Introduction
Encephalomyocarditis virus (EMCV), a member of the
genus Cardiovirus in the family Picornaviridae, is a
single-stranded positive-sense RNA virus of ~7.8 kb in length with
a large open reading frame (ORF) (1).
It contains a 5′ untranslated region (5′ UTR) and a 3′ untranslated
region (3′ UTR). The ORF encodes for 11 proteins, including four
structural proteins (VP1, VP2, VP3, VP4), seven non-structural
proteins (2A, 2B, 2C, 3A, 3B, 3C, 3D) and a leader protein. The VP1
region, which is highly variable, is an important protective
antigen of EMCV (2). It can stimulate
the body to produce neutralizing antibodies. It is also the
important region for researching gene vaccine (3).
EMCV has been recognized worldwide as a pathogen
infecting many species, including pigs, cattle, rodents, raccoons,
elephants, marsupials and primates such as baboons, chimpanzees,
monkeys, and even humans (4–7). The virus, which currently affects the
global swine industry, can cause myocarditis, reproductive failure
and high mortality in pregnant sows, fetuses and weaning piglets.
In China, it had been confirmed that EMCV infection occurs in many
pig farms by the methods of etiology and serology (8).
In the current study, the authors used the reverse
transcription-quantitative polymerase chain reaction method to
successfully amplify the full-length VP1 coding sequence. They
subsequently produced the recombinant gene product in the
Escherichia coli expression system, which is commonly used
to produce adequate amounts of protein suitable for the intended
application. E. coli is easy to transform, grows quickly in
simple media, and requires inexpensive equipment for growth and
storage. The produced recombinant VP1 was then used to develop an
indirect ELISA assay with sensitivity and specificity comparable to
virus neutralization tests.
Materials and methods
RNA isolation
EMCV strain BD2 (KF709977) was isolated from three
clinically ill newborn pigs, which exhibited anorexia, rapid
breathing and acute myocarditis, from a commercial pig farm in
Hebei, China, as described previously (9). BHK-21 cells were grown in 75
cm2 plastic flasks, inoculated with 1 ml of virus
supernatant from positive cell cultures. After an extensive
cytopathic effect was observed, frozen and thawed three times and
centrifuged at 13,400 × g for 10 min at 4°C. Genomic RNA of EMCV
BD2 was extracted from the cultural supernatant of infected BHK-21
cells using the EasyPure Viral DNA/RNA kit (Beijing Transgen
Biotech Co., Ltd., Beijing, China) according to the manufacturer's
instructions.
Amplification of the EMCV VP1
gene
The VP1 gene was amplified by RT-qPCR using the
template from the last step. The genome sequence of the BD2 strain
EMCV-VP1 has been uploaded to the NCBI (accession no.
KF709977.1).
The forward primer contained a BamHI restriction
site at the 5′-end of the sequence coding for VP1, while the
reverse primer contained an XhoI site positioned after the stop
codon at the 3′-end. The sequences of the primers were as follows:
forward, 5′-CGCGGATCCGGAGTAGAAAACGCTGAAAAAG-3′ and reverse,
5′-CCGCTCGAGCTCTAGCATCAAGACTCCAGCT-3′. Both primers were purchased
from Sangon Biotech Co., Ltd. (Shanghai, China) and used without
further purification. The PCR reaction mixture included 1 µg DNA
template, 1 µl PrimeSTART HS DNA Polymerase (Takara Biotechnology
Co., Ltd., Dalian, China), 10 µl 5X PrimeSTART buffer (Beyotime
Institute of Biotechnology, Haimen, China), all four dNTPs at final
concentrations of 0.2 mM each, primers at a final concentration of
0.2 µM each, and ddH2O in a total volume of 50 µl. The
cycling conditions included an initial denaturation step at 98°C
for 30 sec, and then 30 cycles consisting of 30 sec denaturation at
98°C, 30 sec annealing at 56°C, and 50 sec extension at 72°C,
followed by a final extension for 10 min at 72°C. The amplification
product was purified on a 1% (w/v) agarose gel in TAE buffer,
stained with ethidium bromide, and extracted using a commercial kit
(QIAquick Gel Extraction kit; Sangon Biotech Co., Ltd.), according
to the manufacturer's instructions.
Construction of recombinant
proteins
Following extraction, the DNA product and the
expression vector pET32a plasmid were each digested with BamHI and
XhoI using standard molecular biology protocols. The VP1 DNA
insert (55 ng) and linearized pET32a (20 ng) were incubated at 16°C
overnight in the presence of 1 µl T4 DNA ligase in a total volume
of 10 µl according to the manufacturer's instructions.
The entire ligation reaction was transformed into
Trans1-T1 chemically competent cells (Beijing Transgen Biotech
Co.). Several colonies were screened for the presence of EMCV VP1
DNA by colony PCR; colonies that were positive were grown in liquid
culture and plasmids were purified using a TIANprep Mini Plasmid
kit (Tiangen Biotech Co., Ltd., Beijing). The presence of EMCV VP1
DNA was confirmed by restriction digests of the purified plasmids,
and plasmids were sequenced to confirm that the correct sequence
had been inserted. In addition, the specificity of VP1 was
confirmed via cross-reactivity using ELISA kits for classical swine
fever virus (cat. no. SL0002Po; Sunglong Biotech Co., Ltd.,
Hangzhou, China), porcine respiratory and reproductive syndrome
virus (cat. no. KA2120; Abnova, Taipei, Taiwan), pseudorabies virus
(cat. no. PRV041117A; Zhejiang Gloria Bioscience, Co., Ltd.,
Hangzhou, China) and porcine circovirus type 2 virus (cat. no.
AE-200150-2; Alpha Diagnostic International, Inc., Texas, USA).
The expression plasmid was then transformed into
BL21(DE3) chemically competent cells (Takara Biotechnology, Co.,
Ltd.). Colonies that were positive were grown in liquid culture.
Following confirmation of the correct sequence, the bacteria were
expanded on a large scale and preserved with 50% glycerinin normal
saline at −20°C.
Protein expression and analysis
A small culture (15 ml) of LB media containing 100
µg/ml ampicillin was inoculated with 150 µl preserved bacteria.
After shaking at 37°C for 2–3 h, the optical density (OD) of the
culture at 600 nm reached 0.6–0.8. At this point, 1ml of the
culture was collected before addition of
isopropyl-β-D-1-thiogalactopyranoside (IPTG) to a final
concentration of 1 mM. Then expression was measured for several
hours at 37°C with shaking, and cultures were harvested at every
hour. The control was the non-carrier of pET32a that was induced by
IPTG for 5 h. All of the cultures were centrifuged for 3 min at
9,000 × g. Following centrifugation, the supernatant was discarded.
Pellets were resuspended in 50 µl 2X SDS buffer and degenerated at
100°C for 10 min. Following centrifugation for 3 min at 9,000 × g,
the supernatant were run on a 12% SDS-PAGE gel. Finally, the
recombinant protein activity was analyzed by western blotting. For
western blotting, the cells were rinsed three times with PBS, lysed
with radioimmunoprecipitation assay lysis buffer (Beyotime
Institute of Biotechnology) and incubated on ice for 30 min. The
lysates were then centrifuged at 13,400 × g for 20 min at 4°C and
the supernatant extracts were quantified for the total protein
using a Bicinchoninic Acid Protein Assay kit (Beyotime Institute of
Biotechnology) with bovine serum albumin as the standard. Aliquots
from each protein lysate sample were separated in 12% SDS-PAGE and
transferred to a polyvinylidene difluoride membrane. The membrane
was blocked with blocking buffer (TBS and 5% skimmed milk) for 1 h
at room temperature and incubated overnight in the presence of
rabbit anti-foot and mouth disease virus polyprotein (VP1 protein)
primary antibody (1:800; cat. no. PL0301542; PL Laboratories, Inc.,
Port Moody, Canada) at 4°C. The membrane was washed three times
with TBS containing 0.05% Tween-20 (TBST) and subsequently reacted
with anti-rabbit immunoglobulin G (IgG; H + L) conjugated to
horseradish peroxidase (HRP; 1:800; cat. no. E030120; EarthOx, LLC,
Millbrae, CA, USA) for 1 h at room temperature. Following thorough
washing with TBST, the immunoreactive bands were developed using
enhanced chemiluminescence detection reagents (EMD Millipore,
Billerica, MA, USA). The OD of the bands on the films was analyzed
using imaging software (ImageQuant Las 4000, version 1.2; GE
Healthcare Life Sciences, Chalfont, UK).
Recombinant protein (VP1) purification
and analysis
According to the optimal condition, a small culture
(10 ml) of LB media containing 100 µg/ml ampicillin was inoculated
with 10 µl preserved bacteria. After shaking overnight at 37°C, the
small culture was added to 500 ml LB media (with antibiotics at the
concentrations used for the small culture) and incubated at 37°C
with agitation until the OD of the culture at 600 nm reached
0.6–0.8. At this point, the culture was allowed to add IPTG to a
final concentration of 1 mM. Expression was allowed for several
hours at 37°C with shaking, and cultures were harvested at the
optimal time by centrifugation for 20 min at 9,000 × g. The pellets
were stored at −80°C until purification took place. In addition,
the purification of VP1 was conducted under a low temperature
environment, a condition of non degeneration.
The pellets (~1.0 g wet weight, representing 500 ml
liquid culture) were thawed on ice and resuspended completely in 20
ml lysis buffer (Beyotime Institute of Biotechnology). The cells
were sonicated on ice using 10 cycles of the following sequence: 1
sec on, 1 sec off for 20 sec; rest on ice for 40 sec. The lysate
was centrifuged at 13,400 × g for 15 min at 4°C, and the pellet was
collected. The solid white pellet was gently and completely
resuspended on ice with 10 ml binding buffer [20
mMTris(2-carboxyethyl)phosphine-HCl pH 7.9, 5 mM imidazole, 0.5 M
NaCl, 8 M urea]. Then centrifuged at 13,400 × g for 20 min at 4°C.
Following centrifugation, the supernatant was collected. The
solution was added to a column containing 1 ml complete His-Tag
Purification resin (CWBIO, Beijing, China) and washed extensively
with binding buffer containing 5 mM imidazole. EMCV VP1 protein was
eluted in elution buffer (Beyotime Institute of Biotechnology); two
elution buffers (10 ml each, containing 200 and 500 mM imidazole,
respectively) were added sequentially to the column, and the elute
was collected in 5 ml fractions. Fractions were then run on a 12%
SDS-PAGE gel.
Development of the indirect
ELISA-VP1
Optimal dilutions of VP1 and sera were determined by
a checkerboard titration test with EMCV positive and negative sera
previously analyzed by virus neutralization tests. The antibody of
VP1 has neutralizing activity, and it was confirmed that the virus
did not exist in the negative sera. The antigen was coated in
96-well ELISA plates and diluted in 0.05 mol/l carbonate buffer (pH
9.6) to 1:100, 1:200, 1:400, 1:800, 1:1,600 and 1:3,200. Reference
positive and negative sera were both diluted in 1:10, 1:20, 1:40
and 1:80, respectively, and tested to determine the optimal serum
dilution. The dilutions that gave the maximum difference in
absorbance at 450 nm between the positive and the negative sera
(P/N) were selected to test the sera panel. The working dilution of
rabbit anti-swine horseradish peroxidase (HRP)-IgG (Beijing
Solarbio Science & Technology Co., Ltd., Beijing, China), the
reaction temperature, time and other conditions also were
optimized.
Carbonate buffer solution (0.05 mol/l, pH 9.6)
served as coating buffer. The antigen was diluted to 0.605 µg/ml
and coated on 96-well microtiter plates with 100 µl, overnight at
4°C. The plates were washed three times for 5 min with PBS and 0.5%
Tween-20 (PBS-T). Then, the plates were blocked with blocking
buffer [Tris-buffered saline (TBS) and 5% skimmed milk] at 37°C for
1 h followed by a washing step. Subsequently, 100 µl sera was
diluted (1:40) in dilution buffer and incubated at 37°C for 1 h
followed by a washing step. A total of volume of 50 µl HRP-labeled
rabbit-anti-swine protein IgG (1:800; cat. no. E030110; EarthOx,
LLC) was added to all wells and incubated at 37°C for 1 h. After
washing again, 100 µl freshly prepared substrate solution (1 mg/ml
tetramethylbenzidine that contained 3 µl 30% hydrogen peroxide) was
dispensed into each well. Color development was in the dark for 15
min, and the reaction was stopped by addition of 50 µl 2 M sulfuric
acid. The absorbance values were read at 450 nm using an absorbance
microplate reader (BioTek Instruments, Inc., Winooski, VT,
USA).
To determine the cut-off value of the ELISA-VP1, 20
samples of SPF swine serum was obtained and the dilution was 1:40.
The test for indirect ELISA is done in the optimal conditions. The
result was analyzed by statistics to achieve the average value, X,
and the standard deviation, S. The samples with OD450≥X +3S were
positive. The samples with OD450<X+ 3S were negative.
Comparison of the indirect ELISA with
the virus neutralization tests
A total of 120 sera samples were obtained from
different swine herds in Baoding, China. They were tested to
evaluate the correlation between the neutralization test and the
indirect ELISA.
Quality assurance and repeatability of
ELISA
The repeatability (intra-plate variability) was
assessed by the same analyst who tested five sera samples, four
times, on the same plate, on the same day. The mean and
coefficients of variation (CVs) were computed using Excel 2003
(Microsoft Corporation, Redmond, WA, USA).
The reproducibility (inter-plate variability) of the
test results was assessed by testing five sera samples 10 times.
The results were obtained by use of distinct lots, on different
days, by different analysts. The mean and CVs were computed.
Field application of VP1-ELISA in
clinical samples
During 2013 and 2014, a total of 265 clinical swine
serum samples were collected from pig farms in surrounding of
Baoding, China. According to the established indirect ELISA method,
they were detected for the EMCV antibody.
Results
PCR amplification and sequencing of
the EMCV VP1 gene
The VP1 DNA was amplified with primers designed to
insert a BamHI restriction site at the 5′-end of the sequence and
an XhoI site at the 3′-end. Analysis of the reaction
products by agarose gel electrophoresis and ethidium bromide
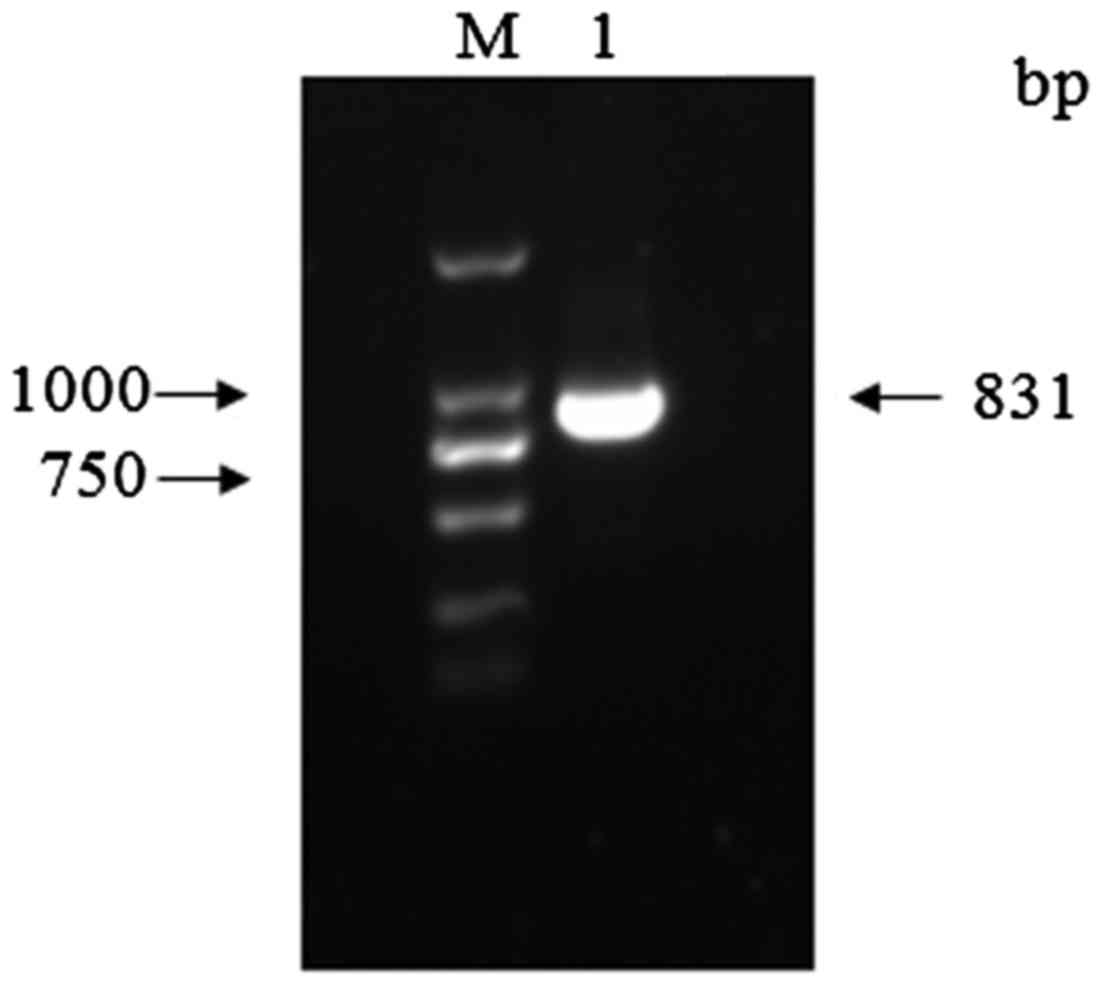
staining revealed a specific amplimer of the expected size, ~831 bp
(Fig. 1).
Expression and purification of
VP1
Following digestion, ligation and sequencing, the
plasmid was transformed into BL21(DE3) chemically competent cells
for expression. Expression was induced at 37°C and was allowed to
continue for 6 h. SDS-PAGE analysis of the samples indicated that
the control of pET32a induced for 5 h and the non-induced
recombinant VP1 protein did not have target bands. The cultures
that were induced by IPTG every hour show that a target band
appeared and the optimal expression time of VP1 protein is 5 h
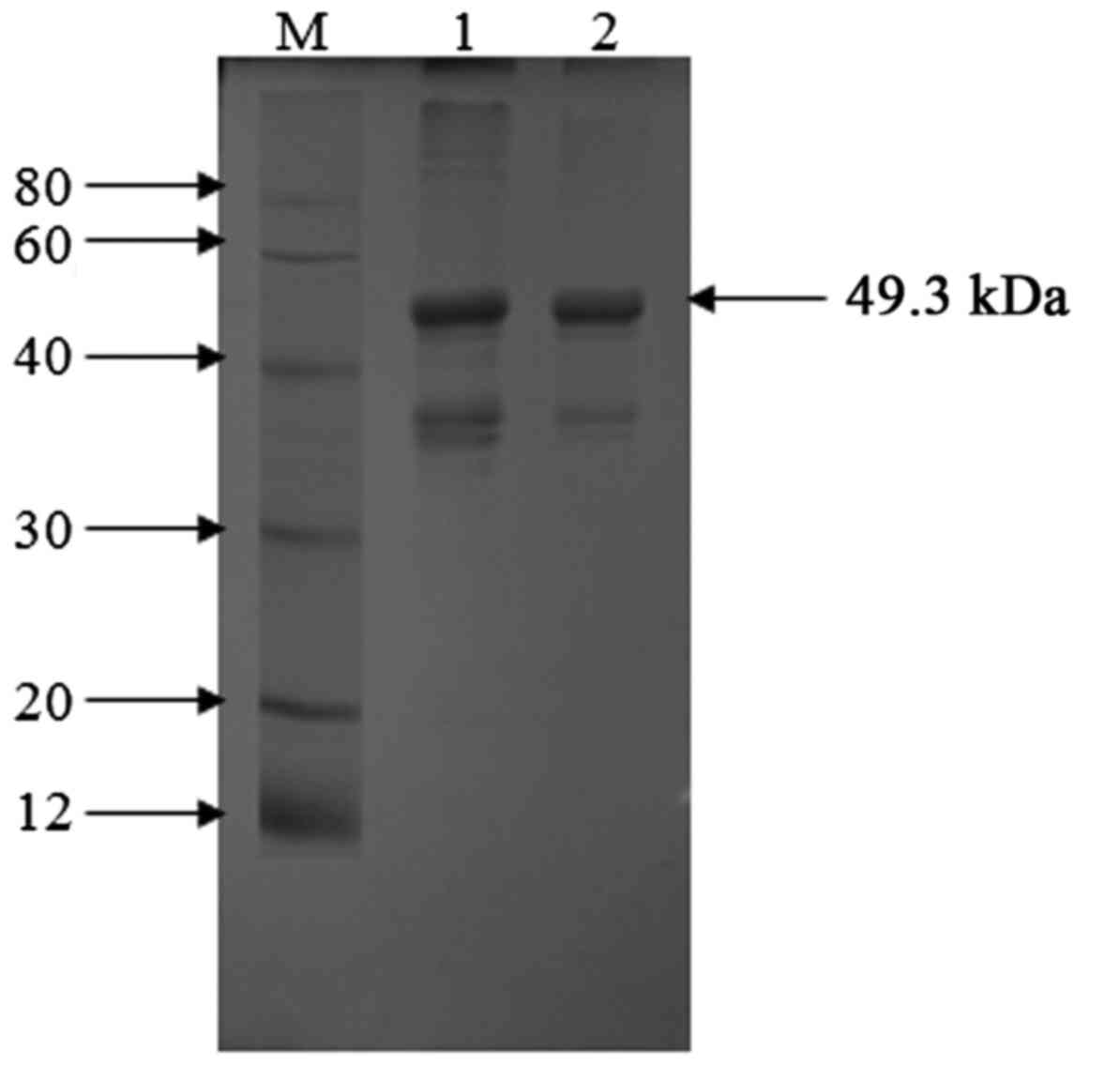
(Fig. 2). Following purification using
Ni-NTA, fractions were run on a 12% SDS-PAGE gel, where the
majority of pure VP1 protein was found to elute at 500 mM imidazole
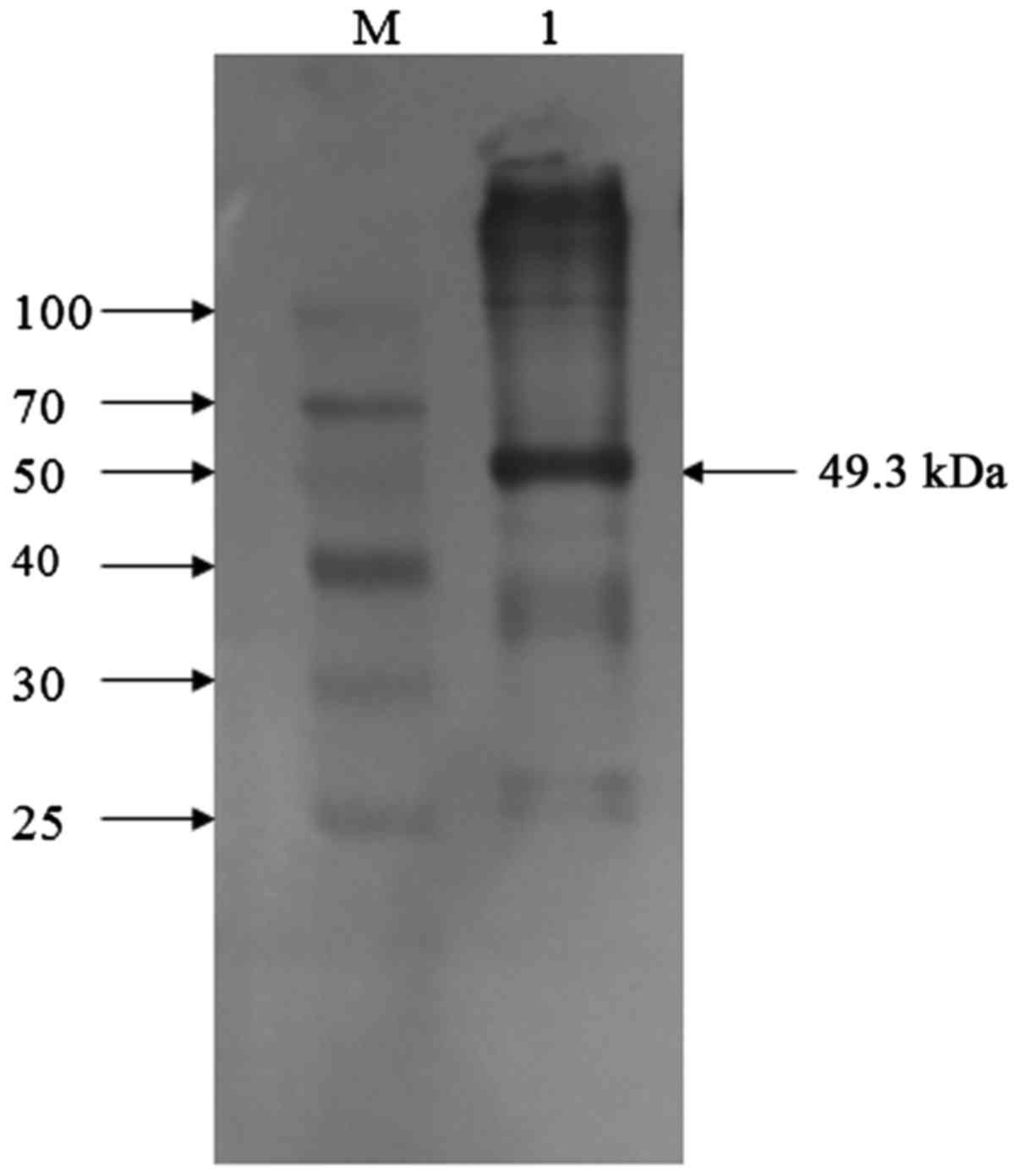
(Fig. 3). Western blotting with an
anti-EMCV VP1 monoclonal antibody revealed a specific band of the
same size (Fig. 4). By purifying,
0.484 mg/ml recombinant VP1 protein was obtained then stored in
−80°C. The positive immunoblotting results suggested that could
recombinant EMCV VP1 expressed and purified in Escherichia
coli could be used to develop an indirect ELISA test.
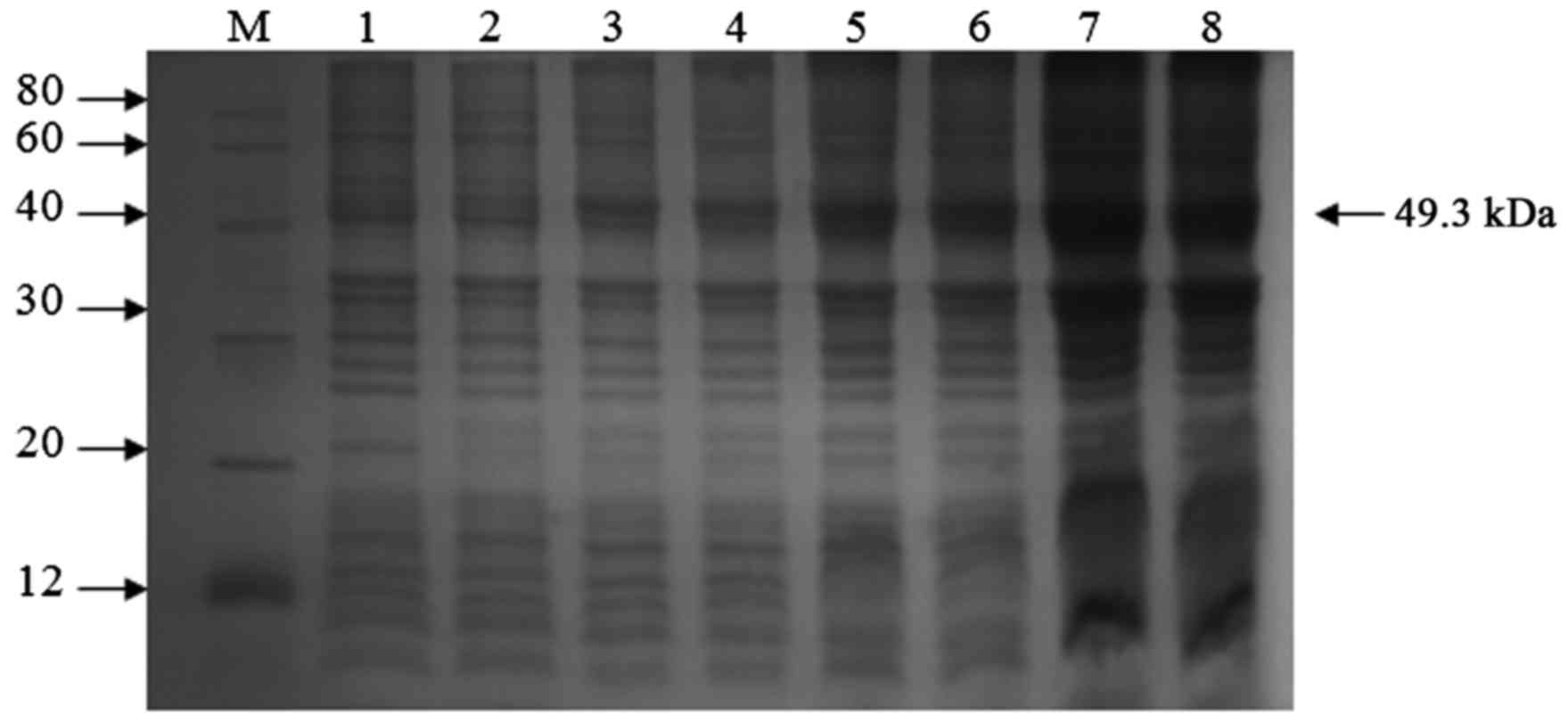 | Figure 2.SDS-PAGE analysis of recombinant VP1
protein expressed in pET32a. Lane 1, pET32a/BL21 induced with
IPTG(5h); lane 2, pET32a- VP1/BL21 without induction with IPTG;
lanes 3–8, pET-32a-VP1/BL21 induced with IPTG for 1, 2, 3, 4, 5 and
6 h, respectively. M, molecular weight marker; IPTG,
isopropyl-β-d-thiogalactoside. |
ELISA-VP1
By using checkerboard titration tests, the OD value
gave the maximum difference between the positive serum and negative
serum (P/N value of 5.62) when the dilutions of antigen and serum
were 1:800 and 1:40, respectively (Table
I). Therefore, the final concentration of coating antigen was
0.605 µg/ml by calculation, and the optimal dilution of the HRP-IgG
was 1:800 (Table II). A total of 20
samples of negative swine serum were tested under optimal
conditions by indirect ELISA. The highest OD450 was 0.194 and the
lowest OD450 was 0.103. The average value is 0.142 and the standard
deviation is 0.02463. X +3S=0.22. Therefore, the cut-off value is
0.22. A serum sample was considered positive if its P/N value was
≥0.22. At this value, the highest efficiency of sensitivity and
specificity was achieved. With this value, four of the 92 positive
samples determined by neutralization test were negative by indirect
ELISA, and two of the 28 negative sera by neutralization test were
tested positive by indirect ELISA. The ELISA gave 95.7% (88/92)
sensitivity and 92.9% (26/28) specificity, respectively (Table III). The result for 265 clinical
swine serum samples from Baoding demonstrated that 217 samples were
positive. The antibody positive rate against EMCV was 81.9%. It
indicated that swine in Baoding infected with EMCV is a serious
issue.
 | Table I.Determination of the coating
concentration of antigen and the dilution of serum samples. |
Table I.
Determination of the coating
concentration of antigen and the dilution of serum samples.
|
| OD of positive serum
with different coating antigen dilutions of recombinant VP1 |
|---|
|
|
|
|---|
| Serum dilution | 1:100 | 1:200 | 1:400 | 1:800 | 1:1,600 | 1:3,200 |
|---|
| 1:20 |
|
|
|
|
|
|
| P | 1.465 | 1.351 | 1.280 | 1.120 | 0.982 | 0.991 |
| N | 0.442 | 0.402 | 0.339 | 0.320 | 0.252 | 0.252 |
| P/N | 3.31 | 3.36 | 3.78 | 3.50 | 3.90 | 3.93 |
| 1:40 |
|
|
|
|
|
|
| P | 1.301 | 1.230 | 1.185 | 1.063 | 0.895 | 0.792 |
| N | 0.377 | 0.334 | 0.299 | 0.189 | 0.170 | 0.154 |
| P/N | 3.45 | 3.68 | 3.96 | 5.62 | 5.26 | 5.14 |
| 1:80 |
|
|
|
|
|
|
| P | 1.086 | 0.982 | 0.931 | 0.883 | 0.802 | 0.647 |
| N | 0.319 | 0.276 | 0.251 | 0.228 | 0.175 | 0.148 |
| P/N | 3.40 | 3.56 | 3.71 | 3.87 | 4.58 | 4.37 |
| 1:160 |
|
|
|
|
|
|
| P | 0.902 | 0.852 | 0.780 | 0.670 | 0.602 | 0.591 |
| N | 0.288 | 0.226 | 0.208 | 0.190 | 0.148 | 0.138 |
| P/N | 3.13 | 3.77 | 3.75 | 3.53 | 4.07 | 4.28 |
 | Table II.Determination of the optimal working
concentration of HRP-labeled rabbit anti-swine IgG. |
Table II.
Determination of the optimal working
concentration of HRP-labeled rabbit anti-swine IgG.
| HRP-labeled
dilution | Positive
serum(Ab) | Negative
serum(Ab) | P/N value |
|---|
| 1:200 | 1.327 | 0.276 | 4.81 |
| 1:400 | 1.269 | 0.214 | 5.93 |
| 1:800 | 1.108 | 0.179 | 6.19 |
| 1:1600 | 0.719 | 0.160 | 4.49 |
 | Table III.Presence of antibodies against EMCV as
determined by iELISA and neutralization test of 120 swine serum
samples from pig farms. |
Table III.
Presence of antibodies against EMCV as
determined by iELISA and neutralization test of 120 swine serum
samples from pig farms.
|
| Neutralization
test |
|
|---|
|
|
|
|
|---|
| iELISAa | Positive (+) | Negative (−) | Total (n) |
|---|
| Positive (+) | 88 | 2 | 90 |
| Negative (−) | 4 | 26 | 30 |
| Total | 92 | 28 | 120 |
Discussion
EMCV mainly causes piglet encephalitis, myocarditis
and sudden mortality. Many countries and regions have reported an
outbreak of the disease in swine herds. For the first time in 2005
in China, EMCV was isolated in dead piglets and aborted fetuses
(10). A serological survey indicated
that EMCV has become a danger to the healthy development of China's
pig industry (8). This research using
the prokaryotic expression vector of pET32a constructed EMCV VP1
gene recombinant expression plasmid pET32a-VP1, and in E.
coli BL21 (DE3) strains successfully expressed protein VP1.
Although the prokaryotic expression system cannot be modified, it
is often used to express high quantities of foreign proteins at low
cost. The current study used the prokaryotic expression system to
demonstrate that, following induction by IPTG, recombinant VP1
protein is highly expressed. The results indicated that the
accumulation of VP1 protein in the cytoplasm may have been due to
an imbalance between the rapid expression of exogenous protein and
its removal from the cell. The cultivation conditions and
availability of the amino acids components of VP1 may have also
been influential factors regarding the level of VP1 expression
(11). Busuttil et al
previously indicated that the soluble protein ratio may be
increased in the following ways: Reduction in temperature;
co-expression with a molecular chaperone; and addition of a
chemical reagent in the process of induction (12). Meanwhile, other experiments by our
group attempted to change several conditions, including
temperature, speed of the shaking table, and the dosage and
frequency of IPTG administration; however, the majority of the
recombinant protein was present in inclusion bodies (data not
shown). To improve the expression of soluble protein, other methods
should be trialed in future studies.
In addition, the present study also identified that
recombinant VP1 was a high activity protein in terms of sensitivity
and specificity. This may form a basis for the development of
clinical diagnostic kits and genetic engineering vaccines.
EMCV is a pathogen of zoonosis and can be diagnosed
by RT-qPCR (13,14). This method has high sensitivity, strong
specificity and a simple operation. It has been widely used in the
diagnosis of diseases of pigs. Nevertheless, this ELISA provided an
alternative, inexpensive and rapid serological detection method
that would be suitable for screening for anti-EMCV antibodies
titers on a large scale. In the current study, the authors
established an indirect ELISA to detect the antibodies against EMCV
with the recombinant pET32a-VP1 epitope protein in swine. By using
checkerboard titration tests, the optimal antigen concentration and
the dilutions of serum was obtained. Meanwhile, the dilution of the
secondary antibody is important as well.
EMCV infections are a worldwide issue. In Panama, a
strain of EMCV was isolated from sick pigs in 1958 for the first
time. By the 1970s, many countries, including Australia, Greece,
Belgium, South Africa, Italy and Japan had reported the disease
(15–17). The positive rate of neutralizing
antibody in serum ranges between 2 and 87% (18). In the Netherlands in 2006, 3,237 sera
from 277 pig breeding farms were analyzed to identify antibody
levels. 9.3% of the serum samples were positive, 58.8% of the sow
was positive (19). In Tianjin, 295
serum samples from 66 pig breeding farms in seven counties were
measured with indirect ELISA for the antibody levels and 84.75% of
the serum were positive (20). In 13
provinces of China, 3,250 serum samples from the 46 farms were
measured with this method for the antibody levels. The rate of
antibody positive is 72% (21).
In conclusion, the indirect ELISA assay was
developed successfully for the detection of antibodies to EMCV with
high levels of sensitivity and specificity. The assay will be
useful test for large-scale serological survey in EMCV infection
and monitoring antibodies titers against EMCV.
Acknowledgements
The present study was supported by the Natural
Science Foundation of Hebei Province in China (grant no.
C2016204191).
Glossary
Abbreviations
Abbreviations:
|
EMCV
|
encephalomyocarditis virus
|
|
ELISA
|
enzyme-linked immunosorbent assay
|
|
IPTG
|
isopropyl-β-d-thiogalactoside
|
References
|
1
|
Palmenberg AC, Kirby EM, Janda MR, Drake
NL, Duke GM, Potratz KF and Collett MS: The nucleotide and deduced
amino acid sequences of the encephalomyocarditis viral polyprotein
coding region. Nucleic Acids Res. 12:2969–2985. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sin JI, Sung JH, Suh YS, Lee AH, Chung JH
and Sung YC: Protective immunity against heterologous challenge
with EMCV by VP1 DNA vaccination: Εffect of coinjection with a
granulocyte-macrophage colony stimulating factor gene. Vaccine.
15:1827–1833. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Suh YS, Ha SJ, Lee CH, Sin JI and Sung YC:
Enhancement of VP1-specific immune responses and protection against
EMCV-K challenge by co-delivery of IL-12 DNA with VP1 DNA vaccine.
Vaccine. 19:191891–1898. 2001. View Article : Google Scholar
|
|
4
|
Carocci M and Bakkali-Kassimi L: The
encephalomyocarditis virus. Virulence. 3:351–367. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gelmetti D, Meroni A, Brocchi E, Koenen F
and Cammarata G: Pathogenesis of encephalomyocarditis experimental
infection in young piglets: A potential animal model to study viral
myocarditis. Vet Res. 37:15–23. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Krylova RI and Dzhikidze EK:
Encephalomyocarditis in monkeys. Bull Exp Biol Med. 139:355–359.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Spyrou V, Maurice H, Billinis C,
Papanastassopoulou M, Psalla D, Nielen M, Koenen F and Papadopoulos
O: Transmission and pathogenicity of encephalomyocarditis virus
(EMCV) among rats. Vet Res. 35:113–122. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ge X, Zhao D, Liu C, Wang F, Guo X and
Yang H: Seroprevalence of encephalomyocarditis virus in intensive
pig farms in China. Vet Rec. 166:145–146. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yuan W, Wang J, Sun M, Zheng Y, Li L,
Zhang X and Sun J: Rapid detection of encephalomyocarditis virus by
one-step reverse transcription loop-mediated isothermal
amplification method. Virus Res. 189:75–78. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ge X, Yang H, Guo X, Lu Y, Wang F, Chen Y
and Cha Z: Isolation and characterization of porcine
encephalomyocarditis virus. Chin J Anim Vet Sci. 38:59–65. 2007.(In
Chinese).
|
|
11
|
Long Y, Liu H and Wang Z: Purification and
renaturation of recombinant inclusion. Heilongjiang Animal Science
and Veterinary Medicine. 16:70–71. 2008.(In Chinese).
|
|
12
|
Busuttil BE, Turney KL and Frauman AG: The
expression of soluble, full-length, recombinant human TSH receptor
in a prokaryotic system. Protein Expr Purif. 23:369–373. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kassimi LB, Gonzague M, Boutrouille A and
Cruciere C: Detection of encephalomyocarditis virus in clinical
samples by immunomagnetic separation and one-step RT-PCR. J Virol
Methods. 101:197–206. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Maurice H, Nielen M, Stegeman JA,
Vanderhallen H and Koenen F: Transmission of encephalomyocarditis
virus (EMCV) among pigs experimentally quantified. Vet Microbiol.
88:301–314. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Koenen F and Vanderhallen H: Comparative
study of the pathogenic properties of a Belgian and a Greek
encephalomyocarditis virus (EMCV) isolate for sows in gestation.
Zentralbl Veterinarmed B. 44:281–286. 1997.PubMed/NCBI
|
|
16
|
Kudo H, Yoshizawa S, Hiroike T and Hirose
O: A retrospective serological survey of the encephalomyocarditis
virus among pigs in Chiba prefecture, Japan. J Vet Med Sci.
57:793–795. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Murnane TG, Craighead JE, Mondragon H and
Shelokov A: Fatal disease of swine due to encephalomyocarditis
virus. Science. 131:498–499. 1960. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Maurice H, Nielen M, Brocchi E, Nowotny N,
Kassimi LB, Billinis C, Loukaides P, O'Hara RS and Koenen F: The
occurrence of encephalomyocarditis virus (EMCV) in European pigs
from 1990 to 2001. Epidemiol Infect. 133:547–557. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Augustijn M, Elbers AR, Koenen F and
Nielen M: Estimation of seroprevalence of encephalomyocarditis in
Dutch sow herds using the virus neutralization test. Tijdschr
Diergeneeskd. 131:40–44. 2006.PubMed/NCBI
|
|
20
|
Dong Y, Huang J, Wang J, Zhao Y and Li L:
Development and application of an indirect ELISA for detection of
antibodies against EMCV. Chin Anim Quar. 25:30–33. 2008.(In
Chinese).
|
|
21
|
Zhang J, Ge X, Ma L, Chen Y, Cha Z, Guo X
and Yang H: Investigation of serum with infected EMCV swine in
large-scale pig farms. Chin J Vet Sci. 43:7–9. 2007.(In
Chinese).
|