:
ATLANTIC-DIP: Prevalence of metabolic syndrome and insulin
resistance in women with previous gestational diabetes mellitus by
International Association of Diabetes in Pregnancy Study Groups
criteria. Acta Diabetol. 52:153–160. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Koning SH, Hoogenberg K, Scheuneman KA,
Baas MG, Korteweg FJ, Sollie KM, Schering BJ, van Loon AJ,
Wolffenbuttel BH, van den Berg PP, et al: Neonatal and obstetric
outcomes in diet- and insulin-treated women with gestational
diabetes mellitus: A retrospective study. BMC Endocr Disord.
16:522016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhao YH, Wang DP, Zhang LL, Zhang F, Wang
DM and Zhang WY: Genomic expression profiles of blood and placenta
reveal significant immune-related pathways and categories in
Chinese women with gestational diabetes mellitus. Diabet Med.
28:237–246. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang L, Roh YS, Song J, Zhang B, Liu C,
Loomba R and Seki E: Transforming growth factor beta signaling in
hepatocytes participates in steatohepatitis through regulation of
cell death and lipid metabolism in mice. Hepatology. 59:483–495.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mauricio D and de Leiva A: Autoimmune
gestational diabetes mellitus: A distinct clinical entity? Diabetes
Metab Res Rev. 17:422–428. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Evangelista AF, Collares CV, Xavier DJ,
Macedo C, Manoel-Caetano FS, Rassi DM, Foss-Freitas MC, Foss MC,
Sakamoto-Hojo ET, Nguyen C, et al: Integrative analysis of the
transcriptome profiles observed in type 1, type 2 and gestational
diabetes mellitus reveals the role of inflammation. BMC Med
Genomics. 7:282014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Binder AM, LaRocca J, Lesseur C, Marsit CJ
and Michels KB: Epigenome-wide and transcriptome-wide analyses
reveal gestational diabetes is associated with alterations in the
human leukocyte antigen complex. Clin Epigenetics. 7:792015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Steinborn A, Saran G, Schneider A, Fersis
N, Sohn C and Schmitt E: The presence of gestational diabetes is
associated with increased detection of anti-HLA-class II antibodies
in the maternal circulation. Am J Reprod Immunol. 56:124–134. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Oben JA, Mouralidarane A, Samuelsson AM,
Matthews PJ, Morgan ML, McKee C, Soeda J, Fernandez-Twinn DS,
Martin-Gronert MS, Ozanne SE, et al: Maternal obesity during
pregnancy and lactation programs the development of offspring
non-alcoholic fatty liver disease in mice. J Hepatol. 52:913–920.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dotta F, Fondelli C and Falorni A: Can NK
cells be a therapeutic target in human type 1 diabetes? Eur J
Immunol. 38:2961–2963. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hauguel-de Mouzon S and Guerre-Millo M:
The placenta cytokine network and inflammatory signals. Placenta.
27:794–798. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lee BC and Lee J: Cellular and molecular
players in adipose tissue inflammation in the development of
obesity-induced insulin resistance. Biochim Biophys Acta.
1842:446–462. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lumeng CN and Saltiel AR: Inflammatory
links between obesity and metabolic disease. J Clin Invest.
121:2111–2117. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hursting SD and Dunlap SM: Obesity,
metabolic dysregulation, and cancer: A growing concern and an
inflammatory (and microenvironmental) issue. Ann N Y Acad Sci.
1271:82–87. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Balsan GA, Vieira JL, Oliveira AM and
Portal VL: Relationship between adiponectin, obesity and insulin
resistance. Rev Assoc Med Bras 1992. 61:72–80. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lebrun P, Baron V, Hauck CR, Schlaepfer DD
and Van Obberghen E: Cell adhesion and focal adhesion kinase
regulate insulin receptor substrate-1 expression. J Biol Chem.
275:38371–38377. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gupta A and Dey CS: PTEN, a widely known
negative regulator of insulin/PI3K signaling, positively regulates
neuronal insulin resistance. Mol Biol Cell. 23:3882–3898. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gupta A, Bisht B and Dey CS: Focal
adhesion kinase negatively regulates neuronal insulin resistance.
Biochim Biophys Acta. 1822:1030–1037. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bisht B and Dey CS: Focal Adhesion Kinase
contributes to insulin-induced actin reorganization into a mesh
harboring Glucose transporter-4 in insulin resistant skeletal
muscle cells. BMC Cell Biol. 9:482008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang L, Cui Y, Fu F, Li Z, Pan X, Li H
and Li L: An insight into the key genes and biological functions
associated with insulin resistance in adipose tissue with
microarray technology. Mol Med Rep. 11:1963–1967. 2015.PubMed/NCBI
|