Introduction
Opioids are wildly administered for treatment and
control of moderate and severe pains. Their efficiency is dependent
on the ability to mimic endogenous peptides on the opioid
receptors. Opioid receptors are classified into three types of
receptor (µ, δ and κ) characterized by molecular cloning and these
receptors have been investigated in numerous pharmacological
studies (1,2). The Mu opioid receptor (MOR) is involved
in morphine-induced analgesia, tolerance and dependence according
to pharmacological studies and analysis of MOR knockout mice
(3–5).
Upon opioid binding, MOR couples with G-protein-coupled receptors,
regulates adenylyl cyclase, intracellular calcium and
mitogen-activated protein kinase, then triggers a cascade of
intracellular events (6). MOR is a
major molecular target of analgesic drugs, morphine, heroin,
methadone and fentanyl (7).
MOR is predominantly expressed in the central
nervous system, and differential expression of MOR is dependent on
receptors of varying densities in different regions (8,9). Individual
human and mouse strains differ in their responses to pain and
opiate drugs (10,11). Although humans and mice exhibit a
different response to opiates, to the best of our knowledge, there
have been no studies on these differential responses. Only one
study indicated that single nucleotide polymorphisms (SNPs) are
associated with mice with differences in morphine preference
(12).
Mouse MOR (mMOR) gene expression is regulated by
distal and proximal promoters (DP and PP, respectively). The two
promoters are similar to housekeeping genes that are lacking a TATA
box. The distal promoter is less active, by 20-fold, than the PP in
adult and embryonic mouse brains as determined using reverse
transcription-polymerase chain reaction (RT-PCR) (13). The proximal core promoter of the mMOR
gene contains the polypyrimidine/polypurine (PPy/u) region, and the
PP of the human MOR (hMOR) gene contains a similar PPy/u region
that is located nearby at transcription initiation site (14,15). The
PPy/u region of the mMOR gene promoter strongly activates the MOR
gene and contains a 26-bp CT-rich region with overlapping single-
and double-stranded DNA sequences, and multiple binding sites for
Sp1, Sp3 and single-stranded binding proteins (16,17).
Regulation of hMOR gene expression in neuronal cells is not well
understood compared with mMOR gene regulation. The hMOR promoter
contains a deferoxamine-response CT-rich region that is located
close to the translational initiation site (18). PPy/u motifs are the common sequence in
eukaryotic cells (19) and possess
special chemical properties, including a non-B DNA conformation
sensitive to S1 nuclease, a triple-stranded forming DNA structure
and guanine-rich guanosine, and a G-quartet structure that is often
observed at the centromere and telomere (19).
In the current study, the structural conformation of
the PPy/u motif and poly(C) binding protein (PCBP1), α-complex
protein 1 (α-CP1) were demonstrated to regulate different
transcriptional activation via the PPy/u motifs on human and mouse
MOR genes. To the best of our knowledge, this is the first
comparative investigation of the mouse and human MOR gene
expression that focuses on the key transcriptional regulatory
element sequence PPy/u motif and the α-CP1 protein. Furthermore,
the reasons for and theoretical backgrounds regarding why humans
and mice exhibit different responses to pain and opiate drugs are
explained.
Materials and methods
Plasmid construction
The mouse promoter construct p336/306 was generated
by ligating an annealed double-stranded oligonucleotide into the
SacI and HindIII sites of a pGL3-basic vector (Promega Corporation,
Madison, WI, USA) using the following oligonucleotide sequences:
Sense, 5′-ATTGAGCTCTCCACTCCTTCTCTCTCCTCCCTCCCCTCTAAAGCTTTTC-3′)
containing a SacI and HindIII site (underlined) and antisense,
5′-GAAAAGCTTTAGAGGGGAGGGAGGAGAGAGAAGGAGTGGAGAGCTCAAT-3′
containing a HindIII and SacI site (underlined). The human promoter
construct p322/292 was generated by ligating an annealed
double-stranded oligonucleotide into the SacI and HindIII sites of
pGL3-basic vector using the following oligonucleotide sequences:
Sense, 5′-ATTGAGCTCTCCACCCCTTTTCCCTCCTCCCTCCCTTCCAAAGCTTTTC-3′
containing a SacI and HindIII site (underlined) and antisense,
GAAAAGCTTTGGAAGGGAGGGAGGAGGGAAAAGGGGTGGAGAGCTCAAT-3′
containing a HindIII and SacI site (underlined). To clone the α-CP1
gene, total RNA was isolated from mouse NS20Y cells obtained from
the American Type Culture Collection (ATCC; Manassas, VA, USA). RNA
was treated with RNase-free DNase (Promega Corporation) according
to the manufacturer's instructions. RT-PCR was performed using the
OneStep RT-PCR kit (Qiagen, Inc., Valencia, CA, USA). PCR was
performed with primers that were designed using the gene sequence
information for each protein: α-CP1 (Gene ID, 13435897): Sense
primer, 5′-CCATGGACGCCGGTGTGACTGA-3′ and antisense primer,
5′-GCTGCACCCCATCCCCTTCTC-3′. The PCR conditions were as follows:
94°C for 3 min; 35 cycles of 94°C for 1 min, 55°C for 1 min, and
72°C for 1 min; and 72°C for 10 min. RT-PCR products were excised
from a 1% agarose gel, purified using a QIAQuick gel extraction kit
(Qiagen, Inc.) and cloned into a pCRII-TOPO vector (Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA). The candidate
plasmids containing inserts of the correct size were confirmed
using restriction enzyme digestion and DNA sequencing on an ABI
3100 sequencer (Applied Biosystems; Thermo Fisher Scientific,
Inc.). For the transient expression studies, the α-CP1 gene was
cloned by digesting the above-mentioned pCRII-TOPO α-CP1 clone with
5′-HindIII and 3′-XhoI into the same sites of a pcDNA4 vector
(Invitrogen; Thermo Fisher Scientific, Inc.), generating a
pcDNA4-α-CP1 plasmid. DNA sequences of all constructs were
confirmed using DNA sequencing. For the protein expression studies
in Escherichia coli, the α-CP1 gene was cloned by digesting the
above-mentioned pcDNA4-α-CP1 plasmid with 5′-HindIII and 3′-XhoI
into the same sites of a pET21b vector (EMD Millipore, Billerica,
MA, USA), generating a pET21b-α-CP1 plasmid. The DNA sequences of
all constructs were confirmed using DNA sequencing.
α-CP1 protein expression
The α-CP1 protein expression was performed as
described previously (20). The
protein was expressed in a Lysogeny broth medium (Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) containing ampicillin (50 µg/ml).
To obtain the protein, several cell growth conditions were
generated by varying the temperature (16, 30 and 37°C) and
isopropyl β-D-1-thiogalactopyranoside (IPTG) concentration (0.1,
0.5 and 1 mM). Typically, 2 ml overnight culture was added to 100
ml medium and incubated with vigorous shaking at ~37°C. When the
culture reached optical density (OD)600=0.5, protein
expression was induced with 1 mM IPTG. Subsequent to induction, the
samples were further incubated at 37°C for 4 h. The cells were
harvested by centrifugation at 10,000 × g for 10 min at 4°C, washed
with TE buffer (10 mM Tris-HCl and 1 mM EDTA; pH 8.0) and stored at
−80°C.
Folding of the α-CP1 protein
The folding of the α-CP1 protein was performed as
described previously (20). The twice
water-washed inclusion bodies were resuspended in 5 volumes of
Buffer C (20 mM Tris-HCl, 1 mM EDTA, 10 mM DTT and 8 M Urea; pH
7.0), stirred at room temperature for 60 min and centrifuged at
10,000 × g for 15 min at room temperature. The pellet was discarded
and the supernatant (5–10 mg/ml) was collected in a fresh tube. The
refolding experiments were performed using protein-folding
spin-columns following the manufacturer's recommendation
(ProFoldin, Hudson, MA, USA).
Preparation of inclusion bodies and
purification of recombinant α-CP1 protein
The preparation of inclusion bodies and purification
of recombinant of the α-CP1 protein were performed as described
previously (20). The cell pellet was
resuspended in 30 ml Buffer A (20 mM Tris-HCl, 100 mM NaCl and 1 mM
PMSF; pH 7.0) and sonicated at 4°C with 5 cycles. The lysate was
centrifuged at 10,000 × g for 15 min at 4°C. The pellet was
resuspended in 5 volumes of Buffer A, stirred at room temperature
for 5 min and centrifuged at 10,000 × g for 15 min at 4°C. The
inclusion bodies were then washed three times with 10 volumes of 20
mM Tris-HCl containing 100 mM NaCl at pH 7.0. The inclusion body
pellet was resuspended in 30 ml Buffer B [50 mM
NaH2PO4, 300 mM NaCl (pH 8.0) and 8 M urea]
to solubilize the inclusion bodies. Sonication was necessary to
suspend the pellet. The suspension was centrifuged at 10,000 × g
for 20 min and the supernatant was transferred to fresh clean
tubes. The supernatant was then added to an equilibrated Ni-NTA
column (Qiagen, Inc.) and allowed to drain. The column was washed
with Buffer B, and the 6x His-tagged α-CP1 was eluted using an
elution buffer [50 mM NaH2PO4, 300 mM NaCl,
250 mM imidazole (pH 8.0) and 8 M urea]. Anti-His antibodies were
purchased from Sigma-Aldrich; Merck KGaA. To determine which
fractions contain the His-tagged α-CP1, an aliquot of each sample
was analyzed using 10% SDS-PAGE.
SDS-PAGE, in-gel tryptic digestion and
matrix-assisted laser desorption ionization-time of flight
(MALDI-TOF) mass spectrometric analysis of α-CP1
The purified α-CP1 protein was resolved on a 10%
SDS-PAGE gel. The Coomassie blue-stained gel was destained, and a
gel slice containing the band of interest was subjected to in-gel
tryptic digestion as described previously (20,21). The
tryptic peptides were extracted with 5% acetic acid, followed by 5%
acetic acid and 50% acetonitrile. The samples were dissolved in 5%
acetic acid and desalted using ZipTip™ C18 reverse-phase desalting
Eppendorf tips (EMD Millipore). The peptides were eluted with 2%
acetonitrile containing 0.1% TFA in a volume of 20 µl. The samples
were analyzed using a MALDI-TOF mass spectrometer (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The masses of the
monoisotopic peaks were compared with a theoretical digestion of
the protein by trypsin. Mascot database searching software (Matrix
Science; http://www.matrixscience.com) was
used to identify the α-CP1 protein.
DNA electrophoretic mobility shift
assay (EMSA)
The EMSA was performed as described previously
(20,22). The polypyrimidine/polypurine (PPy/u)
oligonucleotide, single-stranded probe
(5′-TCCACTCCTTCTCTCTCCTCCCTCCCCTCTA-3′) was end-labeled with
[γ-32P] dATP. The free nucleotides were separated using
centrifugation at 1,100 × g for 4 min at room temperature through a
Sephadex G-25 column (Roche Diagnostcs, Indianapolis, IN, USA). The
end-labeled ssDNA probes were incubated with recombinant α-CP1 (0.5
µg) in a final volume of 20 µl EMSA buffer [10 mM Tris (pH 7.5), 5%
glycerol, 1 mM EDTA, 50 mM NaCl, 1 mM DTT, 0.1 mg/ml poly (dI-dC)]
at room temperature for 20 min. For the oligonucleotide competition
analyses, a 100-fold molar excess of a cold competitor
oligonucleotide was added to the mixture prior to adding the probe.
The reactions were then incubated at 4°C for 30 min. The reaction
mixtures were electrophoresed at 160 V for 2 h on a non-denaturing
4% polyacrylamide gel in 0.5X TBE (45 mM Tris-borate and 1 mM EDTA)
at 4°C and visualized using autoradiography.
S1 nuclease sensitivity assay
The pGL-basic plasmids, p322/292 and p336/306, were
digested with various quantities of the S1 nuclease (Promega
Corporation) in S1 nuclease buffer for 15 min at 37°C as described
previously (20). The digestion was
terminated using phenol/chloroform extraction and the plasmids were
recovered by precipitation. The resulting S1-treated plasmids were
digested further using XbaI (Promega Corporation) and the products
were resolved using electrophoresis on a 1% agarose gel at 100 V
for 1 h.
Transient transfection and reporter
gene assays
Mouse neuroblastoma NS20Y cells and human neuronal
NMB cells obtained from the ATCC were grown in Dulbecco's modified
Eagle's medium supplemented with 10% heat-inactivated fetal bovine
serum (GE Healthcare Life Sciences, Chalfont, UK) at 37°C in a
humidified atmosphere of 5% CO2. NS20Y cells were plated
in 6-well dishes at a concentration of 0.5×106
cells/well and cultured overnight before transfection. Equimolar
concentrations of various plasmids were transfected using the
Effectene transfection reagent (Qiagen, Inc.) as described
previously (20,23). Briefly, for the luciferase analysis of
the p336/306 and p322/292 promoters, 0.5 µg of the reporter
plasmids was combined with the Effectene transfection reagent for
10 min before being added to the NS20Y cells. Forty-eight h after
transfection, the cells that were grown to confluence were washed
once with phosphate-buffered saline and lysed with lysis buffer
(Promega Corporation). To correct for differences in transfection
efficiency, a one-fifth molar ratio of pCH110 (GE Healthcare Life
Sciences) containing the β-galactosidase gene under the SV40
promoter was included in each transfection for normalization. The
luciferase (Promega Corporation) and β-galactosidase (Promega
Corporation) activities of each lysate were determined according to
the manufacturer's recommendations.
RT-PCR and heterologous expression of
α-CP1
Total RNA was isolated using TRI Reagent (Molecular
Research Center, Inc., Cincinnati, OH, USA) according to the
supplier's protocol. For RT-PCR, 2 µg total RNA and the OneStep
RT-PCR reagent (Qiagen, Inc.) were used. The PCR cycle conditions
consisted of 95°C for 1 min, 60°C for 1 min and 72°C for 1 min
followed by a 10-min extension at 72°C. Mouse-specific primers were
as follows: 5′-CATCAAAGCACTGATCACGATTCC-3′ and
5′-TAGGGCAATGGAGCAGTTTCTGC-3′ for MOR; 5′-TGGCCTTAGGGTGCAGGGGG-3′
and 5′-GTGGGCCGCTCTAGGCACCA-3′ for β-actin. The human-specific
primers were as follows: 5′-CCTTCCTGGGCATGGAGTCCTG-3′ and
5′-TACAGCGAGGCCAGGATGG-3′ for β-actin; 5′-CTGGAAGGGCAGGGTACTGGTG-3′
and 5′-CTGCCCCCACGAACGCCAGCAAT-3′ for MOR.
Statistical analysis
All data were presented as the mean ± standard
deviation. Data were analyzed using Student's t-test. P<0.05 was
considered to indicate a statistically significant difference and
GraphPad Prism 5 Software (GraphPad Software, Inc., La Jolla, CA,
USA) was used to perform the analyses.
Results
Promoter structure and comparison of
the minimum PP sequence of human and mouse MOR gene
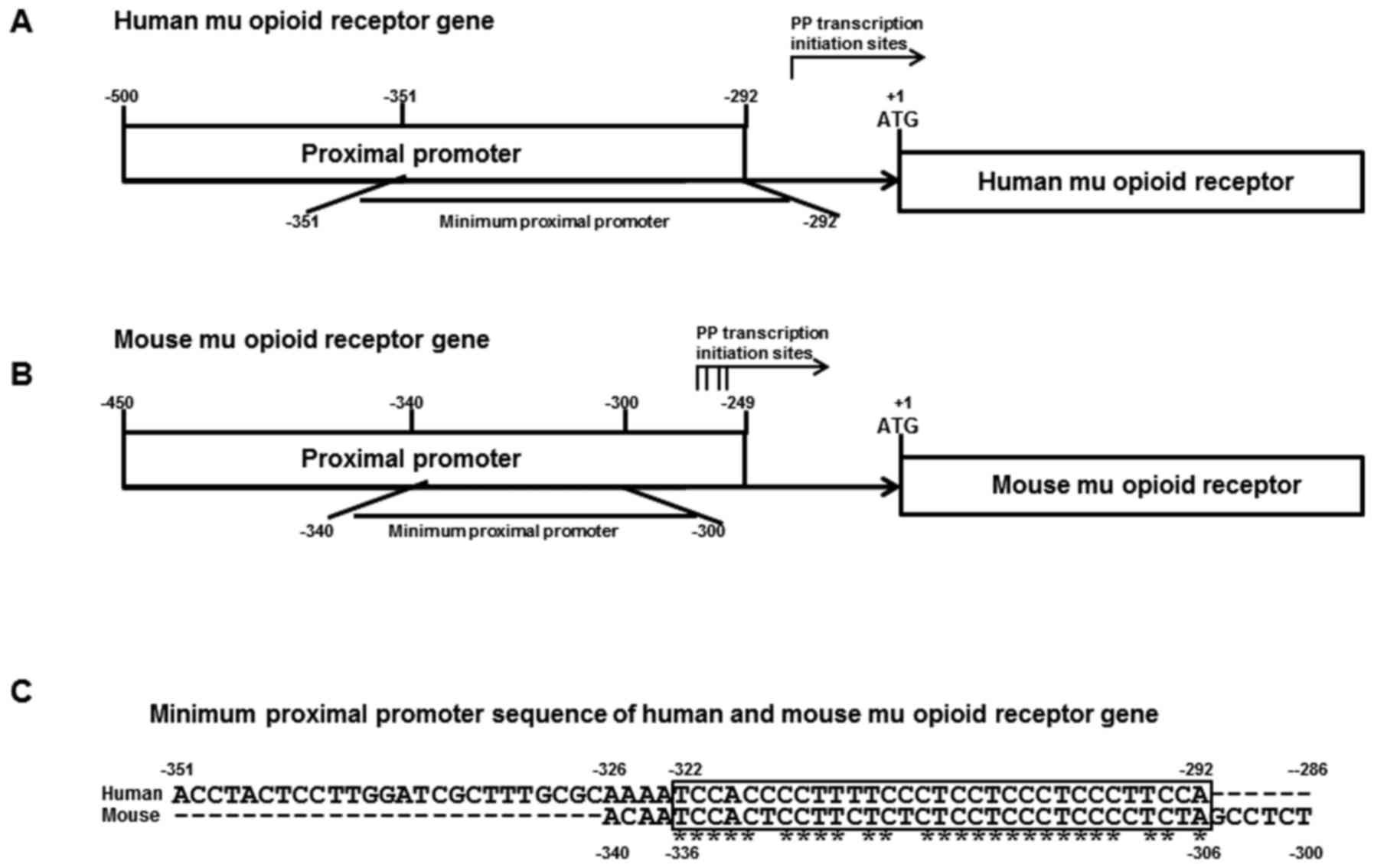
Transcription of the mMOR gene starts at four sites
located between −291 and −268 of the MOR gene using two promoters:
The PP (−450 to −249) and the DP (−1,326 to +1; Fig. 1B). The PP was responsible for major MOR
gene activity (~95%) in the mouse brain. The regulatory elements of
the PP contained PPy/u and a canonical Sp1 binding site. The PPyy/u
exhibited an ssDNA conformational structure (15). Transcription of the hMOR gene starts at
the −256 site and the hMOR gene also uses two promoters: The PP
(−500 to −292) and the DP (−2,388 to +1; Fig. 1A). The regulatory elements of the human
PP contained PPy/u and a canonical Sp1/3 binding site. Structural
analysis of MOR PPy/u indicated that mMOR PPy/u is highly
homologous to hMOR PPy/u (84%; Fig.
1C).
Differential S1 nuclease sensitivity
and promoter activity of human and mouse MOR promoters containing
PPy/u motifs
ssDNA structures derived from the non-B DNA form or
intracellular triple helix structures are sensitive to low
concentrations of S1 nuclease (15).
In order to analyze the structural differences of human and mouse
MOR PPy/u motifs, an S1 nuclease treatment was performed in the
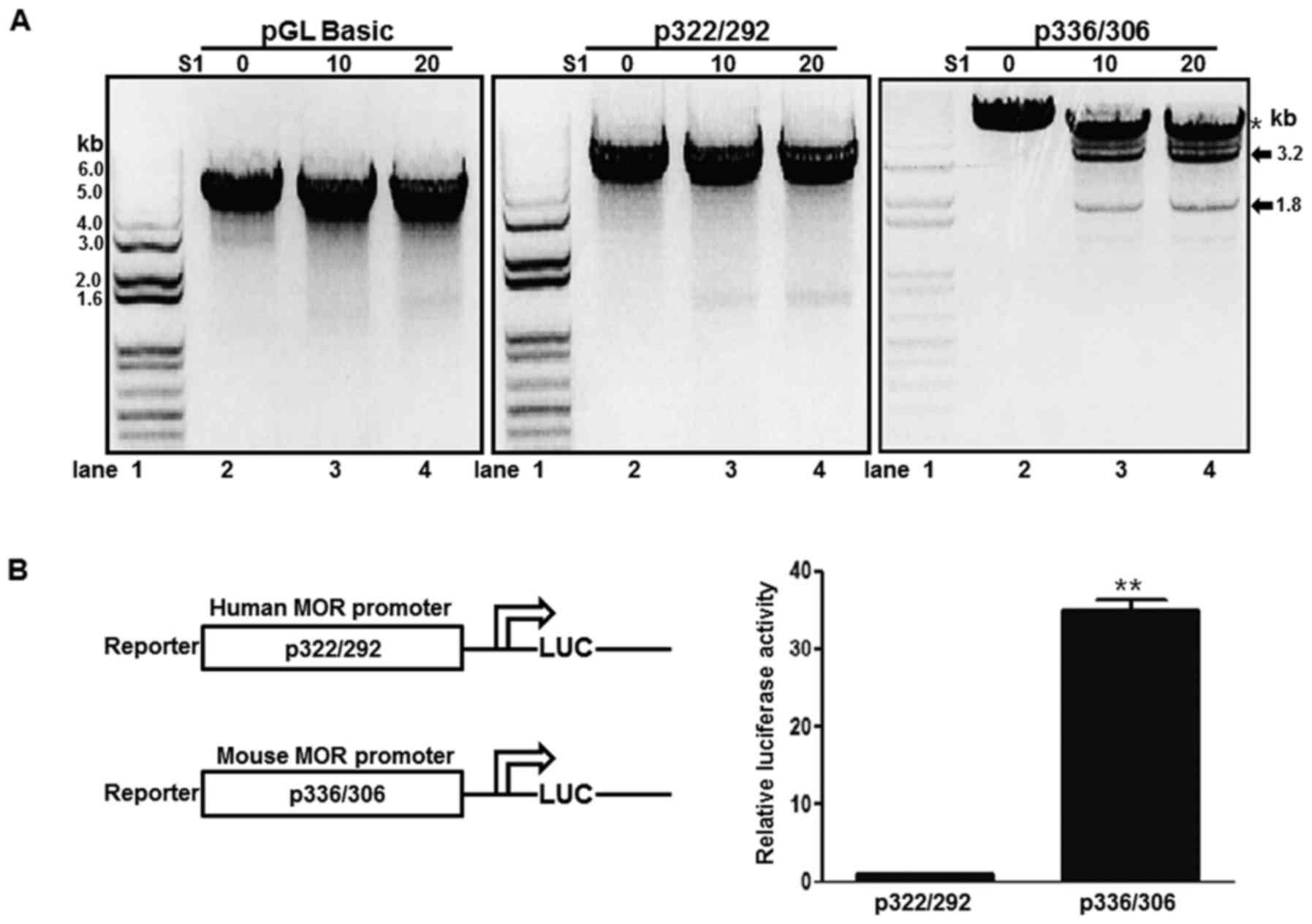
current study. A p322/292 plasmid containing the human PPy/u motif
was treated with S1 nuclease and digested with XbaI. In the
presence of S1 nuclease, the XbaI treatment produced a 5-kb linear
DNA (Fig. 2A, middle panel). In
addition, the pGL-basic plasmid demonstrated a similar result
(Fig. 2A, left panel). A p336/306
plasmid containing the mouse PPy/u motif was treated with S1
nuclease and digested with XbaI. Two DNA bands, 3.2 and 1.8 kb,
were produced and the band density was increased with increasing
quantities of S1 nuclease (Fig. 2A,
right panel). These results indicate that the mouse PPy/u motif is
an ssDNA structure, whereas the human motif is a double-stranded
DNA, as determined according to the S1 nuclease assay. To confirm
the association between structure and gene expression, a reporter
assay was used. The mouse construct in the p336/306 plasmid
exhibited strong promoter activity when compared with the human
construct p322/292 promoter activity in mouse NS20Y cells (Fig. 2B).
Expression, folding and purification
of α-CP1
α-CP1 is a poly(C) binding protein, which is an
ssDNA binding protein. The mouse α-CP1 gene was cloned into the
pET21b vector and the recombinant α-CP1 protein contained a
C-terminal 6X His tag. To obtain the optimal condition for
expressing the soluble α-CP1 protein in E. coli BL21 (DE3), various
conditions, including temperature for cell growth, cell culture
media and induction times were evaluated. However, all conditions
produced insoluble α-CP1. To obtain the maximum production of
insoluble α-CP1, the expression conditions were optimized using a
variety of options including temperatures and IPTG concentrations.
Under optimal conditions, production of insoluble α-CP1 was ~30%
(Fig. 3A). To optimize the α-CP1
protein folding conditions, a spin-column protein folding screening
kit was used, which included nine different protein-folding columns
that represent the nine most promising folding conditions. The #8
column from the kit was selected as the optimal folding condition
of the denatured α-CP1 protein (Fig.
3B). Using Ni-NTA His-binding resin, denatured α-CP1 protein
was purified with 8 M urea. The purified α-CP1 protein was
subsequently folded using the spin-column protein folding screening
kit #8 column. The purified and folded α-CP1 protein was confirmed
using 10% SDS-PAGE and Coomassie staining (Fig. 3C).
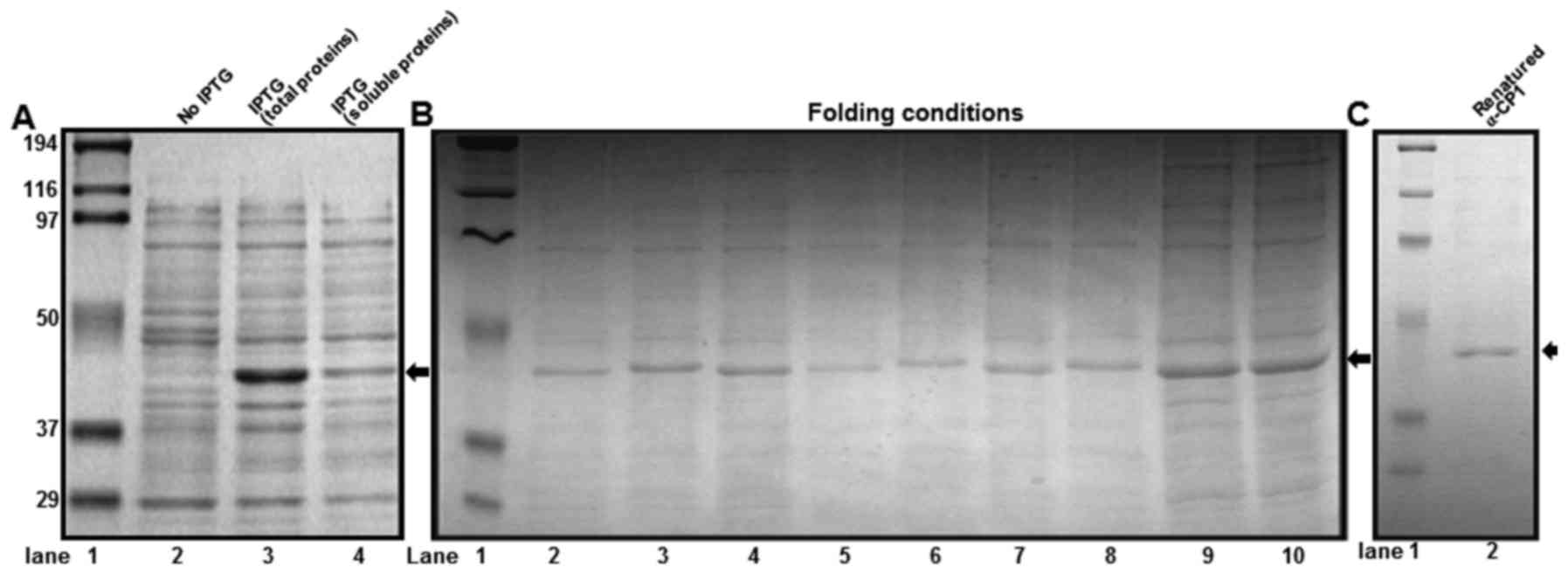 | Figure 3.Expression, purification and folding
conditions of recombinant α-CP1 protein. (A) 10% SDS-PAGE analysis
of recombinant mouse α-CP1 protein expressed by an Escherichia coli
(E. coli)-induced expression system (1 mM IPTG at 37°C). Lane 1,
protein molecular weight markers; lane 2, 10 µl total protein from
E. coli BL21 (DE3)/pET21b-α-CP1 before induction; lane 3, 10 µl
total protein from E. coli BL21 (DE3)/pET21b-α-CP1 after induction;
lane 4, 10 µl soluble protein from E. coli BL21 (DE3)/pET21b-α-CP1
after induction. (B) The optimization of folding conditions for the
purified recombinant mouse α-CP1. The solubilized inclusion bodies
(5–10 mg/ml) were processed using a protein-folding spin-column
screening kit. Lane 1, protein molecular weight markers; lanes
2–10, eluates from spin-columns #1-9. (C) 10% SDS-PAGE analysis of
the affinity-purified renatured recombinant mouse α-CP1. Lane 1,
protein molecular weight markers; lane 2, 5 µl refolded and
purified α-CP1 protein. α-CP1, α-complex protein 1; IPTG, isopropyl
β-D-1-thiogalactopyranoside. |
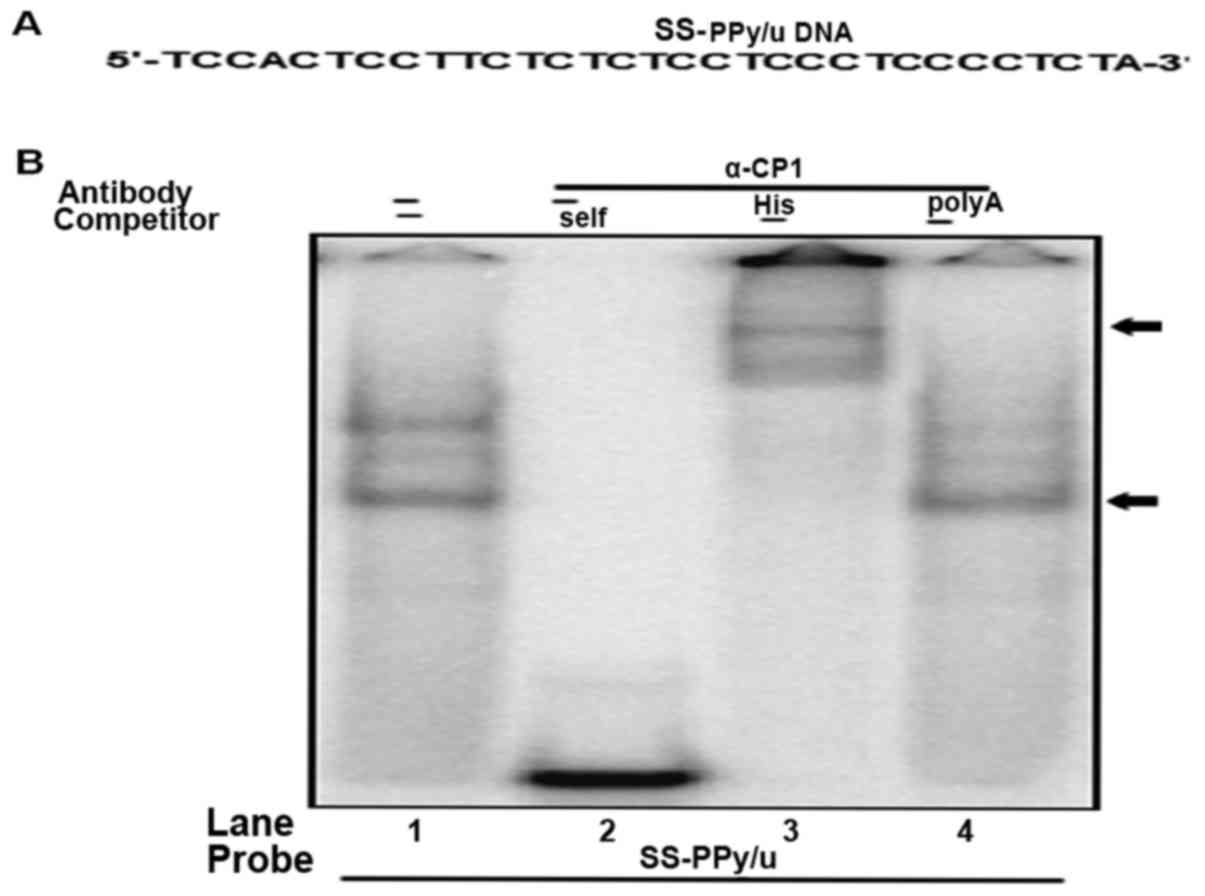
DNA binding property of α-CP1
To determine the physical interaction of purified
α-CP1 protein and single-stranded PPy/u, DNA EMSA was performed
using purified α-CP1 protein and 32P-labeled
single-stranded PPy/u oligonucleotide (Fig. 4A). The specificity of α-CP1
protein/single-stranded PPy/u was verified using an unlabeled
excess self-competitor (Fig. 4B, lane
2) and poly A competitor (Fig. 3D,
lane 4). Furthermore, an anti-His antibody for DNA EMSA was used.
The formation of the α-CP1 protein/single-stranded-PPy/u complex
was abolished by the addition of a His antibody and a super shift,
indicating a specific interaction between α-CP1 protein and
single-stranded PPy/u (Fig. 4B, lane
3).
Differential promoter activity and
endogenous transcription regulation between mouse and human MOR
genes via α-CP1
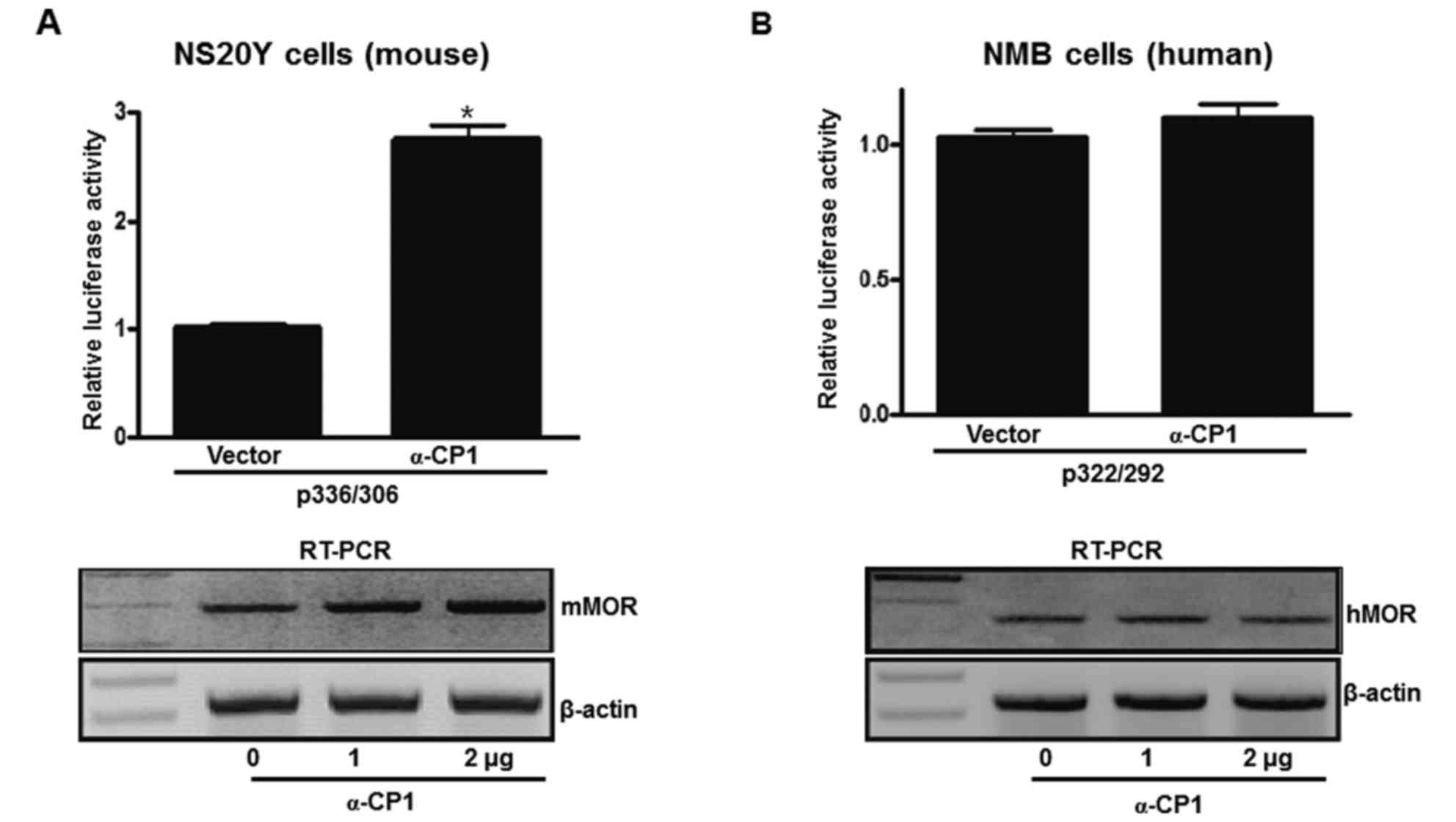
To examine differential promoter activity between
mouse and human MOR genes via the α-CP1 gene, an α-CP1 expression
plasmid and mouse/human PPy/u sequence-containing luciferase
plasmids were co-transfected into mouse neuronal cells (NS20Y).
α-CP1 activated ~280% of p336/306 activity. Additionally, an α-CP1
expression plasmid and human PPy/u sequence-containing luciferase
plasmid was co-transfected into human neuronal cells (NMB). The
α-CP1 did not activate the p322/292 promoter (Fig. 5A and B). To estimate whether
transiently overexpressed α-CP1 results in the upregulation of
endogenous MOR transcripts, RT-PCR analysis using MOR-specific
primers was performed with total RNA from the NS20Y and NMB cells
transfected with varying quantities (0–2 µg) of pcDNA4-αCP1, as
well as with the pcDNA4 vector control. α-CP1 upregulated
endogenous mMOR gene expression in a dose-dependent manner.
However, the hMOR gene was not upregulated by the α-CP1 gene. These
results indicate that α-CP1 acts as a transcriptional activator of
the mMOR gene dependent on the ssDNA structure. In addition, the
α-CP1 protein is important in the regulation of mMOR gene
expression. The human and mouse MOR genes contain a similar PPy/u
sequence, but exhibit differential MOR gene regulation.
Discussion
Comparing two genomic sequences from mice and humans
provides strong resolving power. The conserved sequences of
associated species, namely human and mouse, exhibited similar
functions and gene regulation. The similar sequences offer the
opportunity of using the mouse as an animal model to investigate
human disease and biology (16,24).
Understanding MOR gene expression is particularly important to
establish its analgesic function in humans. Transcriptional
regulation of the MOR gene is predominantly investigated in mice,
and numerous transcription factors [Sp1, Sp3, PCBP, RE-1 silencing
transcription factor and poly (ADP-ribose) polymerase 1] are
involved in mMOR gene regulation (25). In the present study, the PPy/u region,
a key element of MOR gene expression in humans and mouse was
investigated. Species-specific PPy/u motifs differentially confer
S1 nuclease hypersensitivity under acidic pHs and exhibited
transcription regulation. For example, the PPy/u motif of cystic
fibrosis, the transmembrane conductance regulatory gene, is species
specific (26). The mouse PPy/u
element of the MOR gene is highly homologous to its human element
(84%) (Fig. 1C) and the mMOR reporter
exhibited 35-fold increased luciferase activity when compared with
the hMOR reporter (Fig. 2B). The
structural analysis of reporter plasmids using S1 nuclease
indicates that the mouse PPy/u element has a special conformational
structure, namely an ssDNA region (Fig.
2A). The current study demonstrates that the underlying
mechanism of MOR gene activation by the PPy/u motif in mice differs
from that of humans based on different DNA conformations. A
previous study indicated that the mouse PPy/u motif, a single
stranded cis-regulatory element, and PCBP1, an α-CP1 trans-acting
protein, are important for MOR PP activity (15). The present study demonstrated that
α-CP1 enhanced MOR promoter activity and endogenous MOR
transcription via α-CP1 binding to the ssDNA element (17). To the best of our knowledge, this is
the first study to solubilize, fold, purify and produce a
functionally active α-CP1 for DNA EMSA analysis using the E.
coli protein expression system.
In the current study, differential promoter activity
and endogenous transcription regulation of the mouse and human MOR
gene by α-CP1 were investigated. A similar sequence of the PPy/u
motif in the human and mouse MOR promoter exhibited a different
pattern of promoter activity and endogenous transcription
regulation (Fig. 5). Generally, the
promoter of the PPy/u sequence is sensitive to S1 nuclease and its
plasmid is regulated by single-stranded binding proteins (for
example, heterogeneous ribonucleoprotein K and PCBP1-3). However,
the hMOR promoter containing the PPy/u sequence is insensitive to
S1 nuclease and its plasmid was not regulated by single-stranded
binding protein α-CP1. The present study hypothesized that plasmids
containing human PPy/u do not have a single-stranded DNA
conformation.
In conclusion, the differing function of α-CP1 in
humans and mice is determined by its localization in the cell. The
post-transcriptional regulator α-CP1 is localized in the cytosol,
whereas the transcriptional regulator α-CP1 is localized in the
nucleus. Furthermore, transcriptional regulation of the MOR gene is
regulated by α-CP1 localization. To the best of our knowledge, the
present study is the first to compare the human and mouse MOR genes
based on PPy/u motif and α-CP1. The results partially can explain
why MOR gene expression in humans and mice have different responses
to painful stimuli and morphine.
Acknowledgements
The study was supported by the Basic Science
Research Program through National Research Foundation of Korea
(NRF) funded by the Ministry of Education (grant nos.
NRF-2015R1D1A1A01058724, NRF-2011-0006924 and
2016R1A6A1A03012862).
References
|
1
|
Min BH, Augustin LB, Felsheim RF, Fuchs JA
and Loh HH: Genomic structure analysis of promoter sequence of a
mouse mu opioid receptor gene. Proc Natl Acad Sci USA.
91:9081–9085. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wei LN and Loh HH: Regulation of opioid
receptor expression. Curr Opin Pharmacol. 2:69–75. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kieffer BL: Recent advances in molecular
recognition and signal transduction of active peptides: Receptors
for opioid peptides. Cell Mol Neurobiol. 15:615–635. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kieffer BL: Opioids: First lessons from
knockout mice. Trends Pharmacol Sci. 20:19–26. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Law PY, Loh HH and Wei LN: Insights into
the receptor transcription and signaling: Implications in opioid
tolerance and dependence. Neuropharmacology. 47:(Suppl 1).
S300–S311. 2004. View Article : Google Scholar
|
|
6
|
Law PY, Wong YH and Loh HH: Molecular
mechanisms and regulation of opioid receptor signaling. Annu Rev
Pharmacol Toxicol. 40:389–430. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Matthes HW, Maldonado R, Simonin F,
Valverde O, Slowe S, Kitchen I, Befort K, Dierich A, Le Meur M,
Dollé P, et al: Loss of morphine-induced analgesia, reward effect
and withdrawal symptoms in mice lacking the mu-opioid-receptor
gene. Nature. 383:819–823. 1996. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mansour A, Fox CA, Akil H and Watson SJ:
Opioid-receptor mRNA expression in the rat CNS: Anatomical and
functional implications. Trends Neurosci. 18:22–29. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Uhl GR, Sora I and Wang Z: The mu opiate
receptor as a candidate gene for pain: Polymorphisms, variations in
expression, nociception and opiate responses. Proc Natl Acad Sci
USA. 96:7752–7755. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mogil JS: The genetic mediation of
individual differences in sensitivity to pain and its inhibition.
Proc Natl Acad Sci USA. 96:7744–7751. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Korostynski M, Kaminska-Chowaniec D,
Piechota M and Przewlocki R: Gene expression profiling in the
striatum of inbred mouse strains with distinct opioid-related
phenotypes. BMC Genomics. 7:1462006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Doyle GA, Sheng XR, Schwebel CL, Ferraro
TN, Berrettini WH and Buono RJ: Identification and functional
significance of polymorphisms in the mu-opioid receptor gene (Oprm)
promoter of C57BL/6 and DBA/2 mice. Neurosci Res. 55:244–254. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ko JL, Chen HC and Loh HH: Differential
promoter usage of mouse mu-opioid receptor gene during development.
Brain Res Mol Brain Res. 104:184–193. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ko JL, Liu HC, Minnerath SR and Loh HH:
Transcriptional regulation of mouse mu-opioid receptor gene. J Biol
Chem. 273:27678–27685. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ko JL and Loh HH: Single-stranded
DNA-binding complex involved in transcriptional regulation of mouse
mu-opioid receptor gene. J Biol Chem. 276:788–795. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Choe CY, Dong J, Law PY and Loh HH:
Differential gene expression activity among species-specific
polypyrimidine/polypurine motifs in mu opioid receptor gene
promoters. Gene. 471:27–36. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Choi HS, Song KY, Hwang CK, Kim CS, Law
PY, Wei LN and Loh HH: A proteomics approach for identification of
single strand DNA-binding proteins involved in transcriptional
regulation of mouse mu opioid receptor gene. Mol Cell Proteomics.
7:1517–1529. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cook RJ, Karch C, Nahar P, Rivera A and Ko
JL: Effects of desferoxamine-induced hypoxia on neuronal human
mu-opioid receptor gene expression. Biochem Biophys Res Commun.
398:56–61. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schroth GP and Ho PS: Occurrence of
potential cruciform and H-DNA forming sequences in genomic DNA.
Nucleic Acids Res. 23:1977–1983. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kang DH, Song KY, Wei LN, Law PY, Loh HH
and Choi HS: Novel function of the poly(c)-binding protein α-CP2 as
a transcriptional activator that binds to single-stranded DNA
sequences. Int J Mol Med. 32:1187–1194. 2013.PubMed/NCBI
|
|
21
|
Patterson SD and Aebersold R: Mass
spectrometric approaches for the identification of gel-separated
proteins. Electrophoresis. 16:1791–1814. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hwang CK, Wu X, Wang G, Kim CS and Loh HH:
Mouse mu opioid receptor distal promoter transcriptional regulation
by SOX proteins. J Biol Chem. 278:3742–3750. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Choi HS, Hwang CK, Kim CS, Song KY, Law
PY, Wei LN and Loh HH: Transcriptional regulation of mouse mu
opioid receptor gene: Sp3 isoforms (M1, M2) function as repressors
in neuronal cells to regulate the mu opioid receptor gene. Mol
Pharmacol. 67:1674–1683. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hardison RC: Comparative genomics. PLoS
Biol. 1:E582003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wei LN and Loh HH: Transcriptional and
epigenetic regulation of opioid receptor genes: Present and future.
Annu Rev Pharmacol Toxicol. 51:75–97. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vuillaumier S, Dixmeras I, Messaï H,
Lapouméroulie C, Lallemand D, Gekas J, Chehab FF, Perret C, Elion J
and Denamur E: Cross-species characterization of the promoter
region of the cystic fibrosis transmembrane conductance regulator
gene reveals multiple levels of regulation. Biochem J. 327:651–662.
1997. View Article : Google Scholar : PubMed/NCBI
|