Introduction
Hyperplasia of mammary gland (HMG) is also termed
mammary dysplasia. It does not involve inflammation or tumor
proliferative lesions. It is a disease characterized by basic
pathological changes in breast acinar and ductal epithelial cells,
as well as fibrous connective tissue hyperplasia. The peak age of
breast hyperplasia is 35–45 years (1). Mammary gland hyperplasia is a common
disease in women of child-bearing period, accounting for 75% of all
breast diseases, is considered to be precancerous lesions of the
breast. In China, HMG is increasing annually and regarded as a
precancerous lesion, which causes widespread concern. According to
modern medicine, breast tissue is the subject organ of a variety of
hormone actions. The physiological function of the breast tissue is
controlled by the hypothalamic pituitary ovary axis. Increased
sensitivity of any hormone, or breast tissue to hormones, is the
primary cause of breast hyperplasia (2).
MicroRNA (miRNA) is a class of non-encoding RNA
(length, ~22 nt), which regulates gene expression at the
post-transcriptional level (3). After
the stem loop structure of precursor miRNA is spliced by two RNase
III enzymes, Drosha and Dicer, the mature functional miRNA is
formed (4). The mature miRNAs are
incorporated into protein-coding transcripts and represses the
transcript translation or degrades the mRNA in mammals (5). Consequently, this regulates various
biological processes, including development, reproduction,
apoptosis, proliferation, pathogenesis, and lipometabolism
(6,7).
Thus, the abnormal expression of miRNAs may cause various diseases,
such as cardiac disease and cancer (8). A strong association between miRNA and
breast cancer has been reported in the literature. However, in
reviewing the literature, little data was identified regarding the
association between HMG and miRNAs. The aim of the present study
was to use a microRNA array to analyze the role of microRNAs in
rats exhibiting hyperplasia of the mammary glands.
Materials and methods
Sample preparation
A total of six Sprague Dawley specific pathogen-free
female rats (weight, ~240 g; age, 12 weeks) that were not pregnant
were used in the present study. The rats were obtained from Beijing
HFK Bio-science Co. Ltd. (Beijing, China) and reared in the animal
center of Liaoning University of Traditional Chinese Medicine
(Shenyang, China). The rats were housed at 21–25°C and 35–50%
humidity under a 12-h light/dark cycle and were provided feed and
sterilized water in a 1:2 ratio twice a day. All animals were fed
with a standard laboratory diet and tap water was provided ad
libitum. The animals were placed in the experimental room 24 h
before behavioral testing for acclimatization. The rats were housed
for five days to allow adaptation to the laboratory conditions. On
the sixth day, intramuscular 0.5 mg/kg/d estradiol benzoate (Wuhan
Amyjet Scientific, Inc., Wuhan, China) for 20 days, and 5 mg/kg/d
progesterone (An Apoptosis and Epigenetics Company, A8509) was
injected at day 26 for 5 days. The model cycle was 25 days, which
resulted in induction of the mammary gland hyperplasia model. After
modeling successfully, the rats were divided into two groups. A
control (MA) group was treated with physiological saline (2 ml)
once a day for 4 weeks, while a treatment (GA) group underwent 4
weeks of daily treatment with 0.15625g Jiedu Capsule (Radix
Bupleuri 15 g, Astragalus 15 g, honeysuckle 15 g, forsythia 15 g,
cantharidin 15 g; 0.5 g powder/capsule) per 1 kg body weight
dissolved in 2 ml distilled water by intragastric administration.
Following the treatment period, experimental samples were obtained
by surgery. The animals were anesthetized by intraperitoneal
injection with 10% chloral hydrate aqueous solution according to a
0.3 ml/100 g dose. The mammary glands of rats were numbered
according to order from head to tail. The whole third pair of
papillae was excised and placed in an Eppendorf tube and stored at
−80°C for RNA isolation. The study protocol was approved by Animal
Ethical Inspection Review Board of Liaoning University of
Traditional Chinese Medicine, and was conducted according to their
ethical principles of animal welfare.
RNA isolation and preparation
Total RNA was extracted from the mammary papilla
samples using an mirVana™ miRNA Isolation kit (Thermo Fisher
Scientific Inc., Waltham, MA, USA) according to the manufacturer's
instructions. The total RNA quantity and purity were analyzed using
a Bioanalyzer 2100 and RNA 6000 Nano LabChip kit (Agilent
Technologies, Inc., Santa Clara, CA, USA) with RNA integrity number
≥5.0 and an SSP-3000 Nanodrop spectrophotometer (Infinigen Biotech,
Inc., Hacienda Heights, CA, USA) at a wavelength of 260 nm.
Microarray hybridization and data
analysis
The Mouse & Rat miRNA OneArrayV6 (Phalanx
Biotech Group, Hsinchu, Taiwan) contains a total of 4,104 probes,
including 144 experimental control probes, 1,907 unique mouse miRNA
probes and 728 rat miRNA probes. Each probe on the microarray chip
has a chemically modified segment encoding nucleotides
complementary to the target based on miRBase version 17 (http://www.mirbase.org/). Rat genome-wide miRNA
microarray analysis was performed by Beijing Ori-Gene Science and
Technology Corp., Ltd. (Beijing, China). Briefly, fluorescent
targets were prepared from 2.5 µg total RNA using the miRNA ULS
Labeling kit (Kreatech Diagnostics, Amsterdam, The Netherlands).
Labeled miRNA targets were enriched using NanoSep 100K (Pall
Corporation, Port Washington, NY, USA), enriched products were
hybridized to the Mouse & Rat miRNA One Array v3 in Phalanx
hybridization buffer using the One Array Hybridization Chamber, and
hybridized overnight at 37°C, non-specifically bound targets were
removed by three washing steps (first wash, at 37°C for 5min;
second wash, at 37°C for 5min and at 25°C for 5 min; and third wash
included 20 rinses at 37°C). Then, the slides were dried and
scanned using an Axon 4000B scanner (Molecular Devices, LLC,
Sunnyvale, CA, USA).
Multiple sample analyses were conducted, including
normalization, data adjustment, analysis of variance and
clustering. The fluorescence signal intensity of Cy5 of each point
was analyzed by GenePix 4.1 software (Molecular Devices, LLC) and
processed using the R program. The data were initially analyzed by
subtracting the background, and then spots for which the flag was
<0 were filtered out, and spots that passed these criteria were
normalized using a 75% media scaling normalization method.
Normalized spot intensities were converted into gene expression
log2 ratios for the MA and GA groups. Spots with log2 ratios ≤-1 or
≥1 and P<0.05 were selected for further analysis. The
differential expression was analyzed using average linkage
algorithm to cluster and Pearson correlation as a measure of
similarity.
Results
Upregulated miRNA targets in
microarray analysis
In order to investigate the difference of gene
expression between the GA and MA groups, the differences in the
distribution of all detected gene probe signals were statistically
analyzed. The screening conditions of differentially expressed
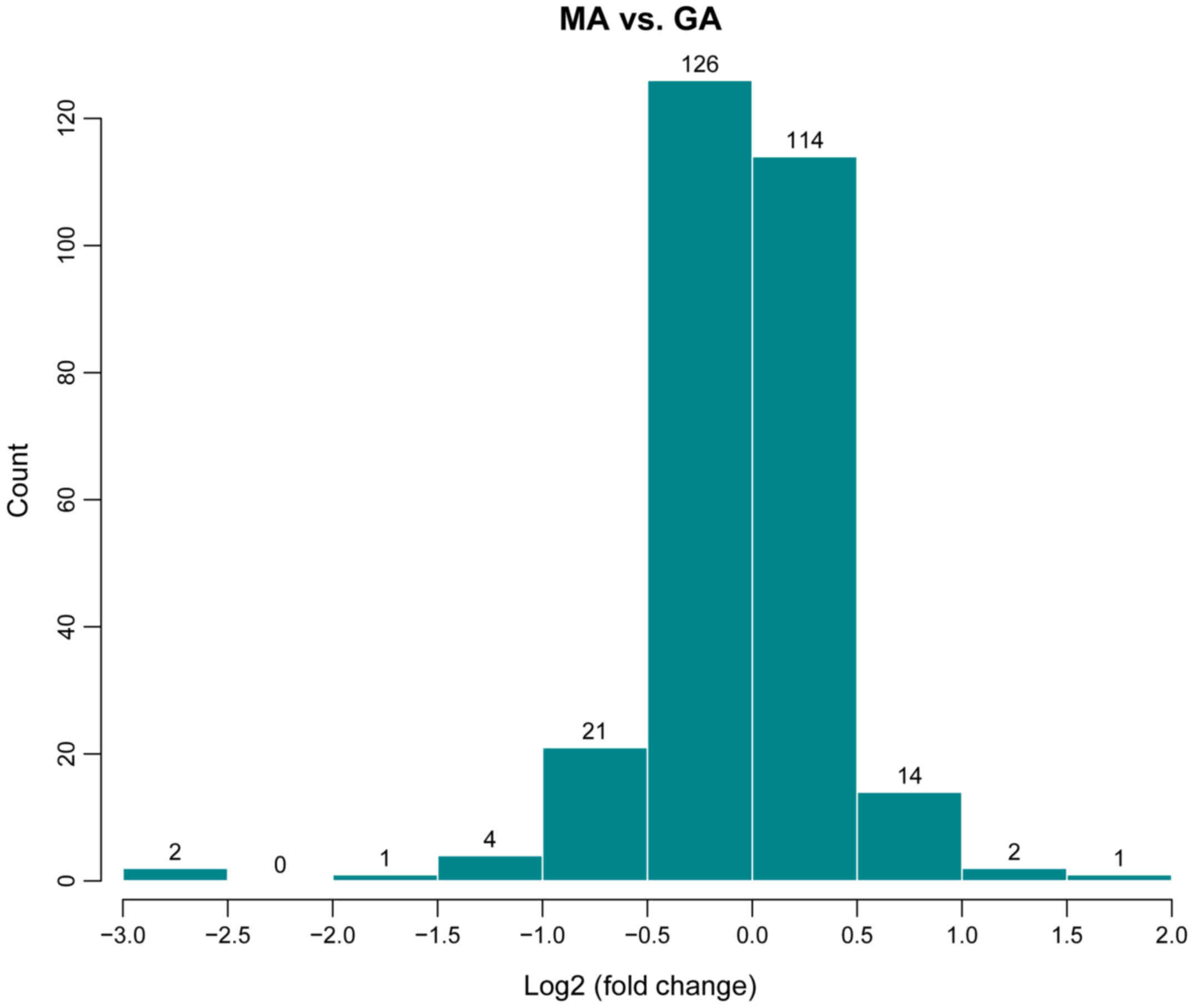
genes are log2 fold-change ≥0.585 and P<0.05 (9). The results of the miRNA microarray
(Fig. 1) indicated 13 upregulated
miRNAs (rno-let-7b-5p, rno-let-7c-5p, rno-miR-31a-5p, rno-miR-297,
rno-miR-466c-5p, rno-miR-30c-1-3p, rno-miR-672-5p,
rno-miR-125b-1-3p, rno-miR-32-3p, rno-miR-3588, rno-miR-466d,
rno-miR-494-3p and rno-miR-6315) and 20 downregulated miRNAs
(rno-miR-30c-2-3p, rno-miR-1-3p, rno-miR-292-5p, rno-miR-133a-3p,
rno-miR-877, rno-miR-615, rno-miR-133b-3p, rno-miR-185-5p,
rno-miR-135a-3p, rno-miR-678, rno-miR-711, rno-miR-296-5p,
rno-miR-298-5p, rno-miR-326-5p, rno-miR-211-3p, rno-miR-290,
rno-miR-3564, rno-miR-3573-5p, rno-miR-423-5p and rno-miR-652-5p)
in the drug treatment group (GA).
Principal component analysis
(PCA)
A PCA method (10,11) was
used to assess differences between the transcriptomes of rats
treated with Jiedu Capsule and controls. PCA1 and PCA2 represented
the top two dimensions of the genes exhibiting differential
expression among the samples, which accounted for 93.41 and 3.21%
of the differentially expressed genes, respectively (Fig. 2).
Cluster analysis of differentially
expressed genes
Cluster analysis is a method of simplifying data by
data modeling, the purpose is to obtain the distribution of data
and the characteristics of each cluster, and the association
between sample clusters is described by the distance index. An
in-depth analysis of differentially expressed genes was conducted,
which were identified with the criteria of P<0.05 and expression
fold change >2.0. Unsupervised hierarchical clustering analysis
was used to investigate the similarity of the whole gene expression
between the experimental samples. Cluster analysis identified that
all the six samples were clustered into two groups (Fig. 3); the 3 samples in the control group
were clustered into one class while those in the treatment group
were clustered into another class. This indicates that the trend of
gene expression is consistent.
Discussion
Hyperplasia of the breast is a common disease in
women aged 20–55 years and is associated with an increased risk of
developing cancer. Breast cancer is the second most common type of
cancer in women, and is an important cause of mortality in most
frequent cancer of cancer-related of death worldwide and generates
a large economic burden (12). A
previous study identified that women with atypical hyperplasia (AH)
had a four- to five-fold increase in breast cancer risk (13). Furthermore, a strong association
between AH and breast cancer was identified (14).
In the present study, miRNA microarray scanning was
performed on the breast tissue of rats that had undergone drug
treatment. It was identified that the expression levels of the 13
miRNAs in the GA and MA groups were markedly different; the
expression levels of 13 miRNAs in group GA was greater than in
group MA, and the expression levels of 20 miRNAs was significantly
decreased in the GA group, and the difference was statistically
significant.
The experimental results and numerous previous
studies reported that miR-31 is over expressed in metastatic breast
cancer (15). Rasheed et al
(16) and Valastyan et al
(17) identified that the primary
function of miR-31 promotes multiple oncogenic phenotypes,
including proliferation, motility and invasion of cancer cells.
Another notable finding was that manipulation of miR-31 expression
levels may be used to modulate senescence-associated pathological
conditions, such as cancer and the ageing process (18). The current study demonstrated that the
expression level of miR-31 in the GA group was significantly
greater than in the MA group.
By contrast, the expression level of miR-30 in the
GA group was lower than that in the MA group. The current study
indicated that breast cancer cells grown under non-attachment
conditions display a unique pattern of miRNA expression, compared
with the parental cells, as highlighted by a marked low expression
of miR-30 family members. Ouzounova et al (19) showed that miR-30a regulates
non-attachment growth. Furthermore, it was demonstrated that
miR-30c may regulate the GATA binding protein 3 gene in breast
cancer at the transcriptional level (20).
Thus, certain genes found in the present study are
in agreement with the existing studies. However, further studies
are required to verify other miRNAs.
In conclusion, the present study identified the
expression profile of circulating miRNAs in the model group and GA
group. Following a scan of the chip, 13 upregulated and 20
downregulated miRNAs were identified, and the functions of these
miRNAs were described. It was demonstrated that the expression
levels of miRNAs are closely associated with biological processes,
such as mammary gland development and cell proliferation. The
present study provides a basis upon which to further investigate
the molecular mechanism of HMG. However, there was no validation of
the differentially expressed miRNAs, and future work is required to
confirm their function. The results of the present study also
suggested that hyperplasia of mammary glands occurs as a result of
complex and dynamic processes regulated by numerous factors,
including multiple miRNAs. Therefore, the functions and
significance of the differentially expressed miRNAs identified in
this study require further investigation. Future work should aim to
functionally analyze miRNAs associated with hyperplasia of mammary
glands and their corresponding target genes or gene regulatory
networks, in an expanded sample size to further verify results.
Acknowledgements
The current study was partially supported by the
Science Public Welfare Research Foundation of Liaoning Province,
China (2014001021), the Key Laboratory of Ministry of Education for
TCM Viscera-State Theory and Applications Foundation (zyzx1609),
the Natural Science Foundation of Liaoning Province, China
(20170540612), the Educational Commission of Liaoning Province,
China (L201717), the Science and Technology Planning Project of
Shenyang, Liaoning, China (18–013-0-11) and the College Students'
innovation and entrepreneurship training program project, Liaoning
University of Traditional Chinese Medicine, (201610162034 and
201710162000088).
References
|
1
|
Chang XJ, Zhou J, Zhang S, Chen J, Chen
CM, Wang ZZ and Xiao W: Effects of guizhi fuling capsule on sex
hormone levels and breast issue morphology of mammary gland
hyperplasia model rats. Zhongguo Zhong Yao Za Zhi. 39:4139–4142.
2014.(In Chinese). PubMed/NCBI
|
|
2
|
Liu B and Guan ZH: Radix bupleuri Extract
Inhibits Hyperplasia of Mammary Gland in Rats. Trop J Pharm Res.
15:2932016. View Article : Google Scholar
|
|
3
|
Ekmekci C, Mousses S and Tuzmen S:
MicroRNA (miRNA), involvement in aberrant promoter methylation
facilitating tumor progression. Cancer Res. 67:1046.
2007.PubMed/NCBI
|
|
4
|
Rahimi G, Jafari N, Khodabakhsh M, Shirzad
Z and Dogaheh HP: Upregulation of microRNA processing enzymes
Drosha and Dicer in gestational diabetes mellitus. Gynecol
Endocrinol. 31:156–159. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mallick B and Ghosh Z: Probing
Evolutionary Biography of MicroRNAs and Associated Factors. Curr
Genomics. 13:144–152. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bae S, Lee EM, Cha HJ, Kim K, Yoon Y, Lee
H, Kim J, Kim YJ, Lee HG, Jeung HK, et al: Resveratrol alters
microRNA expression profiles in A549 human non-small cell lung
cancer cells. Mol Cells. 32:243–249. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Martins IJ: Induction of NAFLD with
Increased Risk of Obesity and Chronic Diseases in Developed
Countries. Open J Endocr Metab Dis. 04:90–110. 2014. View Article : Google Scholar
|
|
9
|
Badri H, Monsieurs P, Coninx I, Nauts R,
Wattiez R and Leys N: Temporal Gene Expression of the
Cyanobacterium Arthrospira in Response to Gamma Rays. PLoS One.
10:e01355652015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vyas S and Kumaranayake L: Constructing
socio-economic status indices: How to use principal components
analysis. Health Policy Plan. 21:459–468. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee C, Abdool A and Huang CH: PCA-based
population structure inference with generic clustering algorithms.
BMC Bioinformatics. 10(Suppl 1): S732009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Venkatadri R, Muni T, Iyer AKV, Yakisich
JS and Azad N: Role of apoptosis-related miRNAs in
resveratrol-induced breast cancer cell death. Cell Death Dis.
7:e21042016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dupont WD and Page DL: Risk factors for
breast cancer in women with proliferative breast disease. N Engl J
Med. 312:146–151. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dupont WD, Parl FF, Hartmann WH, Brinton
LA, Winfield AC, Worrell JA, Schuyler PA and Plummer WD: Breast
cancer risk associated with proliferative breast disease and
atypical hyperplasia. Cancer. 71:1258–1265. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cho JH, Dimri M and Dimri GP: MicroRNA-31
is a transcriptional target of histone deacetylase inhibitors and a
regulator of cellular senescence. J Biol Chem. 290:10555–10567.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rasheed SA, Teo CR, Beillard EJ, Voorhoeve
PM, Zhou W, Ghosh S and Casey PJ: MicroRNA-31 controls G protein
alpha-13 (GNA13) expression and cell invasion in breast cancer
cells. Mol Cancer. 14:672015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Valastyan S, Reinhardt F, Benaich N,
Calogrias D, Szász AM, Wang ZC, Brock JE, Richardson AL and
Weinberg RA: A pleiotropically acting microRNA, miR-31, inhibits
breast cancer metastasis. Cell. 137:1032–1046. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cho JH, Dimri M and Dimri GP: MicroRNA-31
is a transcriptional target of histone deacetylase inhibitors and a
regulator of cellular senescence. J Biol Chem. 290:10555–10567.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ouzounova M, Vuong T, Ancey PB, Ferrand M,
Durand G, Le-Calvez Kelm F, Croce C, Matar C, Herceg Z and
Hernandez-Vargas H: MicroRNA miR-30 family regulates non-attachment
growth of breast cancer cells. BMC Genomics. 14:1392013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bockhorn J, Dalton R, Nwachukwu C, Huang
S, Prat A, Yee K, Chang YF, Huo D, Wen Y, Swanson KE, et al:
MicroRNA-30c inhibits human breast tumour chemotherapy resistance
by regulating TWF1 and IL-11. Nat Commun. 4:13932013. View Article : Google Scholar : PubMed/NCBI
|