Introduction
Lung cancer is one of the most common types of
malignancy and it had the highest mortality rate globally of all
types of cancer among males (1). In
2011, >1.6 million novel lung cancer cases were diagnosed and
~1.3 million patients with lung cancer succumbed to the disease
worldwide (2). Although therapeutic
advances, including surgical resection and chemoradiotherapy, have
improved the prognosis of patients with lung cancer, the overall
survival rates and quality of life remain unsatisfactory. The
migration and invasion of cancer cells that results in metastasis
presents the greatest challenge to clinical therapy (3). Hence, investigation of the molecular
mechanisms of lung cancer cell invasion and migration may provide
novel evidence to improve clinical therapy.
ATP-binding cassette transporter E1 (ABCE1) is a
member of the ATP-binding cassette transporter family (4). Binding of the ABCE1 protein with the
translation initiation factor, prolongation factor or termination
factor serves an important function in promoting the recycling of
ribosomes (5,6). A number of studies have confirmed that
ABCE1 is associated with the proliferation, apoptosis, migration
and invasion of lung cancer cells (4,7).
However, the mechanism of ABCE1 in the occurrence of lung cancer
remains unknown, to the best of our knowledge.
Acetylation modification of proteins participates in
a number of biological processes, including genetic transcription,
protein-protein interaction, protein stability, and cell
proliferation, migration and differentiation (8). Acetylation of proteins may influence
their capacity to bind with DNA and their activity and stability,
thus resulting in the occurrence of cancer and changing the
biological behaviors of tumor cells (9,10).
Acetylation or deacetylation of enzymes may affect their
transcriptional activity and result in various dysfunctions of
genes and proteins, inducing diseases associated with the migration
and invasion of cancer cells (11).
In a previous study, it was demonstrated that the acetylation rate
of ABCE1 was associated with the TNM stage and lymph node
metastasis (12). However, whether
the acetylation of the ABCE1 gene is associated with the occurrence
of lung cancer remains unknown to the best of our knowledge.
The HIV-1 Tat interactive protein (Tip60) is a
lysine acetyltransferase and a member of the MYST family of histone
acetyltransferases (13). A previous
study revealed that the Tip60-histone acetyltransferase complex has
a critical role in DNA repair and apoptosis and that Tip60 binds to
histone H3 trimethylated at lysine 9 to activate ataxia
telangiectasia mutant kinase (14).
Tip60 was demonstrated to be significantly associated with 5-year
disease-free survival of patients with primary melanoma or
metastatic melanoma (15). In
addition, it has been demonstrated that the Tip60
haploinsufficiency observed in breast and prostate cancer allows
Tip60 to function as an oncogene (16). The aim of the present study was to
investigate how Tip60 may interact with ABCE and whether Tip60
could be a valuable diagnostic and prognostic biomarker and
potential therapeutic agent for lung cancer.
Materials and methods
Cell culture and cell lines
The human lung cancer A549 cell line and the
immortalized human bronchial epithelial HBE cell line (as normal
control cells) were obtained from the American Type Culture
Collection (Manassas, VA, USA). The cells were cultured in
RPMI-1640 with 10% fetal bovine serum (FBS; both Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA), 100 U/ml penicillin and
100 µg/ml streptomycin and maintained in an incubator at 37°C and
5% CO2.
Bioinformatics analysis
In order to confirm which enzyme is responsible for
the acetylation of ABCE1, an acetylation set enrichment based
(ASEB) computer program (http://cmbi.bjmu.edu.cn/huac/predict) (17) was used. ASEB can predict not only the
acetylation state, but also the responsible lysine
(K)-acetyl-transferase (KAT) family. The prediction method was
initially established based on the sequence similarity principle.
To predict whether lysine sites within a given protein sequence can
be acetylated by specific KAT families, the KAT family of interest
was initially selected.
Transfection
The sequence of the target gene Tip60-small
interfering RNA (siRNA) was as follows:
5′-GGGCACCATCTCCTTCTTTdTdT-3′. A549 cells were seeded in 6-well
plates at a density of ~3×105 cells/well. A single cell
suspension was prepared using the cells in the logarithmic growth
phase and they were washed with RPMI-1640 without FBS twice. The
cells were incubated without FBS for 4 h at 37°C. The
lentivirus-based Tip60-siRNA expression system (Si-1) and the
negative scrambled control (Si-N; 5′-CAACAAGATGAAGAGCACCdTdT-3′)
were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China).
Cell infection was performed using the riboFECT™ CP Transfection
kit (Guangzhou RiboBio Co., Ltd.) according to the manufacturer's
protocol. Briefly, a total of 5 µl 20 µM siRNA was added into 120
µl 1X riboFECT™ CP Buffer, and the mixture incubated for 5 min at
37°C. Subsequently, 12 µl riboFECT™ CP Reagent was added into the
mixture and it was incubated for 15 min at 37°C. The riboFECT™ CP
mixture was diluted with 1,863 µl RPMI-1640 to a total volume of 2
ml. The cells were cultured for a further 48 h, at which point the
subsequent experiments were performed. The blank control group (N)
consisted of untransfected A549 cells.
Co-immunoprecipitation (Co-IP)
A549 and HBE cells were lysed on ice with
radioimmunoprecipitation assay (RIPA) buffer (Thermo Fisher
Scientific, Inc.) and equal amounts of protein (500 µg) from the
whole cell extracts were pre-cleared with protein A/G agarose beads
(Thermo Fisher Scientific, Inc.) at 4°C for 2 h. Subsequently, the
protein was incubated with Immunoglobulin G (control antibody,
dilution, 1:1,000; cat. no. ab172730; Abcam, Cambridge, MA, USA) or
ABCE1 antibody (dilution, 1:500; cat. no. ab185548; Abcam) for 24 h
at 4°C with gentle shaking. The beads were washed 3 times with 1 ml
RIPA buffer, and the immune complexes were eluted by boiling at
100°C for 5 min in 2X SDS-containing sample buffer. The beads were
then washed 3 times using PBS, and 50 µg protein per lane was
resolved using SDS-PAGE (10% gel) followed by immunoblotting with
anti-acetyl lysine antibody (dilution, 1:5,000; cat. no. ab185548;
Abcam) overnight at 4°C.
Western blotting
Cells were lysed on ice for 20 min with RIPA buffer
and the protein concentration was assessed using a BCA kit
(Beyotime Institute of Biotechnology, Haimen, China). The cell
lysates (Si-1, Si-N and N) and immunocomplexes were resolved using
12% SDS-PAGE and electronically transferred onto polyvinylidene
fluoride membranes (EMD Millipore, Billerica, MA, USA). Following
blocking at 4°C overnight with 5% non-fat milk in Tris-buffered
saline with 0.1% Tween-20, the membranes were incubated with
primary antibodies against ABCE1 (dilution, 1:5,000; cat. no.
ab185548; Abcam), GAPDH (dilution, 1:10,000; cat. no. AP0063;
Bioworld Technology, Inc., St. Louis Park, MN, USA),
cyclin-dependent kinase inhibitor 1A (dilution, 1:3,000; cat. no.
ab109520; Abcam), caspase 3 (dilution, 1:5,000; cat. no. ab13847;
Abcam), MDM2 proto-oncogene (dilution, 1:3,000; cat. no. ab38618;
Abcam) and B cell leukemia/lymphoma 2 (dilution, 1:500; cat. no.
ab32124; Abcam) at 4°C overnight. Subsequently, the membranes were
washed three times with TBST buffer. The membranes were then
incubated with anti-rabbit (dilution, 1:10,000; cat. no. ab150077;
Abcam) or anti-mouse (dilution, 1:10,000; cat. no. ab150113; Abcam)
secondary antibodies conjugated to horseradish peroxidase (EMD
Millipore) at room temperature for 2 h. The bands were visualized
using enhanced chemiluminescence (Thermo Fisher Scientific, Inc.)
Densitometry was calculated using Quantity One software 4.6.2
(Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Cell proliferation assay
Cells (Si-1, Si-N and N) were seeded in a 96-well
plate at a density of 5×103 cells/well. After 12, 24, 48
and 72 h, 50 µl 1X MTT (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) solution was added to each well and they were incubated
for 4 h at 37°C. Following centrifugation for 5 min at 1,000 × g,
the supernatant was eliminated and 150 µl dimethyl sulfoxide
(Beijing Solarbio Science & Technology Co., Ltd, Beijing,
China) was added. The wells were incubated at room temperature for
3 h. Subsequently, the optical density was detected at 570 nm to
form a cell proliferation curve.
Flow cytometric assays of cell
apoptosis
The apoptosis of the three groups of cells (Si-1,
Si-N and N) was assessed using flow cytometry. The cells were
collected following 48 h of transfection, digested by trypsin
without EDTA, and then PBS was used to wash them three times.
Subsequently, the cells were collected and fixed in 70% ethanol
overnight at 4°C, and then they were centrifuged for 5 min at 300 ×
g three times in 4°C. Cells (5×105) were suspended in
100 µl 1X Binding Buffer (part of the Annexin V FITC Apoptosis
Detection kit II; BD Biosciences, San Jose, CA, USA) and mixed with
5 µl annexin V-fluorescein isothiocynate (Annexin V FITC Apoptosis
Detection kit II; BD Biosciences) and 5 µl propidium iodide (PI;
Becton, Dickinson and Company, Franklin, Lakes, NJ, USA) at room
temperature. The mixture was then incubated for 15 min at 25°C.
Subsequently, 400 µl 1X Binding Buffer (Annexin V FITC Apoptosis
Detection Kit II) was added to each tube and the samples were
evaluated using flow cytometry. The data were analyzed with
CellQuest software version 5.1 (BD Biosciences).
Wound healing assay
Cells of the three groups (Si-1, Si-N and N) were
plated in 6-well plates (5×103 cells/ml per well) with
RPMI-1640 and 10% FBS. After 24 h, the cells were serum deprived
and scratched with a 200-µl plastic pipette tip to create a linear
wound. The monolayer was washed twice with PBS to remove debris and
detached cells, and then the cells were exposed to serum-free
medium at 37°C and 5% CO2. The wound was observed under
an inverted microscope (Leica Microsystems GmbH, Wetzlar, Germany)
at ×100 magnification at 0, 24 and 48 h. The results are expressed
as a spread index, which was determined as follows: Distance
migrated by the Si-1-treated cells relative to the distance
migrated by the Si-N-treated cells.
Transwell experiments
Migration assays of the A549 cells were performed
using a Transwell chamber (8-µm pore size; Corning, Inc., Corning,
NY, USA). A549 cells of the three groups (Si-1, Si-N and N) were
seeded (2.5×105 cells per chamber) into the upper
chamber with 1% FBS RPMI-1640 medium. The lower chamber was loaded
with RPMI-1640 medium supplemented with 10% FBS. Following
incubation for 24 h at 37°C in 5% CO2, the cells on the
upper surface of the filters were removed using cotton swabs. All
of the migrated cells were fixed at 25°C with 4% paraformaldehyde
for 15 min and then stained for 5 min at room temperature with 1%
crystal violet solution. Cell migration was quantified by counting
the number of stained cells per field in 10 random fields for each
chamber under an epifluorescence inverted microscope
(magnification, ×100). Identical chambers coated with Matrigel were
used to determine the invasive potential.
Animal study
A total of 12 female NOD/Scid mice (age, 6–8 weeks;
weight range, 18–22 g) were purchased from Beijing Vital River
Laboratory Animal Technology Co., Ltd. (Beijing, China) and housed
at a humidity of 55% and a temperature of 26°C with a 12-h
light/dark cycle and ad libitum access to food and water.
The mice were maintained in a pathogen-free facility. The mice were
randomly divided into Si-1, Si-N and N groups (n=4 per group). A549
cells (5×104 cells) infected with Si-1 or Si-N or
untransfected cells were injected into the right buttock of each
mouse. The tumor volumes were measured every 4 days. Following 1
month, the mice were sacrificed by cervical dislocation and the
tumors were dissected and weighed. All animal experiments were
performed in accordance with the AVMA guidelines and the guidelines
established by China Medical University (Shenyang, China). The
present protocol was approved by the Animal Welfare and Research
Ethics Committee of China Medical University.
Statistical analysis
Statistical analysis was performed using SPSS
software version 19.0 (IBM Corp., Armonk, NY, USA). The measurement
data are presented as the mean ± standard deviation or standard
error. Two-group continuous variable comparisons were performed
using an unpaired Student's t-test and multiple group comparisons
were performed using one-way analysis of variance followed by
Student-Newman-Keuls test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Tip60 regulates ABCE1 acetylation in
HBE and A549 cells
Co-IP and western blotting were used to confirm the
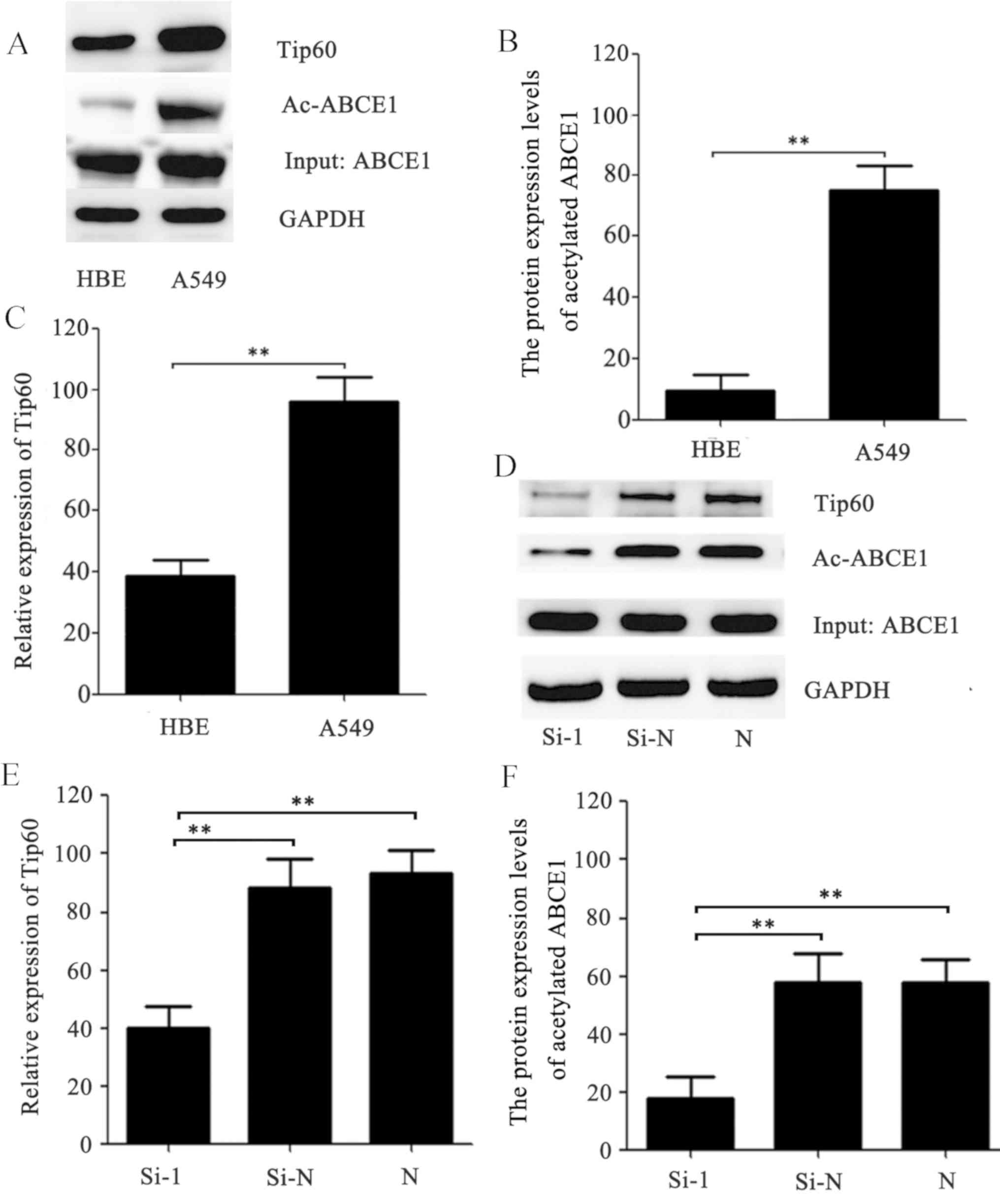
acetylation of ABCE1 in A549 and HBE cells. As presented in
Fig. 1A and B, the acetylation of
ABCE1 was significantly upregulated in the A549 cells compared with
the HBE cells (P<0.01). In order to investigate the enzyme that
regulates the acetylation of ABCE1, ASEB was used to predict that
Tip60 has the potential to bind to ABCE1 (data not shown). Western
blot analysis and Co-IP were subsequently used to determine that
the protein expression levels of Tip60 were significantly reduced
in the HBE cells compared with those in the A549 cells (P<0.01;
Fig. 1A and C). RNA interference was
then used to knockdown Tip60 in the A549 cells. The results
demonstrated that the Tip60 expression levels were significantly
reduced in the cells transfected with Si-1 siRNA compared with
those in the cells transfected with the negative control Si-N siRNA
(P<0.01; Fig. 1D and E).
Consistent with a previous study (18), the Si-1 siRNA significantly inhibited
Tip60 expression and reduced ABCE1 acetylation compared with the
Si-N and N groups (P<0.01; Fig. 1D
and F). Collectively, the results indicate that Tip60 directly
regulates the acetylation of ABCE1.
Tip60-siRNA suppresses A549 cell
proliferation, invasion and migration and induces cancer cell
apoptosis
In order to assess the influence of Tip60 on cell
proliferation in lung cancer, an MTT assay was performed. The cells
were transfected with siRNA against Tip60 and a negative control.
The proliferative ability of the Si-1-transfected cells was lower
compared with that of the Si-N-transfected cells at 48 and 72 h.
The MTT assay demonstrated that A549 cell proliferation was
markedly suppressed by Si-1 siRNA at 72 h (P<0.05; Fig. 2A). Wound healing and Transwell assays
were performed to evaluate the suppressive effect of Tip60-siRNA in
lung cancer cells. As expected, there was a marked reduction in the
wound healing, migration and invasion of the A549 cells transfected
with Si-1 siRNA when compared with the control cells (P<0.01;
Fig. 2B-D). Downregulation of Tip60
expression and acetylation of ABCE1 inhibits tumor growth. In order
to further elucidate how Tip60 and ABCE1 acetylation suppress tumor
growth, flow cytometry combined with annexin V-FITC/PI staining was
used to analyze cell apoptosis. Notably, the results indicated that
cell apoptosis was significantly increased in the cells transfected
with Si-1 siRNA compared with the control groups (P<0.05;
Fig. 2E and F).
Tip60-siRNA activates the apoptotic
pathway
Western blotting was used to investigate the genes
involved in the apoptotic pathway. Downregulating the expression of
Tip60 in the A549 cells markedly inhibited MDM2 and BCL2 expression
compared with the control cells (Fig.
3). Furthermore, downregulation of Tip60 increased the
expression levels of the apoptosis-associated protein p21 and
cleaved-caspase 3 in the A549 cells (Fig. 3). Collectively, the results indicate
that downregulation of Tip60 expression directly activates the
apoptotic pathway to suppress tumor cell proliferation, invasion
and migration.
 | Figure 3.Tip60-siRNA activates the apoptotic
pathway. Western blot analysis of Tip60, MDM2, P21, BCL2, caspase
3, cleaved caspase-3 and GAPDH in the A549 cells following
transfection with Si-1 or Si-N. Tip60, Tat interactive protein 60
kDa; MDM2, MDM2 proto-oncogene; p21, cyclin-dependent kinase
inhibitor 1A; BCL2, B cell lymphoma 2; Si-1, cells transfected with
Tip60-siRNA; Si-N, cells transfected with negative control siRNA;
N, untransfected cells; siRNA, small interfering RNA. |
Tip60-siRNA inhibits lung cancer
growth in vivo
To confirm whether Tip60 siRNA may serve the same
function in the suppression of tumor growth in vivo, a
subcutaneous xenograft model was used. Consistent with the in
vitro results, downregulation of Tip60 expression significantly
reduced the tumor weight (P<0.01) and markedly decreased the
tumor volume compared with the mice injected with control cells
(Fig. 4A and B).
Discussion
Lung cancer is one of the most common types of
tumor, with increasing rates of incidence and mortality globally
(2). Previously, chemotherapy,
radiotherapy and surgery have been used to treat patients with lung
cancer; however, traditional treatments have a low remission rate
for patients with advanced cancer and often result in negative side
effects, including anemia and multiple organ dysfunction syndrome
(19). Now that there is a greater
understanding of oncogenes, targeted therapies have provided a
better choice for patients with advanced cancer. However,
resistance and other problems that emerge from targeted therapies
in lung cancer exist. Furthermore, a greater number of target genes
and mechanisms require elucidation to improve the clinical
treatment for lung cancer (20).
There are various types of protein modifications,
including acetylation, methylation, ubiquitination and
phosphorylation, which heritably regulate gene expression.
Acetylation is involved in the majority of cellular biological
processes (21). The current
evidence indicates that protein acetylation participates in almost
all normal vital processes in a cell and serves a notable role in
initiating a number of events in certain types of cancer by
altering gene expression, including regulation of oncogenes and/or
tumor suppressors (22–25). Acetylation or deacetylation may
influence a wide range of intracellular biological functions,
including chromatin stability, the cell cycle, gene transcription
and DNA damage repair (9,10). The pathways of non-histone
acetylation serve an essential function in tumorigenesis (26). Previous studies have demonstrated
that certain types of acetylase may facilitate growth, invasion and
chemotherapy resistance of prostate, bladder and lung cancer cells
(27,28). As an important member of the MYST
family, Tip60 serves a particular role in the acetylation of
certain transcription factors and non-histone proteins (12). Tip60 may acetylate K73/K76 of the
important transcriptional activator Twist and induce it to bind
with bromodomain containing protein 4, which is a member of the
bromodomain and extra-terminal family of proteins. This further
activates the expression of Wnt family member 5A and thereby
upregulates breast cancer cell invasion and tumorsphere formation
(29). In prostate cancer, Tip60
promotes cancer cell proliferation via translocation of the
androgen receptor into the nucleus, and Tip60 silencing arrests the
cell cycle at the G1 phase (18).
Additionally, Tip60 regulates the MYC proto-oncogene and
amplification of bHLH transcription factors, and participates in
events that result in tumorigenesis in various types of human
cancer (30). A previous study
demonstrated that Tip60 inhibitor may induce cell apoptosis to
suppress gastric cancer cell proliferation by increasing the levels
of activated caspase-3 and caspase-9 (11). Additionally, Tip60 inactivation
reduces the non-histone acetylation levels of certain transcription
factors in a DNA-recovered defective nucleus, thus influencing DNA
repair and gene mutation and causing cancer (31). In the present study, it was revealed
that downregulation of Tip60 may inhibit cancer cell migration and
invasion, which is consistent with the results of a previous study
(19).
ABCE1 exerts a broad range of functions, which are
mainly involved in the translocation of substrates across
membranes, but are also involved in translation, DNA repair and
chromosome maintenance (5). It has
been confirmed that ABCE1 is involved in cell proliferation and
anti-apoptotic activity and serves an important role in the
translation process (32). Based on
the functions of the ABCE1 gene, a previous study suggested that
the ABCE1 gene is associated with tumor formation, prognosis and
metastasis (7). It has been
demonstrated that when the ABCE1-silencing gene is transfected into
tumor cells, there is a substantial change in their proliferation
and apoptosis (33). A previous
study revealed that the expression levels of the ABCE1 protein in
lung carcinoma tissues and metastatic lymph nodes are higher
compared with those in normal tissues (7). However, how the ABCE1 gene functions as
a tumor-associated gene that induces the invasion and migration of
tumor cells in lung adenocarcinoma remains unknown. As Tip60
regulates a number of other genes, including EPC1 and DMAP1, and
participates in cancer recurrence, further studies are required to
fully elucidate the mechanism (34).
The present study, to the best of our knowledge, is
the first to demonstrate that ABCE1 is acetylated in lung
adenocarcinoma cells and that acetylation of ABCE1 is rare in
normal bronchial epithelial cells, indicating that the occurrence,
development and migration of lung adenocarcinoma may be closely
associated with acetylation of the ABCE1 protein. Furthermore, a
Co-IP assay demonstrated that downregulation of Tip60 reduces ABCE1
acetylation significantly. The results suggest that ABCE1
acetylation may be regulated by the catalytic effect of Tip60 and
thereby influence the biological behaviors of tumor cells. An MTT
assay and xenograft model were used to assess whether knockdown of
Tip60 expression and ABCE1 acetylation were able to inhibit lung
cancer cell proliferation and growth in vitro and in
vivo. A wound healing and Transwell assay demonstrated that the
migration and invasion abilities of lung adenocarcinoma cells were
significantly reduced when ABCE1 acetylation was decreased by the
downregulation of Tip60. Additionally, western blotting and flow
cytometric analysis indicated that Tip60 performs functions via
activating the apoptotic pathway. However, although the present
study demonstrated that the acetylation of ABCE1 is regulated by
Tip60, further studies are required in order to identify the
binding site of Tip60. Furthermore, the mechanism by which ABCE1
acetylation regulates the apoptotic pathway of tumor cells remains
to be elucidated.
The present study is, to the best of our knowledge,
the first to suggest that occurrence and development of lung cancer
are associated with the acetylation of the ABCE1 protein.
Furthermore, the present study revealed that downregulation of
Tip60 reduces ABCE1 acetylation and induces apoptosis of tumor
cells. Tip60 may be a key molecule regulating ABCE1 acetylation in
lung cancer cells and therefore may be a promising therapeutic
target in advanced lung cancer.
Acknowledgements
The authors would like to thank the China Medical
University Experimental Technology Center (Shenyang, China) for the
provision of laboratory equipment.
Funding
The present study was supported by the Natural
Science Foundation of China (grant no. 30973502).
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
ZYL designed the study and wrote the manuscript. ZYL
and QY performed the experiments. DLT provided the reagents and
mice, and provided technical support and conceptual advice. HTJ
analyzed and interpreted the data and DLT reviewed and edited the
manuscript. All authors read and approved the manuscript.
Ethics approval and consent to
participate
All experimental procedures conformed to the Guide
for the Care and Use of Laboratory Animals published by the US
National Institutes of Health (NIH Publication No. 85-23, revised
1996) and the study was approved by the Institutional Animal Ethics
Committee of China Medical University (Shenyang, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fiala O, Šorejs O, Pešek M and Fínek J:
Immunotherapy in the treatment of lung cancer. Klin Onkol. 30
(Supplementum3):S22–S31. 2017.(In Czech). View Article : Google Scholar
|
|
4
|
Ren Y, Li Y and Tian D: Role of the ABCE1
gene in human lung adenocarcinoma. Oncol Rep. 27:965–970. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen ZQ, Dong J, Ishimura A, Daar I,
Hinnebusch AG and Dean M: The essential vertebrate ABCE1 protein
interacts with eukaryotic initiation factors. J Biol Chem.
281:7452–7457. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Becker T, Franckenberg S, Wickles S,
Shoemaker CJ, Anger AM, Armache JP, Sieber H, Ungewickell C,
Berninghausen O, Daberkow I, et al: Structural basis of highly
conserved ribosome recycling in eukaryotes and archaea. Nature.
482:501–506. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang B, Gao Y, Tian D and Zheng M: A
small interfering ABCE1-targeting RNA inhibits the proliferation
and invasiveness of small cell lung cancer. Int J Mol Med.
25:687–693. 2010.PubMed/NCBI
|
|
8
|
Creppe C, Malinouskaya L, Volvert ML,
Gillard M, Close P, Malaise O, Laguesse S, Cornez I, Rahmouni S,
Ormenese S, et al: Elongator controls the migration and
differentiation of cortical neurons through acetylation of
alpha-tubulin. Cell. 136:551–564. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhuang S: Regulation of STAT signaling by
acetylation. Cell Signal. 25:1924–1931. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang W, Wang Z and Lei QY: Acetylation
control of metabolic enzymes in cancer: An updated version. Acta
Biochim Biophys Sin (Shanghai). 46:204–213. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shi M, Lu XJ, Zhang J, Diao H, Li G, Xu L,
Wang T, Wei J, Meng W, Ma JL, et al: Oridonin, a novel lysine
acetyltransferases inhibitor, inhibits proliferation and induces
apoptosis in gastric cancer cells through p53- and
caspase-3-mediated mechanisms. Oncotarget. 7:22623–22631.
2016.PubMed/NCBI
|
|
12
|
Teicher BA and Fricker SP: CXCL12
(SDF-1)/CXCR4 pathway in cancer. Clin Cancer Res. 16:2927–2931.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang X, Wu J and Luan Y: Tip60: Main
functions and its inhibitors. Mini Rev Med Chem. 17:675–682. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau
LA, Whetstine JR and Price BD: Histone H3 methylation links DNA
damage detection to activation of the tumour suppressor Tip60. Nat
Cell Biol. 11:1376–1382. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen G, Cheng Y, Tang Y, Martinka M and Li
G: Role of Tip60 in human melanoma cell migration, metastasis, and
patient survival. J Invest Dermatol. 132:2632–2641. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Brown JA, Bourke E, Eriksson LA and Kerin
MJ: Targeting cancer using KAT inhibitors to mimic lethal
knockouts. Biochem Soc Trans. 44:979–986. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang L, Du Y, Lu M and Li T: ASEB: A web
server for KAT-specific acetylation site prediction. Nucleic Acids
Res 40 (Web Server issue). W376–W379. 2012. View Article : Google Scholar
|
|
18
|
Shiota M, Yokomizo A, Masubuchi D, Tada Y,
Inokuchi J, Eto M, Uchiumi T, Fujimoto N and Naito S: Tip60
promotes prostate cancer cell proliferation by translocation of
androgen receptor into the nucleus. Prostate. 70:540–554.
2010.PubMed/NCBI
|
|
19
|
Socinski MA, Obasaju C, Gandara D, Hirsch
FR, Bonomi P, Bunn PA Jr, Kim ES, Langer CJ, Natale RB, Novello S,
et al: Current and emergent therapy options for advanced squamous
cell lung cancer. J Thorac Oncol. 13:165–183. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zappa C and Mousa SA: Non-small cell lung
cancer: Current treatment and future advances. Transl Lung Cancer
Res. 5:288–300. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Martínez-Reyes I and Chandel NS:
Acetyl-CoA-directed gene transcription in cancer cells. Genes Dev.
32:463–465. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lovinsky-Desir S and Miller RL:
Epigenetics, asthma, and allergic diseases: A review of the latest
advancements. Curr Allergy Asthma Rep. 12:211–220. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Garagnani P, Pirazzini C and Franceschi C:
Colorectal cancer microenvironment: Among nutrition, gut
microbiota, inflammation and epigenetics. Curr Pharm Des.
19:765–778. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang K, Tian S and Fan E: Protein lysine
acetylation analysis: Current MS-based proteomic technologies.
Analyst. 138:1628–1636. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shanmugam MK, Arfuso F, Arumugam S,
Chinnathambi A, Jinsong B, Warrier S, Wang LZ, Kumar AP, Ahn KS,
Sethi G and Lakshmanan M: Role of novel histone modifications in
cancer. Oncotarget. 9:11414–11426. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hirano G, Izumi H, Kidani A, Yasuniwa Y,
Han B, Kusaba H, Akashi K, Kuwano M and Kohno K: Enhanced
expression of PCAF endows apoptosis resistance in
cisplatin-resistant cells. Mol Cancer Res. 8:864–872. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shiota M, Yokomizo A, Tada Y, Uchiumi T,
Inokuchi J, Tatsugami K, Kuroiwa K, Yamamoto K, Seki N and Naito S:
P300/CBP-associated factor regulates Y-box binding protein-1
expression and promotes cancer cell growth, cancer invasion and
drug resistance. Cancer Sci. 101:1797–1806. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shi J, Wang Y, Zeng L, Wu Y, Deng J, Zhang
Q, Lin Y, Li J, Kang T, Tao M, et al: Disrupting the interaction of
BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like
breast cancer. Cancer Cell. 25:210–225. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Patel JH, Du Y, Ard PG, Phillips C,
Carella B, Chen CJ, Rakowski C, Chatterjee C, Lieberman PM, Lane
WS, et al: The c-MYC oncoprotein is a substrate of the
acetyltransferases hGCN5/PCAF and TIP60. Mol Cell Biol.
24:10826–10834. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gorrini C, Squatrito M, Luise C, Syed N,
Perna D, Wark L, Martinato F, Sardella D, Verrecchia A, Bennett S,
et al: Tip60 is a haplo-insufficient tumour suppressor required for
an oncogene-induced DNA damage response. Nature. 448:1063–1067.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huang B, Zhou H, Lang X and Liu Z:
siRNA-induced ABCE1 silencing inhibits proliferation and invasion
of breast cancer cells. Mol Med Rep. 10:1685–1690. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Qu X and Zhang L: Effect of
ABCE1-silencing gene, transfected by electrotransfer, on the
proliferation, invasion, and migration of human thyroid carcinoma
SW579 cells. Genet Mol Res. 14:14680–14689. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Judes G, Rifaï K, Ngollo M, Daures M,
Bignon YJ, Penault-Llorca F and Bernard-Gallon D: A bivalent role
of TIP60 histone acetyl transferase in human cancer. Epigenomics.
7:1351–1363. 2015. View Article : Google Scholar : PubMed/NCBI
|