Introduction
Cervical cancer is the third most commonly diagnosed
type of cancer and fourth leading cause of cancer mortalities in
females worldwide (1). Furthermore,
with more than half a million new cases and 265,700 mortalities per
year, cervical cancer continues to constitute a major public health
problem, particularly in developing countries, such as China
(2). The development of cervical
cancer is a multistep process involving a precursor preinvasive
stage (3). Typically, it takes several
years, even decades, to progress from pre-cancer to invasive
cervical cancer, which offers many opportunities for intervention.
Therefore, the elucidation of the molecular pathogenesis of
cervical cancer may contribute to reducing the incidence and
mortality rates of cervical cancer (4).
The human papillomavirus (HPV) infection is
essential for the development of cervical cancer and its precursor
lesions (5,6). However, only certain individuals who
remain infected with high-risk HPV develop cervical precancerous
lesions and cervical cancer. Although HPV is important for the
transformation of cervical epithelial cells, it is insufficient for
the development of cervical cancer, and there are a variety of
environmental and heritable genetic conditions that influence the
development of cervical cancer (7,8).
Accumulating evidence has revealed the marked potential of
microRNAs (miRNAs) for treating cervical cancer (9–11).
Various studies have indicated that tumorigenesis
may be caused by regulation disorders of cell cycle-associated
proteins, including cyclins, cyclin-dependent kinases (CDKs) and
CDK inhibitors. CDK6 is a CDK family member located on human
chromosome 7. Its activity first appears in the mid-G1
phase to phosphorylate, and thus regulate, the activity of tumor
suppressor protein retinoblastoma (Rb). By releasing transcription
factor, E2 factor (E2F) into the nucleus, the promoters of
associated genes, which mediate tumorigenesis, are affected
(9,12).
Furthermore, various miRNA components are reported to be involved
in CDK6-mediated tumorigenesis, such as miR-145, miR-320 and miR-29
(9,10,13).
miRNAs are a class of small, noncoding RNAs that
function as post-transcriptional regulators of gene expression by
binding to the 3′-untranslated region (UTR) of target mRNA through
a seed-match region, leading to translational repression or
cleavage of target mRNA (14).
Single-nucleotide polymorphisms (SNPs) located in the 3′-UTRs of
genes may affect interactions with microRNAs (miRNAs), whose
association with tumorigenesis is currently a focus of research
(14,15). One miRNAtargets numerous messenger RNAs
(mRNAs), and one mRNA may be regulated by more than one miRNA.
Therefore, functional variations, such as SNPs located in the
3′-UTRs of cancer-associated genes, may cause differential
regulation of target gene expression and simultaneously alter
numerous molecular pathways that are associated with tumorigenesis.
However, SNPs of the 3′-UTR region of the CDK6 gene have rarely
been investigated.
Under the hypothesis that gene polymorphisms or
haplotypes of the CDK6 gene have an impact on cervical precancerous
lesions, a case-control study was performed to demonstrate whether
SNPs located in the miRNA-binding sites within the 3′-UTR of the
CDK6 mRNAs or haplotypes influence the susceptibility of cervical
precancerous lesions. Furthermore, the potential interactions
between these SNPs and environmental factors in the etiology of
cervical precancerous lesions were investigated in a Chinese
population.
Materials and methods
Subjects
The present study was approved by the Ethics
Committee of the School of Medicine, Jinan University (Guangzhou,
China). Cervical specimens were collected with a Cytobrush (Qiagen,
Inc., Valencia, CA, USA) and placed into a ThinPrep Pap test vial
(Hologic, Inc., Marlborough, MA, USA) containing 20 ml PreservCyt
Solution (Hologic, Inc.). Referral Pap specimens were used for the
ThinPrep cytologic test, which was performed by Kingmed Center for
Clinical Laboratory Corporation (Guangzhou, China) using the
ThinPrep 2000 System (Hologic, Inc.) and evaluated for routine
screening cytology. A Pap smear was positive for a squamous
intraepithelial lesion (SIL) if there was a low (LSIL) or high
(HSIL) grade SIL, as classified according to the Bethesda
Classification System (16). Excluding
cases with other types of uterine disease and without a history of
hysterectomy, a total of 164 cases [120 LSIL (73.2%) and 44 HSIL
(26.8%)] were recruited. A total of 296 control samples without an
intraepithelial lesion or malignancy were recruited from the area
of residence of the cases. The study was explained to each
individual and written consent for their participation was
obtained.
HPV testing
Exfoliated cervical cell samples were collected from
vaginal swabs between May 2013 and November 2014 and conserved in
2.5 ml denaturation buffer (Qiagen, Inc.). Total DNA from cervical
cells was extracted using commercial magnetic beads kit (Chemagen;
PekinElmer, Inc., Waltham, MA, USA) according to the manufacturer's
instructions. Subsequently, 16 HPV genotypes were detected,
including HPV16, HPV18, HPV31, HPV33, HPV35, HPV39, HPV45, HPV51,
HPV52, HPV56, HPV58, HPV59, HPV66, and HPV68 (all high risk HPV),
as well as HPV6 and HPV11 (low risk HPV), using the MassARRAY
(Sequenom, Inc., San Diego, CA, USA) technique based on
matrix-assisted laser desorption/ionization time-of flight
(MALDI-TOF) mass spectrometry) (17,18). These
procedures were performed in the clinical standard laboratory of
the Beijing Genomics Institute (Shenzhen, China).
Genomic DNA extraction and
genotyping
Genomic DNA was extracted from peripheral whole
blood (5 ml; collected from women who attended the Family Planning
Service Stations or nearby medical institutions between May 2013
and November 2014) using a TIANamp blood DNA extraction kit
(TianGen Biotech Co., Ltd., Beijing, China) according to the
manufacturer's instructions. All DNA samples were resuspended in
water and stored at −20°C until use. The DNA concentration was
determined using a spectrophotometer (Nano Drop ND-1000;
PerkinElmer, Inc.). Samples with a mean optical density (OD) 260
nm/OD280 nm of 1.8–2.0 and DNA concentration >20 ng/µl were
considered to be free of contamination.
Subsequently, CDK6 gene targeted by miRNAs was
identified using miRBase (http://www.mirbase.org/) and TargetScan (http://www.targetscan.org/vert_71/). To ensure
the reliability of the predictions, multiple prediction algorithms
were applied to assess the binding capacity of miRNAs, including
RNA22 (https://cm.jefferson.edu/rna22/Interactive/) and
BiBiServ2 (http://bibiserv2.cebitec.uni-bielefeld.de/). SNPs
residing on the miRNA-binding sites were found by an extensive
search in dbSNP (http://www.ncbi.nlm.nih.gov/SNP) using Haploview
(https://www.broadinstitute.org/haploview/haploview)
and HapMap (http://hapmap.ncbi.nlm.nih.gov/). Finally, five SNPs
(rs8179, rs4272, rs42033, rs42035 and rs2377) were identified in
predicted miRNA-binding sites with minor allele frequencies
>0.05 in the Chinese Han population. The selected SNPs were
genotyped using MALDI-TOF within the MassARRAY system) (19). The distribution of SNPs in the CDK6
gene is presented in Table I.
 | Table I.Distribution of SNPs in the CDK6
gene. |
Table I.
Distribution of SNPs in the CDK6
gene.
| SNP ID | Position | Functional
region | Major/minor
alleles | MAF, % | P-value (HWE)
(control) |
|---|
| CDK6 (7q21.2) |
|
|
|
|
|
|
rs8179 | 92606850 | 3′-UTR | G/A |
3.4 | 0.41 |
|
rs4272 | 92607515 | 3′-UTR | A/G | 15.3 | 1.00 |
|
rs42033 | 92608219 | 3′-UTR | A/T |
3.2 | 0.45 |
|
rs42035 | 92610217 | 3′-UTR | T/C | 13.8 | 0.98 |
|
rs42377 | 92614358 | 3′-UTR | G/A | 16.3 | 0.96 |
|
rs8179 | 92606850 | 3′-UTR | G/A |
3.4 | 0.41 |
Statistical analysis
A t-test or χ2 test was used to compare
case and control subjects for the selected demographic
characteristics. The Hardy-Weinberg equilibrium theory
(p2+2pq+q2=1; where p is the frequency of the
wild-type allele and q is the frequency of the variant allele) was
used in controls to calculate the genotype frequencies of all five
SNPs using the χ2 test. In addition, logistic regression
was used to calculate the odds ratios (ORs) and their relative 95%
confidence intervals (CIs) for risk estimation. A χ2
test was also used to evaluate the dependence of the allele
frequencies between the case and control subjects.
The multifactor dimensionally reduction (MDR)
approach (20,21) was used to evaluate high-order gene-gene
and gene-environment interactions in cervical precancerous lesions
risk. To establish the best n-factor model, the data were divided
into 10 sets: One for testing and nine for training. Briefly,
multilocus genotypes were pooled into high- and low-risk groups,
reducing the genotype predictors to one dimension. An MDR
permutation text procedure was used to evaluate the significance of
the selected models by calculating empirical 1,000-fold permutation
tests. The best prediction model was selected based on maximum
testing balance accuracy (TBA) and cross-validation consistency
(CVC). MDR results were considered to be statistically significant
at P<0.05. Based on the MDR results, the interactions of
significance were selected for the logistic model to calculate the
OR.
The additive interaction between the factors was
subsequently evaluated according to the following strategy
(22). Among case and control
subjects, a binary classification was used for the HPV infection
(infection vs. non-infection) and the genotypes (homozygous for the
major allele vs. one or two copies of the minor allele). The risk
for cervical precancerous lesions for a given SNP and HPV infection
status was expressed by ORi, j, where the first index (i) indicated
the HPV infection status, coded as 0 for non-infected subjects and
1 for infected subjects, and the second index (j) indicated the SNP
genotype, which was coded as 0 for subjects homozygous for the
major allele and 1 for subjects bearing one or two copies of the
minor allele. Subjects who were HPV-uninfected and homozygous for
the major allele served as the reference group, and their cervical
precancerous lesion risk was coded as OR00=1. The
relative ORs were obtained by logistic regression. The CIs were
calculated by the regression coefficients and corresponding
covariance matrix (23). Deviation
from an additive model was calculated as the relative excess risk
due to interaction (RERI). Biological interactions in the
regression models were evaluated as departure from additivity.
Based on the adjusted ORs obtained in the logistic regression
models, an Excel spreadsheet (www.epinet.se)
was used to calculate RERI on an additive scale and its
corresponding CIs (23). An RERI value
(95% CI) that does not cross 0 indicates a biological interaction
(24). In addition, RERI>0
indicates a positive interaction or more than additivity and
RERI<0 indicates a negative interaction or less than additivity
(25,26).
Finally, haplotype analysis (haplotype frequency
estimation and linkage dis-equilibrium) between the groups was
performed using online SNPStats software (https://www.snpstats.net/snpstats/). The global score
test was used to estimate the overall differences in haplotype
frequencies between case and control subjects. The estimated
adjusted ORs and 95% CI were also calculated to assess the effect
of individual haplotypes on cervical precancerous lesions.
MDR software v.3.0.2 and MDR permutation testing
software (version 1.0 beta 2) were used in the current study and
were freely available online (www.epistasis.org). All other statistical analyses
were performed using SPSS software v.16.0 (SPSS, Inc., Chicago, IL,
USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
Characteristics of the study
population
The frequency distributions of selected demographic
characteristics of the case and control subjects are presented in
Table II. Increased risk factors for
cervical precancerous lesions were observed for a smoking history,
shorter time from menarche to the first intercourse (TMI), higher
number of pregnancies and HPV infection. No significant differences
were observed between case and control subjects in terms of age,
body mass index (BMI), age at menarche and age at first
intercourse.
 | Table II.Demographic and clinical
characteristics of cervical precancerous lesion patients (n=164)
and control subjects (n=296). |
Table II.
Demographic and clinical
characteristics of cervical precancerous lesion patients (n=164)
and control subjects (n=296).
|
| Controls, n
(%) | Cases, n (%) | OR (95% CI) | P-value |
|---|
| Age (mean ± SD),
years | 42.92±7.35 | 41.49±7.97 | N/A |
0.053a |
| Body mass index
(kg/m2) | 22.44±3.20 | 22.14±2.82 | N/A |
0.312a |
| Tobacco
smoking |
| No | 253 (85.5) | 120 (73.2) | Ref. |
0.001b,c |
|
Yes | 43 (14.5) | 44 (26.8) |
2.16(1.34–3.46) |
|
| Age at menarche
(mean ± SD), years | 14.85±1.63 | 15.12±1.96 | N/A |
0.113a |
| Age at first
intercourse (mean ± SD), years | 22.91±2.89 | 22.35±2.92 | N/A |
0.050a |
| TMI (mean ± SD),
years | 8.05±3.21 | 7.23±3.09 | N/A |
0.008b,c |
| Number of
pregnancies (mean ± SD) | 2.48±1.19 | 2.77±1.46 | N/A |
0.020b,c |
| High risk human
papillomavirus infection |
| No | 208 (70.3) | 32 (19.5) | Ref. |
<0.001b,c |
|
Yes | 88 (29.7) | 132 (80.5) | 9.75
(6.16–15.44) |
|
Association between SNPs located
within the 3′-UTR of the CDK6 gene and cervical precancerous lesion
susceptibility
Five SNPs in the 3′-UTR of the CDK6 gene were
selected in the current study. The genotype distributions of the
five SNPs in the control subjects were in Hardy-Weinberg
equilibrium (Table I). As presented in
Table III, significant differences
were identified in the rs8179 and rs42033 alleles, as well as the
genotype distributions between the case and control subjects.
Logistic regression analysis revealed that, following adjustment
for HPV infection, age, BMI, tobacco smoking, age at menarche, age
at first intercourse and number of pregnancies, the GA genotype in
rs8179 was associated with a 0.17-fold decreased cervical
precancerous lesions risk compared with the AA genotype (95% CI,
0.05–0.57; P=0.004) and the AT genotype in rs42033 was associated
with a 0.18-fold decreased cervical precancerous lesions risk
compared with the AA genotype (95% CI, 0.05–0.59; P=0.005). While
the other three SNPs were not observed to be relevant to the risk
of cervical precancerous lesions.
 | Table III.Allele frequencies and genotype
distributions of cyclin-dependent kinase 6 SNPs in cervical
precancerous lesion patients (n=164) and control subjects
(n=296). |
Table III.
Allele frequencies and genotype
distributions of cyclin-dependent kinase 6 SNPs in cervical
precancerous lesion patients (n=164) and control subjects
(n=296).
| SNP | Variants | Controls, n
(%) | Cases, n (%) | OR (95% CI) | P-value | Oradjusted (95%
CI)a | P-value |
|---|
| rs8179-Allele | G | 565 (95.4) | 324 (98.8) | 1 (Ref.) |
|
|
|
|
| A | 27 (4.6) | 4
(1.2) | 0.26
(0.09–0.75)b | 0.007b |
|
|
| Genotype | GG | 269 (90.9) | 160 (97.6) | 1 (Ref.) | N/A | 1 (Ref.) | N/A |
|
| GA | 27 (9.1) | 4
(2.4) | 0.25
(0.09–0.73)b | 0.011b | 0.17
(0.05–0.57)b | 0.004b |
|
| AA | 0
(0.0) | 0
(0.0) | – | – | – | – |
| rs4272-Allele | A | 501 (84.6) | 278 (84.8) | 1 (Ref.) |
|
|
|
|
| G | 91
(15.4) | 50
(15.2) | 0.99
(0.68–1.44) | 0.959 |
|
|
| Genotype | AA | 212 (71.6) | 117 (71.3) | 1 (Ref.) | N/A | 1(Ref.) | N/A |
|
| GA | 77
(26.0) | 44
(26.8) | 1.04
(0.67–1.60) | 0.875 | 1.26
(0.75–1.12) | 0.390 |
|
| GG | 7
(2.4) | 2
(1.8) | 0.78
(0.20–3.06) | 0.718 | 0.59
(0.12–3.05) | 0.532 |
| Recessive | AA+GA | 289 (97.6) | 161 (98.2) | 1 (Ref.) | N/A | 1 (Ref.) | N/A |
|
| GG | 7
(2.4) | 3
(1.8) | 0.77
(0.20–3.02) | 0.695 | 0.56
(0.11–2.82) | 0.478 |
| Dominant | AA | 212 (71.6) | 117 (71.3) | 1 (Ref.) | N/A | 1 (Ref.) | N/A |
|
| GA+GG | 84
(28.4) | 47
(28.7) | 1.02
(0.67–1.55) | 0.949 | 1.19
(0.72–1.98) | 0.501 |
| rs42033-Allele | A | 567 (95.8) | 324 (98.8) | 1 (Ref.) |
|
|
|
|
| T | 25
(4.21) | 4
(1.2) | 0.28
(0.10–0.81)b | 0.013b |
|
|
| Genotype | AA | 271 (91.5) | 160 (97.6) | 1 (Ref.) | N/A | 1 (Ref.) | N/A |
|
| AT | 25 (8.4) | 4
(2.4) | 0.27
(0.09–0.79)b | 0.017b | 0.18
(0.05–0.59)b | 0.005b |
|
| TT | 0
(0.0) | 0
(0.0) | – | – | – | – |
| rs42035-Allele | T | 508 (85.8) | 285 (86.9) | 1 (Ref.) |
|
|
|
|
| C | 84
(14.2) | 43
(13.1) | 0.91
(0.62–1.36) | 0.649 |
|
|
| Genotype | TT | 218 (73.7) | 122 (74.4) | 1 (Ref.) | N/A | 1 (Ref.) | N/A |
|
| TC | 72
(24.3) | 41
(25.0) | 1.02
(0.65–1.59) | 0.939 | 0.87
(0.51–1.47) | 0.591 |
|
| CC | 6
(2.0) | 1
(0.6) | 0.30
(0.04–2.50) | 0.265 | 0.40
(0.04–3.86) | 0.429 |
| Recessive | TT+TC | 290 (98.0) | 163 (99.4) | 1 (Ref.) | N/A | 1 (Ref.) | N/A |
|
| CC | 6
(2.0) | 1
(0.6) | 0.30
(0.04–2.48) | 0.262 | 0.42
(0.04–3.99) | 0.447 |
| Dominant | TT | 218 (73.7) | 122 (74.4) | 1 (Ref.) | N/A | 1 (Ref.) | N/A |
|
| TC+CC | 78
(26.4) | 42
(25.6) | 0.96
(0.62–1.49) | 0.862 | 0.83
(0.50–1.40) | 0.493 |
| rs42377-Allele | G | 494 (83.4) | 276 (84.1) | 1 (Ref.) |
|
|
|
|
| A | 98
(16.6) | 52
(15.9) | 0.95
(0.66–1.37) | 0.78 |
|
|
| Genotype | GG | 206 (69.9) | 116 (70.7) | 1 (Ref.) | N/A | 1 (Ref.) | N/A |
|
| GA | 82
(27.7) | 44
(26.8) | 0.95
(0.62–1.47) | 0.826 | 1.12
(0.66–1.87) | 0.682 |
|
| AA | 8
(2.7) | 4
(2.4) | 0.89
(0.26–3.01) | 0.849 | 0.77
(0.18–3.35) | 0.726 |
| Recessive | GG+GA | 288 (97.3) | 160 (97.6) | 1 (Ref.) | N/A | 1 (Ref.) | N/A |
|
| AA | 8
(2.7) | 4
(2.4) | 0.90
(0.27–3.04) | 0.865 | 0.75
(0.17–3.22) | 0.693 |
| Dominant | GG | 206 (69.6) | 116 (70.7) | 1 (Ref.) | N/A | 1 (Ref.) | N/A |
|
| GA+AA | 90
(30.4) | 48
(29.3) | 0.95
(0.63–1.44) | 0.799 | 1.08
(0.65–1.79) | 0.764 |
Gene-environment interaction for
cervical precancerous lesions susceptibility
According to the MDR selection model, the best model
was HPV infection, which had the maximum CVC (10/10) and highest
TBA, 75.38%; significance test, P=0.001; permutation test,
P=0.000–0.001. The second was a three-factor interaction model,
containing HPV infection, the number of pregnancies and rs8179,
with maximum CVC (8/10) and highest TBA, 76.47%; significance test,
P=0.001; permutation test, P=0.000–0.001) (Table IV). Additionally, the interactive
effects of the HPV infection and each SNP based on an additive
scale were evaluated. According to the RERI indexes, a significant
antagonistic interaction was identified between CDK6 rs8179,
rs42033 and HPV infection (RERI=−9.00, 95% CI, −14.69 to −3.30;
P=0.002; RERI=−8.84, 95% CI, −14.45 to −3.22; P=0.002) as preseted
in Table V.
 | Table IV.Multifactor dimensionality reduction
models of the cyclin-dependent kinase 6 gene and environmental
factors of cervical precancerous lesions. |
Table IV.
Multifactor dimensionality reduction
models of the cyclin-dependent kinase 6 gene and environmental
factors of cervical precancerous lesions.
| Best models | Training balanced
accuracy | Testing balanced
accuracy | Cross-validation
consistency |
P-valuea |
|---|
| HPV infection | 0.7538 | 0.7538 | 10/10 | 0.000–0.001 |
| HPV infection,
rs8179 | 0.7615 | 0.7582 |
8/10 | 0.000–0.001 |
| HPV infection,
rs8179 and number of pregnancies | 0.7647 | 0.7582 |
8/10 | 0.000–0.001 |
 | Table V.Results for gene-environment
interaction analysis for each candidate single nucleotide
polymorphism and HPV infection. |
Table V.
Results for gene-environment
interaction analysis for each candidate single nucleotide
polymorphism and HPV infection.
|
| Deviation from
additive model |
|---|
|
|
|
|---|
| Interaction
group | Gene | Relative excess
risk due to interaction (95% confidence interval) |
P-valuea |
|---|
| 1 | rs42035*HPV
infection | −4.64
(−11.36 to 2.08) | 0.176 |
| 2 | rs42033*HPV
infection |
−9.00 (−14.69 to
−3.30) | 0.002 |
| 3 | rs42377*HPV
infection | −0.09 (−7.26 to
7.09) | 0.981 |
| 4 | rs4272*HPV
infection | 1.04
(−6.89 to 8.99) | 0.796 |
| 5 | rs8179*HPV
infection |
−8.84 (−14.45 to
−3.22) | 0.002 |
Three-way interaction between HPV
infection, rs311678 status and number of pregnancies with the risk
for cervical precancerous lesions
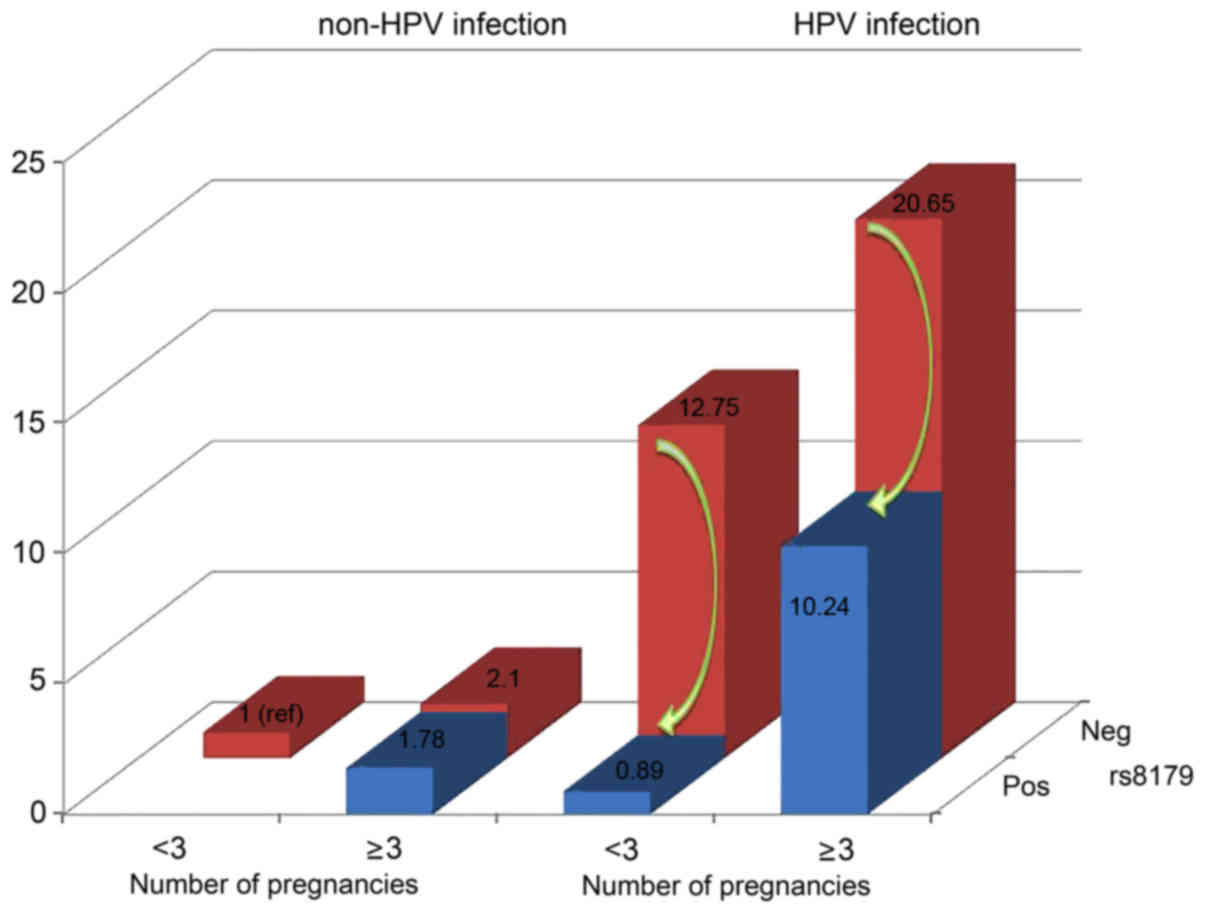
Based on the MDR model, a risk analysis of different
combinations was performed among the three factors (Table VI). The combination without any risk
factors [including non-HPV-infected, wild-type for rs8179 and a
reduced number of pregnancies (<3) served as a reference group.
The individuals with a combination of three factors had a
10.24-fold greater risk of developing cervical precancerous lesions
(95% CI, 1.25–84.07; P=0.030), the OR for cervical precancerous
lesions in the presence of HPV infection, wild-type for rs8179 and
reduced number of pregnancies (<3) was greater (OR=12.75; 95%
CI, 6.03–26.97; P<0.001), while the highest OR for cervical
precancerous lesions was in the presence of HPV infection,
wild-type for rs8179 and a greater number of pregnancies (≥3)
(OR=20.65; 95% CI, 9.60–44.44; P<0.001).
 | Table VI.Risk group analysis with three risk
factors: HPV infection, number of pregnancies and rs8179. |
Table VI.
Risk group analysis with three risk
factors: HPV infection, number of pregnancies and rs8179.
| HPV infection | Number of
pregnancies | rs8179 | Cases | Controls | Odds ratio (95%
confidence interval) |
P-valuea |
|---|
| – | – | – | 11 | 106 | 1 |
|
| – | + | – | 20 | 85 | 2.10
(0.94–4.70) |
0.070 |
| – | – | + | 0 | 12 | – | – |
| – | + | + | 1 | 5 | 1.78
(0.19–16.92) |
0.616 |
| + | – | – | 62 | 46 | 12.75
(6.03–26.97) | <0.001 |
| + | + | – | 67 | 32 | 20.65
(9.60–44.44) | <0.001 |
| + | – | + | 1 | 8 | 0.89
(0.10–8.10) |
0.916 |
| + | + | + | 2 | 2 | 10.24
(1.25–84.07) |
0.030 |
Haplotype analysis
Strong linkage disequilibrium values were observed
between rs8179, rs4272, rs42033 and rs42377 (D'≥0.86,
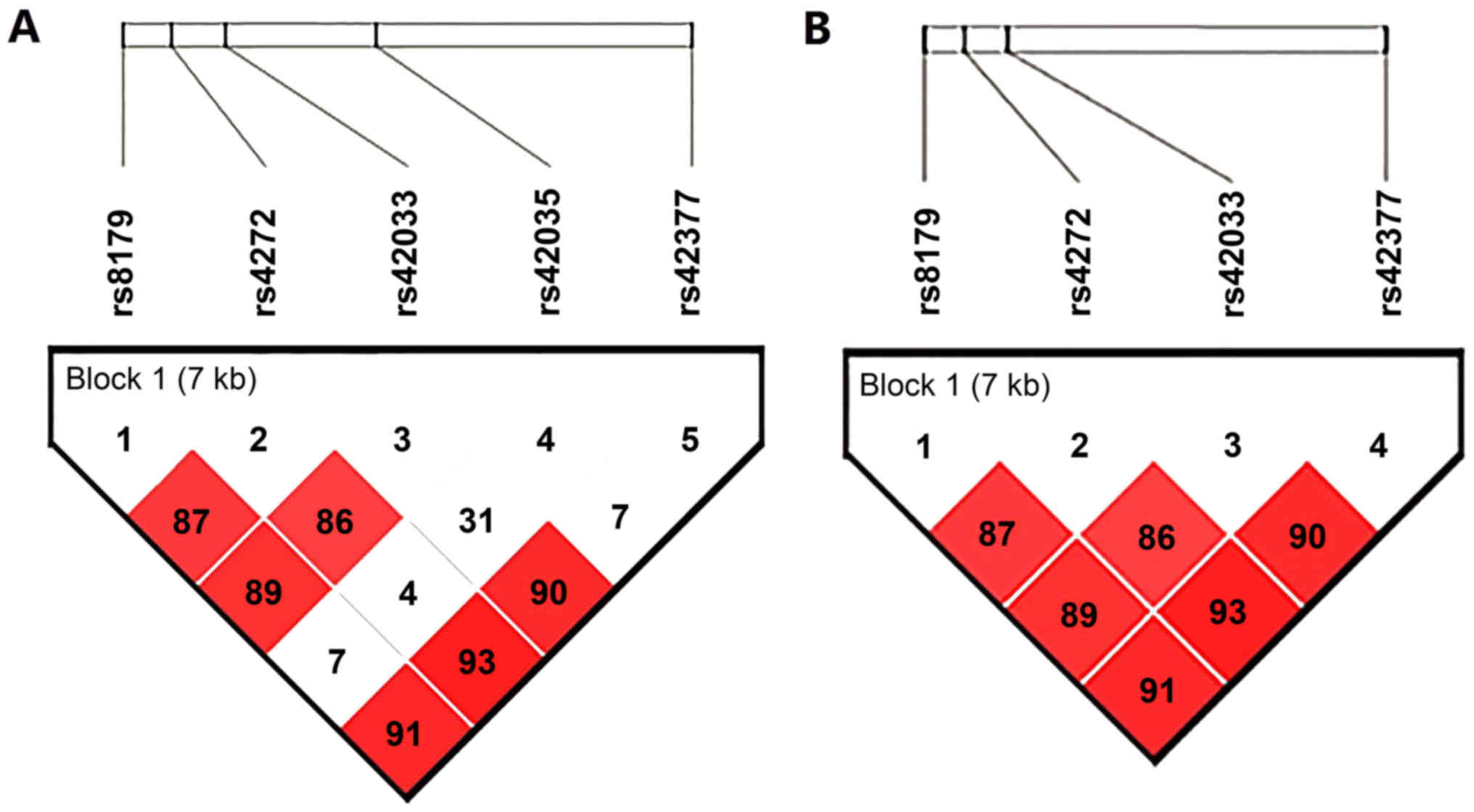
r2=0.36–0.90; Fig. 1).
Haplotypes were constructed based on four CDK6 polymorphisms
(rs8179, rs4272, rs42033 and rs42377). The overall global test
identified a difference in the frequency of haplotypes between case
and control subjects for rs8179-rs4272-42033-42377 (P=0.002). The
results indicated that the haplotype AGTA significantly correlated
with a reduced risk of cervical precancerous lesions (OR=0.21, 95%
CI, 0.06–0.75; P=0.016; Table VII)
compared with the highest frequency haplotype GAAG after adjusting
for HPV infection, age, BMI, tobacco smoking, age at menarche, age
at first intercourse and number of pregnancies. No other frequency
difference was observed in these haplotypes between cervical
precancerous lesion patients and control subjects.
 | Table VII.Haplotype frequencies of CDK6
polymorphisms and cervical precancerous lesions. |
Table VII.
Haplotype frequencies of CDK6
polymorphisms and cervical precancerous lesions.
|
| Haplotype
frequency |
|
|
|
|
|---|
|
|
|
|
|
|
|
|---|
|
Haplotypea | Control (%) | Cases (%) | ORb (95%CI) | P-value | ORb,c
(95% CI) |
P-valuec,e |
|---|
| GAAG | 81.90 | 83.84 | 1.00 | – | 1.00 | – |
| GGAA | 10.46 | 13.72 | 1.29
(0.85–1.98) | 0.240 | 1.67
(0.99–2.84) | 0.057 |
| AGTA |
3.72 |
1.22 | 0.32
(0.11–0.95) | 0.040 | 0.21
(0.06–0.75) | 0.016 |
| GAAA |
1.71 |
0.92 | 0.64
(0.17–2.46) | 0.520 | 0.45
(0.10–2.13) | 0.320 |
| Rared |
1.54 |
0.31 | 0.21
(0.03–1.53) | 0.120 | 0.17
(0.02–1.49) | 0.110 |
Discussion
To the best of our knowledge, this is the first
study to investigate the interactive effects of SNPs in the 3′-UTR
of the CDK6 gene and HPV infection on the risk of cervical
precancerous lesions. To investigate whether SNPs in the CDK6 gene
affected the occurrence of cervical precancerous lesions, the
associations between variants of the CDK6 gene and cervical
precancerous lesions were comprehensively analyzed.
CDK6 is a critical regulatory cancer-associated gene
in tumorigenesis and is highly expressed in the majority of cancer
types, such as breast, ovarian, gastric, pancreatic and cervical
cancers (27–31). CDK6 is ubiquitously expressed in human
tumor cells, and is involved in cell cycle control and
differentiation by promoting the G1/S transition
(12,32,33). CDK6
regulates the progression of the G1 phase by combining
with cyclin D to promote phosphorylation of the tumor suppressor
gene, Rb (pRb), and by releasing transcription factor, E2F, into
the nucleus to affect the promoters of the associated genes may
affect tumorigenesis (12).
To date, various studies have investigated the
association between SNPs in miRNA binding sites and cervical cancer
risk, three of which have involved SNPs located in miRNA target
genes (i.e., LAMB3-rs2566, BCL2-rs3744935 and
TNFAIP8-rs11064) (34–36). These studies indicated that SNPs in the
3′-UTR of target genes may be significant to establishing the
cancer risk of certain individual. However, to the best of our
knowledge, no studies have yet demonstrated a link between genetic
variations of the 3′UTR of the CDK6 gene and susceptibility of
Chinese women to developing cervical precancerous lesions. In the
present results, the minor A allele in CDK6-rs8179 reduced the risk
of cervical precancerous lesions and had an antagonistic
interaction with the HPV infection, as did the minor T allele in
CDK6-rs42033. Considering than functional variations, such as SNPs
located in the miRNA seed-match region, affect the binding affinity
of miRNAs to target mRNAs, causing differential regulation of
target gene expression and altering numerous molecular pathways
simultaneously, rs8179 and rs42033 in the 3′UTR of the CDK6 gene
may affect miRNA-mediated regulatory function. This leads to
changes in the level of CDK6 expression, which may affect the
development of cervical precancerous lesions. The underlying
mechanism requires further investigation in functional studies.
In addition, one study indicated that an increased
CDK6 expression level was significantly associated with the
histological grade of cervical carcinogenesis and that CDK6 was
positively associated with E6/E7 (10). The authors indicated that CDK6 may
serve as a candidate HPV-induced oncogene in cervical cancer and
HR-HPVs may gain an advantage in inducing carcinogenesis by
reshaping the environment of the cellular miRNA composition and
target gene expression to benefit the virus (10). Therefore, the present study
hypothesized that there is an interaction between HPV and each SNP
in the CDK6 gene, which is associated with the risk of cervical
precancerous lesions. Additionally, the current study demonstrated
an interactive effect between rs8179, rs42033 and the HPV infection
on an additive scale. Furthermore, the MDR model indicated that the
interaction between the HPV infection, rs8179 and the number of
pregnancies was important at the population level; among
HPV-infected individuals who had <3 pregnancies, women carrying
the minor A allele in CDK6-rs8179 had a 50% reduced risk of
cervical precancerous lesions compared with individuals who were
wild type at rs8179 (Fig. 2).
Additionally, using haplotype analysis, it was identified that the
haplotype AGTA was associated with a decreased risk of cervical
precancerous lesions. In addition to the HPV infection, cervical
precancerous lesions are associated with various environmental
factors. The current results demonstrated that women with a smoking
history, shorter TMI and higher number of pregnancies were at risk
of cervical precancerous lesions. Smoking reduces cervical
immunity, which enhances the persistence of the HPV infection
(37). Various studies have
demonstrated that women with cervical precancerous lesions had a
shorter TMI (38,39). It was hypothesized that this group may
exhibit delayed maturation of the cervical transformation zone and,
thus, an increased area of immature epithelium, suggesting that a
subpopulation of adolescents with cervical precancerous lesions
have a biologically immature cervix, which may increase the
vulnerability of the cervical epithelium to HPV infection and
neoplastic change may, therefore, increase the risk of cervical
carcinoma, as it retains the transformation zone on the exocervix
for many years, facilitating direct exposure to HPV (38–40).
There were various limitations of the current study.
The sample size for genotyping was relatively small, resulting in a
low statistical power of the genetic aspect of this study. Larger
studies involving individuals from different ethnic populations are
required to verify the findings, and this may facilitate the
elucidation of the intrinsic mechanism. In addition, these data
demonstrated the protective effect of the polymorphisms of the CDK6
gene in cervical precancerous lesions; however, the precise
molecular mechanism underlying the effect remains to be elucidated,
which would provide a basis for further studies (for example, where
mutations are cloned in luciferase reporter constructs to
characterize the binding specificity of miR-29). Protein expression
studies of a similar group of patients are being considered to
determine whether the polymorphisms are associated with cervical
precancerous lesions.
In conclusion, the current data provide novel
evidence that the rs8179 and rs42033 polymorphisms and haplotype
AGTA in CDK6 may influence the development of cervical precancerous
lesions in Chinese women. Furthermore, the present study
demonstrates two- and three-way gene-environment interactions in
the etiology of cervical precancerous lesions. Novel
epidemiological clues regarding the protective role of the CDK6
gene in cervical precancerous lesions and further insights into the
interactions involved in the etiology of cervical precancerous
lesions were also presented. These results provide further evidence
that introns should no longer be considered nonsense. In addition,
the possibility that high risk HPVs may have an advantage in the
induction of carcinogenesis in the environment of the cellular
miRNA composition and target gene expression to the benefit of the
virus.
Acknowledgements
The present study was supported in part by China
Population Welfare Foundation (grant no. [2011]31), Science and
Technology Planning Project of Guangdong Province (grant no.
2013B032000001), Family Planning Foundation of Guangdong Province
(grant no. 2012004), National Natural Science Foundation of China
(grant nos. 30901249, 81101267 and 81541070), the Guangdong Natural
Science Foundation (grant nos. 10151063201000036 and
S2011010002526), Guangdong Province Medical Research Foundation
(grant no. A2014374 and A2015310) and Project from Jinan university
(grant no. 21612-426, 21615427). The authors would like to thank
all of the collaborators, Yang Liu, Baohuan Zhang, Man Wang, Na
Zhang and the staff for their hard work in running the study and
the women who participated in this study.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ambros RA and Kurman RJ: Current concepts
in the relationship of human papillomavirus infection to the
pathogenesis and classification of precancerous squamous lesions of
the uterine cervix. Semin Diagn Pathol. 7:158–172. 1990.PubMed/NCBI
|
|
4
|
Xiao D, Huang W, Ou M, Guo C, Ye X, Liu Y,
Wang M, Zhang B, Zhang N, Huang S, et al: Interaction between
susceptibility loci in cGAS-STING pathway, MHC gene and HPV
infection on the risk of cervical precancerous lesions in Chinese
population. Oncotarget. 7:84228–84238. 2016.PubMed/NCBI
|
|
5
|
Nieves-Ramirez ME, Partida-Rodriguez O,
Alegre-Crespo PE, Tapia-Lugo Mdel C and Perez-Rodriguez ME:
Characterization of single-nucleotide polymorphisms in the tumor
necrosis factor alpha promoter region and in lymphotoxin α in
squamous intraepithelial lesions, precursors of cervical cancer.
Transl Oncol. 4:336–344. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bodily J and Laimins LA: Persistence of
human papillomavirus infection: Keys to malignant progression.
Trends Microbiol. 19:33–39. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Castle PE, Schiffman M, Wheeler CM and
Solomon D: Evidence for frequent regression of cervical
intraepithelial neoplasia-grade 2. Obstet Gynecol. 113:18–25. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kobayashi A, Weinberg V, Darragh T and
Smith-McCune K: Evolving immunosuppressive microenvironment during
human cervical carcinogenesis. Mucosal Immunol. 1:412–420. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang J, Wang L, Li B, Huo M, Mu M, Liu J
and Han J: miR-145 downregulates the expression of cyclin-dependent
kinase 6 in human cervical carcinoma cells. Exp Ther Med.
8:591–594. 2014.PubMed/NCBI
|
|
10
|
Li Y, Wang F, Xu J, Ye F, Shen Y, Zhou J,
Lu W, Wan X, Ma D and Xie X: Progressive miRNA expression profiles
in cervical carcinogenesis and identification of HPV-related target
genes for miR-29. J Pathol. 224:484–495. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang X, Meyers C, Guo M and Zheng ZM:
Upregulation of p18Ink4c expression by oncogenic HPV E6 via
p53-miR-34a pathway. Int J Cancer. 129:1362–1372. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sridhar J, Akula N and Pattabiraman N:
Selectivity and potency of cyclin-dependent kinase inhibitors. AAPS
J. 8:E204–E221. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tadano T, Kakuta Y, Hamada S, Shimodaira
Y, Kuroha M, Kawakami Y, Kimura T, Shiga H, Endo K, Masamune A, et
al: MicroRNA-320 family is downregulated in colorectal adenoma and
affects tumor proliferation by targeting CDK6. World J Gastrointest
Oncol. 8:532–542. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee AR, Park J, Jung KJ, Jee SH and
Kim-Yoon S: Genetic variation rs7930 in the miR-4273-5p target site
is associated with a risk of colorectal cancer. Onco Targets Ther.
9:6885–6895. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zheng N, Yang P, Wang Z and Zhou Q:
OncomicroRNAs-mediated tumorigenesis: Implication in cancer
diagnosis and targeted therapy. Curr Cancer Drug Targets. 17:40–47.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nayar R and Wilbur DC: The Pap test and
Bethesda 2014. Cancer Cytopathol. 123:271–281. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yi X, Li J, Yu S, Zhang A, Xu J, Yi J, Zou
J, Nie X, Huang J and Wang J: A new PCR-based mass spectrometry
system for high-risk HPV, part I: Methods. Am J Clin Pathol.
136:913–919. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qu S, Huang J, Zhao J, Zhao X, Deng H,
Yang H, Chen W, Liu L, Zhang L and Gao S: A comparison of
matrix-assisted laser desorption/ionization time-of-flight mass
spectrometry and surface plasmon resonance for genotyping of
high-risk human papillomaviruses. Intervirology. 54:326–332. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jurinke C, van den Boom D, Cantor CR and
Köster H: Automated genotyping using the DNA MassArray technology.
Methods Mol Biol. 187:179–192. 2002.PubMed/NCBI
|
|
20
|
Hahn LW, Ritchie MD and Moore JH:
Multifactor dimensionality reduction software for detecting
gene-gene and gene-environment interactions. Bioinformatics.
19:376–382. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Heidema AG, Feskens EJ, Doevendans PA,
Ruven HJ, van Houwelingen HC, Mariman EC and Boer JM: Analysis of
multiple SNPs in genetic association studies: Comparison of three
multi-locus methods to prioritize and select SNPs. Genet Epidemiol.
31:910–921. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tunesi S, Ferrante D, Mirabelli D, Andorno
S, Betti M, Fiorito G, Guarrera S, Casalone E, Neri M, Ugolini D,
et al: Gene-asbestos interaction in malignant pleural mesothelioma
susceptibility. Carcinogenesis. 36:1129–1135. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Andersson T, Alfredsson L, Källberg H,
Zdravkovic S and Ahlbom A: Calculating measures of biological
interaction. Eur J Epidemiol. 20:575–579. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Muthuri SG, Doherty S, Zhang W, Maciewicz
RA, Muir KR and Doherty M: Gene-environment interaction between
body mass index and transforming growth factor beta 1 (TGFβ1) gene
in knee and hip osteoarthritis. Arthritis Res Ther. 15:R522013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Knol MJ, van der Tweel I, Grobbee DE,
Numans ME and Geerlings MI: Estimating interaction on an additive
scale between continuous determinants in a logistic regression
model. Int J Epidemiol. 36:1111–1118. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Knol MJ, VanderWeele TJ, Groenwold RH,
Klungel OH, Rovers MM and Grobbee DE: Estimating measures of
interaction on an additive scale for preventive exposures. Eur J
Epidemiol. 26:433–438. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dai X, Li L, Liu X, Hu W, Yang Y and Bai
Z: Cooperation of DLC1 and CDK6 affects breast cancer clinical
outcome. G3 (Bethesda). 5:81–91. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xia B, Yang S, Liu T and Lou G: miR-211
suppresses epithelial ovarian cancer proliferation and cell-cycle
progression by targeting Cyclin D1 and CDK6. Mol Cancer. 14:572015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li LP, Wu WJ, Sun DY, Xie ZY, Ma YC and
Zhao YG: miR-449a and CDK6 in gastric carcinoma. Oncol Lett.
8:1533–1538. 2014.PubMed/NCBI
|
|
30
|
Lu X, Fang Y, Wang Z, Xie J, Zhan Q, Deng
X, Chen H, Jin J, Peng C, Li H and Shen B: Downregulation of gas5
increases pancreatic cancer cell proliferation by regulating CDK6.
Cell Tissue Res. 354:891–896. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Arvanitis DA and Spandidos DA:
Deregulation of the G1/S phase transition in cancer and squamous
intraepithelial lesions of the uterine cervix: A case control
study. Oncol Rep. 20:751–760. 2008.PubMed/NCBI
|
|
32
|
Choi YJ and Anders L: Signaling through
cyclin D-dependent kinases. Oncogene. 33:1890–1903. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Costello JF, Plass C, Arap W, Chapman VM,
Held WA, Berger MS, Su Huang HJ and Cavenee WK: Cyclin-dependent
kinase 6 (CDK6) amplification in human gliomas identified using
two-dimensional separation of genomic DNA. Cancer Res.
57:1250–1254. 1997.PubMed/NCBI
|
|
34
|
Zhou X, Chen X, Hu L, Han S, Qiang F, Wu
Y, Pan L, Shen H, Li Y and Hu Z: Polymorphisms involved in the
miR-218-LAMB3 pathway and susceptibility of cervical cancer, a
case-control study in Chinese women. Gynecol Oncol. 117:287–290.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Reshmi G, Surya R, Jissa VT, Babu PS,
Preethi NR, Santhi WS, Jayaprakash PG and Pillai MR: C-T variant in
a miRNA target site of BCL2 is associated with increased risk of
human papilloma virus related cervical cancer-an in silico
approach. Genomics. 98:189–193. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shi TY, Cheng X, Yu KD, Sun MH, Shao ZM,
Wang MY, Zhu ML, He J, Li QX, Chen XJ, et al: Functional variants
in TNFAIP8 associated with cervical cancer susceptibility and
clinical outcomes. Carcinogenesis. 34:770–778. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Giuliano AR, Sedjo RL, Roe DJ, Harri R,
Baldwi S, Papenfuss MR, Abrahamsen M and Inserra P: Clearance of
oncogenic human papillomavirus (HPV) infection: Effect of smoking
(United States). Cancer Causes Control. 13:839–846. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Syrjänen K, Shabalova I, Petrovichev N,
Kozachenko V, Zakharova T, Pajanidi J, Podistov J, Chemeris G,
Sozaeva L, Lipova E, et al: Age at menarche is not an independent
risk factor for high-risk human papillomavirus infections and
cervical intraepithelial neoplasia. Int J STD AIDS. 19:16–25. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Moscicki AB, Winkler B, Irwin CE Jr and
Schachter J: Differences in biologic maturation, sexual behavior,
and sexually transmitted disease between adolescents with and
without cervical intraepithelial neoplasia. J Pediatr. 115:487–493.
1989. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Muñoz N, Franceschi S, Bosetti C, Moreno
V, Herrero R, Smith JS, Shah KV, Meijer CJ and Bosch FX;
International Agency for Research on Cancer, ; Multicentric
Cervical Cancer Study Group, : Role of parity and human
papillomavirus in cervical cancer: The IARC multicentric
case-control study. Lancet. 359:1093–1101. 2002. View Article : Google Scholar : PubMed/NCBI
|