Introduction
Cancer is a leading cause of mortality worldwide,
accounting for 7.6 million fatalities (around 13% of total
fatalities) in 2008. Cancer-associated fatalities worldwide are
projected to continue to rise to >13.1 million by 2030 (1,2). To
date, the commonly adopted approaches to cancer treatment are
surgery, radiotherapy and chemotherapy. However, there are
increasing limitations resulting from poor prognosis and the
instance of negative side-effects (3,4).
Furthermore, carcinogenesis involves multiple genetic and
epigenetic changes; therefore, a single chemopreventive agent may
not be sufficient to prevent these events (5). In addition, it has been observed that
certain effective therapeutic agents act on multiple targets rather
than one specific disease-associated target (6). Therefore, the use of a combination of
agents that have multi-target effects may be an improved treatment
strategy for cancer.
There is increasing evidence that Chinese herbal
medicines are frequently administered to counteract the
side-effects of chemotherapy and radiotherapy in patients who are
being treated for cancer (5,7), and
they have been adopted as adjuvants for cancer therapy in the USA
(8). Furthermore, natural products
have been indicated to be more promising candidates for cancer
treatments than purely synthetic compounds (9). The earliest Chinese medicine book,
Huangdi neijing, referred to symptoms that were comparable
to those of cancer >2,000 years ago (10). Herbal medicines containing numerous
constituents demonstrate variable pharmacological activities; thus,
multi-herb therapy may be an effective, conventional and
complementary medical approach for cancer prevention and control
(11). Therefore, understanding
the molecular mechanisms of Chinese herbal medicines may aid the
modernization of herbal remedies and the discovery of novel cancer
treatments.
Xiao Chai Hu Tang (XCHT), a traditional herbal
formula from the medical treatise Shang Han Lun that was developed
by Zhang in the Eastern Han Dynasty, has been administered for the
treatment of cancer in China (10,11).
Data from recent studies have demonstrated that XCHT treats cancer
by enhancing immune regulation, anti-angiogenesis and the apoptosis
of tumor cells (11–13). However, the underlying molecular
mechanisms of its anticancer effects are poorly understood.
Therefore, the aim of the present study was to elucidate the
multi-target effects of the compounds in XCHT, based on an
established computational pharmacological model (14,15).
The information may aid the development of a combination of agents
for the treatment of cancer.
Materials and methods
Collection and chemical space mapping of
the compounds in XCHT
XCHT consists of seven medicinal herbs, namely
Radix Bupleuri, Scutellaria baicalensis, Panax ginseng, Zizyphi
Fructus, Pinellia ternata, Zingiber officinale and Radix
Glycyrrhizae. A total of 434 non-duplicated compounds present
in these herbs were identified in the Chinese Herbal Drug Database
and the Handbook of the Constituents in Chinese Herb Original
Plants (16,17). The two-dimensional (2D) structures
of the compounds were drawn using ISIS Draw version 2.5 (MDL
Information Systems, Inc., San Leandro, CA, USA), transformed into
3D-molecule models and optimized using Discovery Studio 2.0 (DS
2.0; Accelrys, Inc., San Diego, CA, USA) with a Merck molecular
force field. Using the quantitative structure-activity relationship
module of DS 2.0 (Accelrys, Inc.), 150 diversity descriptors of the
compounds, including 1D-, 2D- and 3D-molecular descriptors
(14), were calculated. Principal
component analysis (PCA) was performed to map the chemical space
distribution of the compounds.
Molecular docking
The crystal structures of the protein-ligand
complexes for the 29 cancer-associated targets were used for the
docking calculations that were performed using the LigandFit module
within the DS 2.0 software (18,19).
They were downloaded from the Research Collaboratory for Structural
Bioinformatics (RCSB) Protein Data Bank (PDB; http://www.rcsb.org/pdb/home/home.do;
Table I). For each crystal
structure, the crystallographic water molecules were removed, the
missing hydrogen atoms were added and the inhibitor from the
crystal structure was used to define the active site. The 434
compounds identified as being present in XCHT were docked into the
protein models and the interactions between the compounds and
proteins were evaluated using a DockScore (20). The 434 docked structures were
sorted according to their DockScore, in readiness for network
construction being undertaken.
 | Table ITwenty-nine key target proteins that
are associated with cancer. |
Table I
Twenty-nine key target proteins that
are associated with cancer.
| Protein | PDB code |
|---|
| Human androgen
receptor | 1E3G |
| Epidermal growth
factor receptor | 1M17 |
| Farnesyl protein
transferase | 1SA4 |
| Fibroblast
collagenase | 2TCL |
| Heat shock protein
90 | 1UYD |
|
Inosine-5′-monophosphate
dehydrogenase | 1ME9 |
| Kinesin spindle
protein | 2FL6 |
| Multidrug
resistance-associated protein 1 | 2CBZ |
| NAD(P)H quinone
oxidoreductase | 1KBQ |
| Poly [ADP-ribose]
polymerase-1 | 1UK0 |
| Protein-tyrosine
phosphatase SHP-1 | 1FPR |
| Receptor
tyrosine-protein kinase erbB-4 | 3BBT |
|
Serine/threonine-protein kinase PLK1 2RKU
VEGFR2 | 3B8R |
| Tyrosine-protein
kinase SRC | 1FMK |
| Bcl-xL | 1YSI |
| Bcl-2 | 2022 |
| CDK2 | 1AQ1 |
| Cytochrome P450
19A1 | 3EQM |
| DNA topoisomerase
II | 1ZXM |
| Angiopoietin-1
receptor | 2OO8 |
| Estrogen
receptor | 1UOM |
| c-Met | 3EFJ |
| Histone deacetylase
4 | 2VQV |
| MMP-7 | 1MMR |
| PPARγ | 1I7I |
| PI3Kγ | 2A5U |
| Protein kinase
B | 3CQU |
| Thymidylate
synthase | 1CI7 |
Network construction and analysis
Cytoscape 2.8.3 (The Cytoscape Consortium, San
Diego, CA, USA) was used to construct the subsequent networks
(21,22). In order to construct the
compound-target (C-T) network, a compound node and a target node
were linked if the DockScore of the compound and the target was in
the top 13 (top 3%) of all the compounds (15). If two compounds shared ≥1 target,
they were linked to create a compound-compound (C-C) association
network. The data were analyzed using Cytoscape plugins (The
Cytoscape Consortium).
Results
Chemical space distribution of the
compounds in XCHT
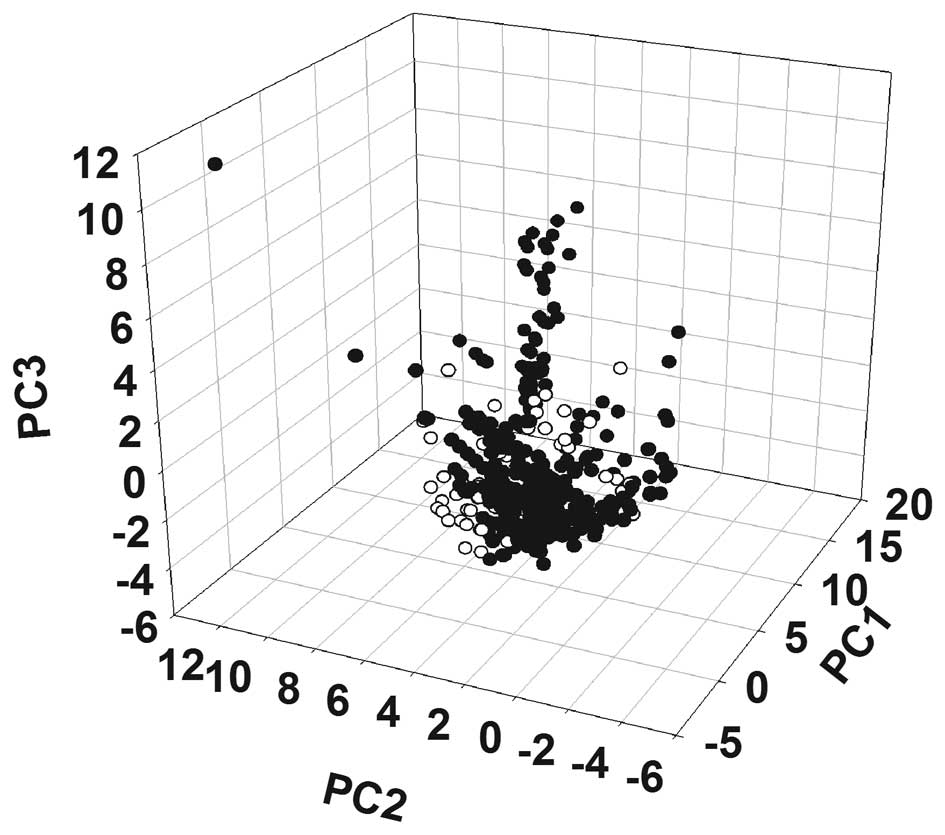
The chemical space of XCHT was mapped based on the
PCA and the results of the mapping are shown in Fig. 1 and Table II. The distribution of XCHT in the
chemical space ranges from dense to loose. Table II shows that the means of the
molecular weight, the number of hydrogen-bond donors, the number of
hydrogen-bond acceptors and AlogP were 376.26, 3.26, 5.94 and 2.71,
respectively. According to Lipinski’s rule of five (23), the majority of the compounds in
XCHT exhibited drug-like properties. To further demonstrate the
drug-like properties of the compounds in XCHT, the map of chemical
space of the known inhibitors that are associated with cancer
targets, according to the Therapeutic Targets Database (TTD) was
also constructed (Fig. 1)
(18). The majority of the known
inhibitors and the compounds present in XCHT were observed to have
clustered at the bottom back corner of the chemical space.
 | Table IIMean, SD, minimum and maximum of the
key molecular descriptors of compounds in Xiao Chai Hu Tang. |
Table II
Mean, SD, minimum and maximum of the
key molecular descriptors of compounds in Xiao Chai Hu Tang.
| Molecular
descriptor | Mean | SD | Minimum | Maximum |
|---|
| No. of carbon
atoms | 20.75 | 13.47 | 2 | 60 |
| No. of nitrogen
atoms | 0.12 | 0.66 | 0 | 7 |
| No. of oxygen
atoms | 5.87 | 6.31 | 0 | 29 |
| Molecular
weight | 376.26 | 272.67 | 58.03 | 1271.44 |
| No. of hydrogen
acceptors | 5.94 | 6.32 | 0 | 29 |
| No. of hydrogen
donors | 3.26 | 3.88 | 0 | 19 |
| AlogP | 2.71 | 2.78 | −7.58 | 19.52 |
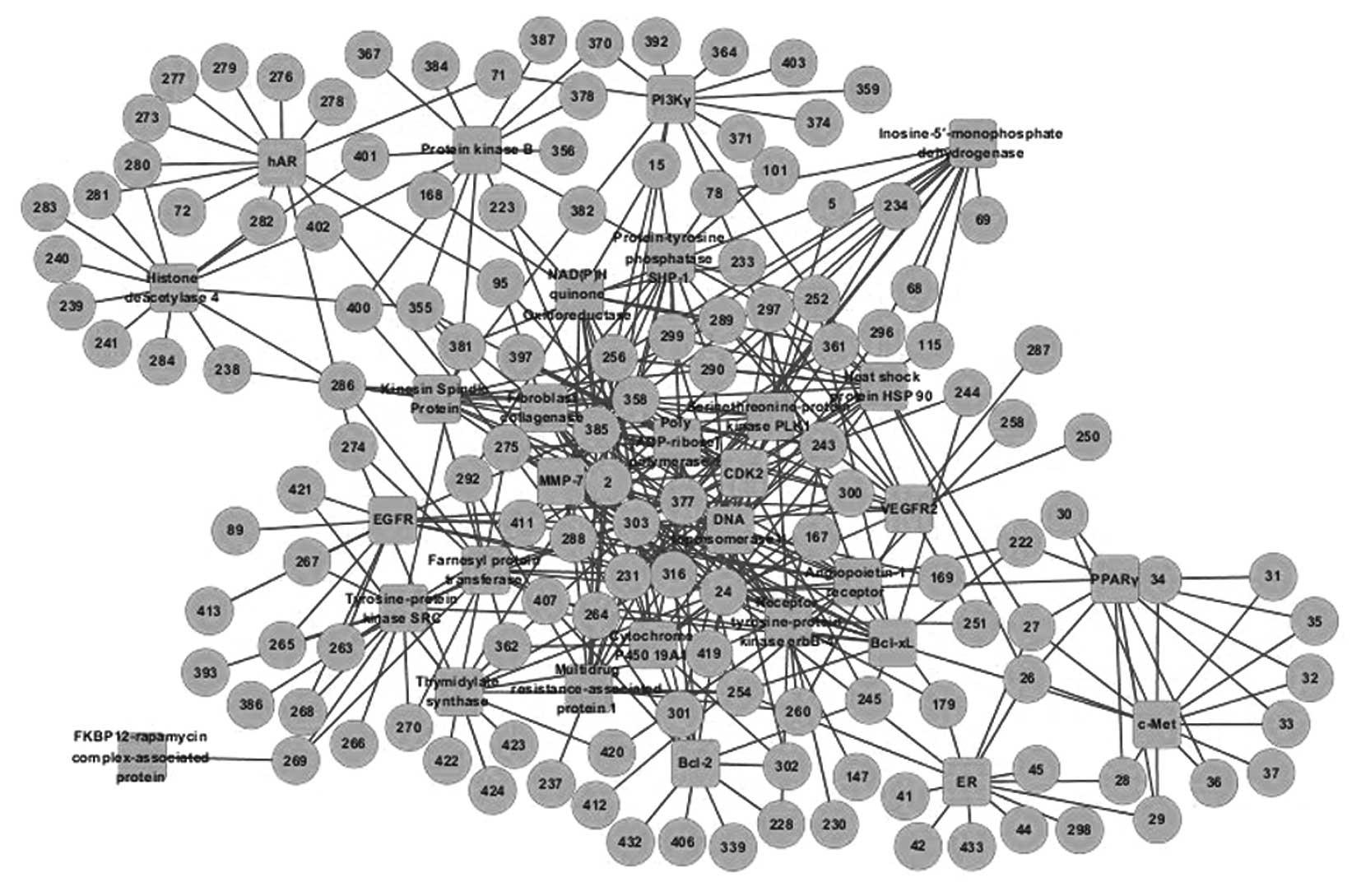
Identification of the multi-target
compounds in XCHT that are associated with cancer therapy
The docking results indicated that >80% of the
potential inhibitors interacted with fewer than three targets;
however, a small number of compounds interacted with a large number
of targets, up to a maximum of 18. Based on the screened compounds
and their corresponding targets, the C-T network was generated
(Fig. 2). The network parameters
and key compounds of the C-T network are listed in Tables III and IV, respectively. The results demonstrate
that certain predicted inhibitors present in XCHT possess the
properties of promiscuous drugs and combination therapies.
 | Table IIINetwork properties of the C-T and C-C
networks. |
Table III
Network properties of the C-T and C-C
networks.
| Parameters | C-T network | C-C network |
|---|
| Network
density | 0.029 | 0.784 |
| Network
heterogeneity | 1.045 | 0.675 |
| Network
centralization | 0.084 | 0.494 |
| Characteristic path
length | 3.687 | 1.986 |
| Average no. of
neighbors | 4.696 | 22.809 |
| Shortest path | 25760 (100%) | 17030 (100%) |
| Cluster
coefficient | 0 | 0.829 |
 | Table IVThe 15 compounds the highest degree
of target interaction in the compound-target network. |
Table IV
The 15 compounds the highest degree
of target interaction in the compound-target network.
| Index | Known
interaction | Chemical name | Degree |
|---|
| 377 | Yes | Scutellarin | 18 |
| 24 | No | Folic acid | 17 |
| 303 | Yes | Vicenin-2 | 15 |
| 385 | No | 5,7,4′-Trihydroxy-
6-C-arabinoside-8- C-glucoside flavone | 15 |
| 358 | No |
Dihydrobaicalin | 13 |
| 316 | No | Kaempferitrin | 13 |
| 2 | No | Adenosine
triphosphate | 13 |
| 275 | Yes | Liquiritin | 8 |
| 231 | No | Gancaonin E | 8 |
| 243 | No |
Glycyrrhisoflavone | 7 |
| 264 | No | Licorice saponin
C2 | 6 |
| 288 | No |
Neoisoliquiritin | 6 |
| 254 | No | Isoliquiritin | 6 |
| 355 | Yes | Baicalein | 5 |
| 252 | Yes |
Isolicoflavonol | 5 |
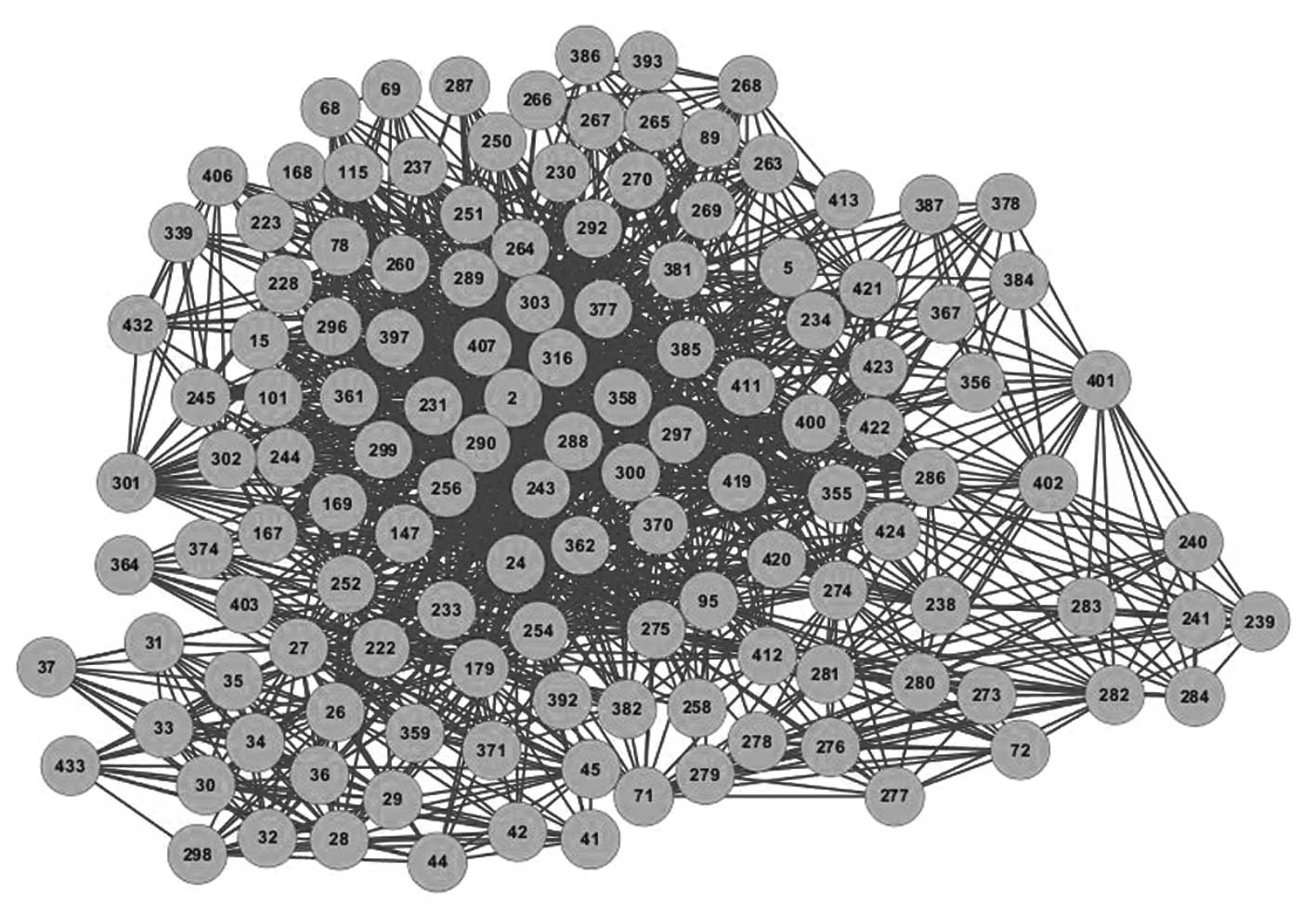
Multi-compound combination therapy of
XCHT for cancer
To understand the association between the potential
inhibitors, the C-C network was constructed (Fig. 3). The network parameters of the C-C
network are listed in Table III.
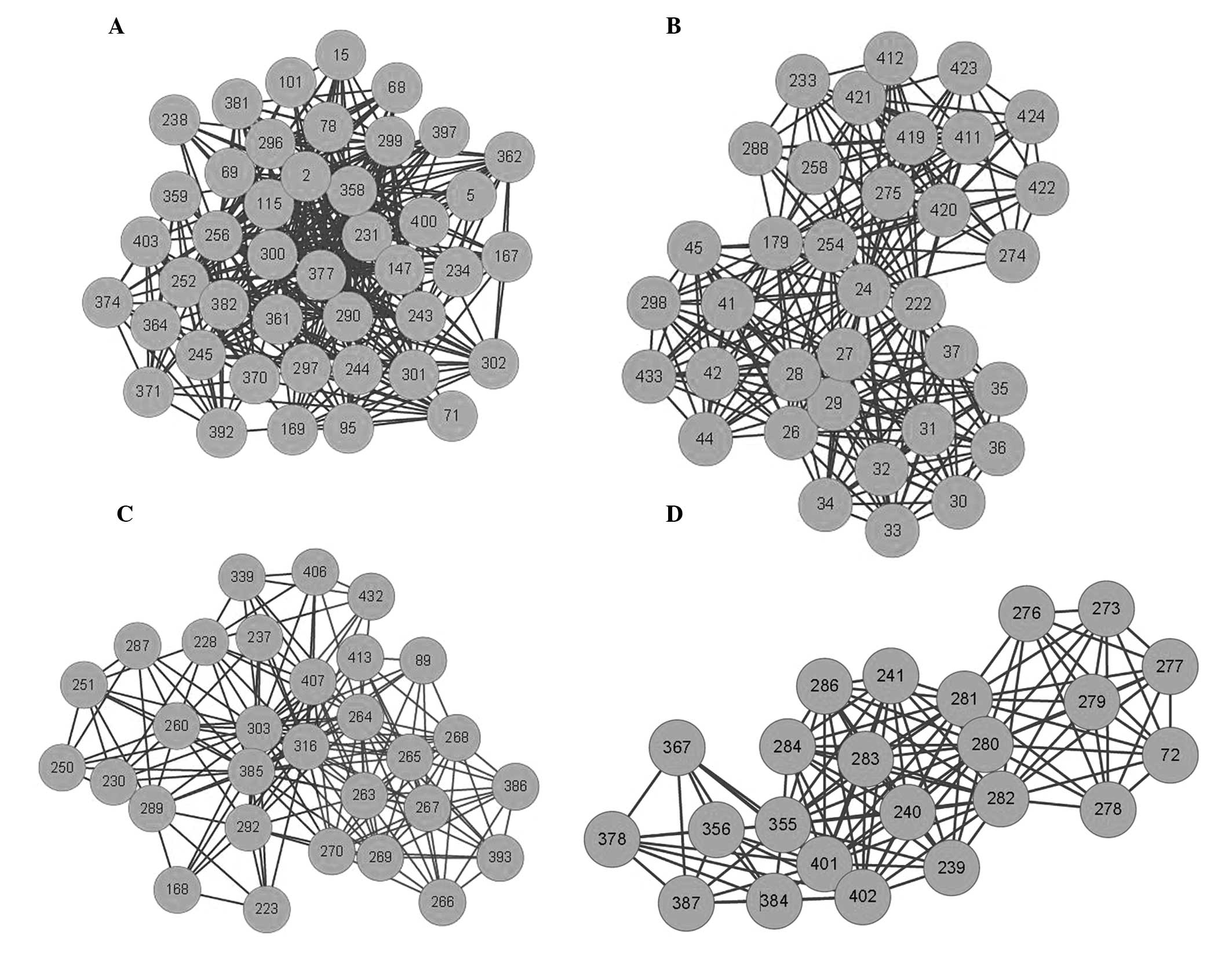
A Cytoscape GLay plugin, which automatically transforms the input
network into a simplified model (22), was used to identify four separate
clusters within the C-C network (Fig.
4; Table V) and to demonstrate
the different multi-compound and multi-herb combinations within
XCHT. Furthermore, the cluster results indicate that different
multi-compound combinations were able to act on different
targets.
 | Table VSignificant information associated
with the categories of the subnetworks. |
Table V
Significant information associated
with the categories of the subnetworks.
| Cluster | Effector
targets | Botanical source of
the compounds |
|---|
| Cluster 1 | VEGFR2,
tyrosine-protein kinase SRC, thymidylate synthase,
serine/threonine-protein kinase PLK1, receptor tyrosine-protein
kinase erbB-4, protein-tyrosine phosphatase SHP-1, protein kinase
B, poly [ADP-ribose] polymerase-1, PI3Kγ, NAD(P)H quinone
oxidoreductase, multidrug resistance-associated protein 1, MMP-7,
kinesin spindle protein, inosine-5′-monophosphate dehydrogenase,
histone deacetylase 4, HSP 90, hAR, fibroblast collagenase,
farnesyl protein transferase, EGFR, DNA topoisomerase II,
cytochrome P450 19A1, CDK2, Bcl-xL, Bcl-2 and angiopoietin-1
receptor. | Scutellaria
baicalensis, Panax ginseng, Zingiber officinale,
Zizyphi fructus, Radix glycyrrhiza |
| Cluster 2 | ER, Bcl-2, Bcl-xL,
CDK2, cytochrome P450 19A1, EGFR, ER, farnesyl protein transferase,
fibroblast collagenase, HSP 90, hAR, kinesin spindle protein,
MMP-7, multidrug resistance-associated protein 1, NAD(P)H quinone
oxidoreductase, PI3Kγ, poly [ADP-ribose] polymerase-1, PPARγ,
protein-tyrosine phosphatase SHP-1, receptor tyrosine-protein
kinase erbB-4, serine/threonine-protein kinase PLK1, thymidylate
synthase and VEGFR2. | Panax
ginseng, Zizyphi fructus, Zingiber officinale,
Scutellaria baicalensis, Pinellia ternata, Radix
glycyrrhizae |
| Cluster 3 | VEGFR2,
tyrosine-protein kinase SRC, thymidylate synthase,
serine/threonine-protein kinase PLK1, receptor tyrosine-protein
kinase erbB-4, protein-tyrosine phosphatase SHP-1, protein kinase
B, poly [ADP-ribose] polymerase-1, PI3Kγ, NAD(P)H quinone
oxidoreductase, multidrug resistance-associated protein 1, MMP-7,
kinesin spindle protein, HSP 90, fibroblast collagenase, EGFR,
farnesyl protein transferase, DNA topoisomerase II, cytochrome P450
19A1, CDK2, Bcl-xL, Bcl-2, angiopoietin-1 receptor and
tyrosine-protein kinase SRC. | Radix
bupleuri, Panax ginseng, Zizyphi fructus,
Zingiber officinale, Scutellaria baicalensis,
Pinellia ternata, Radix glycyrrhizae |
| Cluster 4 | protein kinase B,
farnesyl protein transferase, fibroblast collagenase, hAR, histone
deacetylase 4, kinesin spindle protein, MMP-7, poly [ADP-ribose]
polymerase-1 and serine/threonine-protein kinase PLK1. | Panax
ginseng, Zizyphi fructus, Scutellaria
baicalensis, Radix glycyrrhizae |
Discussion
Traditional Chinese medicine (TCM) has been widely
adopted for cancer care in China and other Asian countries, and is
increasingly used as a complementary therapy by Western cancer
patients (24). However, there are
numerous questions regarding TCM and a lack of modern scientific
language for describing it. In the present study, a computational
pharmacological model was constructed to investigate the molecular
characteristics and cancer therapeutic mode of a common TCM, XCHT,
as an example to demonstrate the potential application of the
model.
Notably, a significant proportion of the compounds
in XCHT were clustered in a specific region of chemical space.
According to the theory of chemical space (25), these compounds possessed similar
functions, which indicates that the compounds present in the herbal
components of XCHT are compatible with each other. It may provide
the scientific basis for the composition of a formulation from a
combination of these herbs or compounds. Furthermore, these
compounds occupy a chemical space that is the same as or close to
that occupied by other known inhibitors that exhibit the
anticarcinogenic drug-like properties of XCHT. A minor proportion
of the compounds within XCHT were dispersed and at intervals from
other compounds, which corresponds with the various functions of
XCHT that have been observed in a clinical setting (26,27).
These may provide the foundation for the screening of suitable
active compounds from XCHT that may be used as therapeutic agents
against cancer.
A C-T network was constructed, based on molecular
docking, to elucidate the therapeutic efficacy of XCHT against
cancer. The results demonstrated that the maximum number of targets
that a single compound was able to act on was 18, which indicated
that XCHT is a broad-spectrum formula that has the ability to
inhibit numerous significant target proteins. Furthermore, the C-T
network (Fig. 2) indicates that
the multiple potentially active compounds in XCHT are able to
interact with various cancer-associated targets and that limited
individual compounds, termed promiscuous drugs, are able to
interact with multiple targets. Table III demonstrates the biological
activities of certain compounds associated with the herbal
components of XCHT that have been reported in the literature
(28–33). Due to the molecular complexity of
cancer, multi-targeted therapies are becoming increasingly
important as, in the long-term, they maximize the therapeutic
effect and overcome the mechanisms of resistance (34,35).
Thus, the properties of multi-targeted therapies that have been
identified in XCHT may be a key explanation for why XCHT is
effective as an anticancer treatment.
To further investigate the therapeutic mode of XCHT
against cancer, a C-C network was constructed and the potentially
active compounds were clustered. As shown in Fig. 4, there were four clusters
containing different compounds that may serve to provide different
combination therapies and pharmacological functions. In addition,
the botanical sources of each cluster were identified to be
different. This may explain why XCHT has utility in the treatment
of pancreatic, stomach, liver and lung cancer (26,27);
furthermore, each cluster interacted with certain common targets.
Generally, the range of the network clustering coefficient is
between 0 and 1; the greater the coefficient, the higher the
clustering property. The clustering coefficient of the C-C network
in the present study was 0.829, which indicates that this
particular network has a high clustering property. The compounds in
high clustering have the similar activity, so they may have the
synergistic combined effects. This indicated that XCHT may have a
synergistic combined effect. In addition, the botanical sources of
the compounds in cluster 3 include all the ingredients of XCHT
(Table V), which further confirms
the rationality of the specific herbal combination used in XCHT. In
addition, it has been identified that XCHT in holistic combinations
possesses various pharmacological properties due to the different
components attacking various targets or different steps in the
pathologic process of cancer (11–13).
Therefore, the components of XCHT, which have different mechanisms
of anti-cancer action, interact primarily in an additive or
synergistic manner.
In conclusion, a computational pharmacological model
was employed to illustrate the multi-compound, multi-target and
multi-combination therapeutic mechanism of XCHT. The results
provide an indication of the polypharmacological anticancer effect
of XCHT and may aid the identification of novel therapeutic
strategies for cancer patients based on the active compounds
present in XCHT.
Acknowledgements
This study was supported by the Developmental Fund
of ChenKeji Integrative Medicine (grant no. CKJ2010032).
Abbreviations:
|
XCHT
|
Xiao Chai Hu Tang
|
|
TTD
|
Therapeutic Targets Database
|
|
hAR
|
human androgen receptor
|
|
ER
|
estrogen receptor
|
|
VEGFR2
|
vascular endothelial growth factor
receptor 2
|
|
MMP
|
matrix metalloproteinase
|
|
PPARγ
|
peroxisome proliferator-activated
receptor-γ
|
|
PI3Kγ
|
phosphoinositide 3-kinase-γ
|
|
CDK2
|
cyclin-dependent protein kinase 2
|
|
C-T network
|
compound-target network
|
|
C-C network
|
compound-compound association
network
|
|
TCM
|
tradition Chinese medicine
|
References
|
1
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
3
|
Carney DN and Hansen HH: Non-small-cell
lung cancer - stalemate or progress? N Engl J Med. 343:1261–1262.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bröker LE and Giaccone G: The role of new
agents in the treatment of non-small cell lung cancer. Eur J
Cancer. 38:2347–2361. 2002.
|
|
5
|
Yang CS, Wang H and Hu B: Combination of
chemopreventive agents in nanoparticles for cancer prevention.
Cancer Prev Res (Phila). 6:1011–1014. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Roth BL, Sheffler DJ and Kroeze WK: Magic
shotguns versus magic bullets: selectively non-selective drugs for
mood disorders and schizophrenia. Nat Rev Drug Discov. 3:353–359.
2004. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xu S, Zhao Y, Zhao YX and Long D:
Influence of maintenance therapy of shenyi capsule on survival of
advanced non-small cell lung cancer patients after chemotherapy.
Journal of Guangzhou University of Traditional Chinese Medicine.
30:337–340. 2013.(In Chinese).
|
|
8
|
Wang CZ, Calway T and Yuan CS: Herbal
medicines as adjuvants for cancer therapeutics. Am J Chin Med.
40:657–669. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Konkimalla VB and Efferth T:
Evidence-based Chinese medicine for cancer therapy. J
Ethnopharmacol. 116:207–210. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li XL and Li HR: Three factors ‘toxicity,
blood stasis, cold congealing’ for pathogenesis exploring of tumor
metastasis. China Journal of Traditional Chinese Medicine and
Pharmacy. 21:440–441. 2006.(In Chinese).
|
|
11
|
Han GX and Feng JZ: Application of xiao
chai hu tang in preventing and treating cancer. Zhejiang Journal of
Traditional Chinese Medicine. 4:687–688. 2010.(In Chinese).
|
|
12
|
Liao HF, Lu MC, Chang HC, Wei CC, Kao CH,
Chen ZH, Huang CC and Li C: Effects of herbal medicinal formulas on
suppressing viral replication and modulating immune responses. Am J
Chin Med. 38:173–190. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li J, Xie M and Gan Y: Effect of
Xiaochaihu decoction and different herbal formulation of component
on inhibiting H22 liver cancer in mice and enhancing immune
function. Zhongguo Zhong Yao Za Zhi. 33:1039–1044. 2008.(In
Chinese).
|
|
14
|
Zheng CS, Xu XJ, Ye HZ, Wu GW, Li XH,
Huang SP and Liu XX: Computational approaches for exploring the
potential synergy and polypharmacology of Duhuo Jisheng Decoction
in the therapy of osteoarthritis. Mol Med Rep. 7:1812–1818.
2013.PubMed/NCBI
|
|
15
|
Gu J, Zhang H, Chen L, Xu S, Yuan G and Xu
X: Drug-target network and polypharmacology studies of a
Traditional Chinese Medicine for type II diabetes mellitus. Comput
Biol Chem. 35:293–297. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qiao X, Hou T, Zhang W, Guo S and Xu X: A
3D structure database of components from Chinese traditional
medicinal herbs. J Chem Inf Comput Sci. 42:481–489. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou JX, Xie GR and Yang XD: Handbook of
the Constituents in Chinese Herb Original Plants. Chemical Industry
Press; Beijing: pp. 1165–1211. 2004, (In Chinese).
|
|
18
|
Zhu F, Shi Z, Qin C, Tao L, Liu X, Xu F,
Zhang L, Song Y, Liu X, Zhang J, et al: Therapeutic target database
update 2012: a resource for facilitating target-oriented drug
discovery. Nucleic Acids Res. 40(Database issue): D1128–D1136.
2012.PubMed/NCBI
|
|
19
|
Venkatachalam CM, Jiang X, Oldfield T and
Waldman M: LigandFit: a novel method for the shape-directed rapid
docking of ligands to protein active sites. J Mol Graph Model.
21:289–307. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Montes M, Braud E, Miteva MA, Goddard ML,
Mondésert O, Kolb S, Brun MP, Ducommun B, Garbay C and Villoutreix
BO: Receptor-based virual ligand screening for the identification
of novel CDC25 phosphatase inhibitors. J Chem Inf Model.
48:157–165. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: new features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Su G, Kuchinsky A, Morris JH, States DJ
and Meng F: GLay: community structure analysis of biological
networks. Bioinformatics. 26:3135–3137. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lipinski CA, Lombardo F, Dominy BW and
Feeney PJ: Experimental and computational approaches to estimate
solubility and permeability in drug discovery and development
settings. Adv Drug Deliv Rev. 46:3–26. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Carmady B and Smith CA: Use of Chinese
medicine by cancer patients: a review of surveys. Chin Med.
6:222011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dobson CM: Chemical space and biology.
Nature. 432:824–828. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sun ML and Zeng BR: Anti-tumor clinical
application of Xiaochaihu-tang and the progress of experimental
study on its disassembled prescriptions. Hunan Journal of
Traditional Chinese Medicine. 29(4): 134–136. 2013.(In
Chinese).
|
|
27
|
Yang JY, Li YZ, Fan ZW, Zheng SM and Xu
JZ: Clinical application overview of Xiaochaihu-tang. Chinese
Medicine Modern Distance Education of China. 9(9): 74–75. 2011.(In
Chinese).
|
|
28
|
Chan JY, Tan BK and Lee SC: Scutellarin
sensitizes drug-evoked colon cancer cell apoptosis through enhanced
caspase-6 activation. Anticancer Res. 29:3043–3047. 2009.PubMed/NCBI
|
|
29
|
Xu H and Zhang S: Scutellarin-induced
apoptosis in HepG2 hepatocellular carcinoma cells via a STAT3
pathway. Phytother Res. 27:1524–1528. 2013.PubMed/NCBI
|
|
30
|
Franek KJ, Zhou Z, Zhang WD and Chen WY:
In vitro studies of baicalin alone or in combination with Salvia
miltiorrhiza extract as a potential anti-cancer agent. Int J
Oncol. 26:217–224. 2005.PubMed/NCBI
|
|
31
|
Nagaprashantha LD, Vatsyayan R, Singhal J,
Fast S, Roby R, Awasthi S and Singhal SS: Anti-cancer effects of
novel flavonoid vicenin-2 as a single agent and in synergistic
combination with docetaxel in prostate cancer. Biochem Pharmacol.
82:1100–1109. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ohno H, Araho D, Uesawa Y, Kagaya H,
Ishihara M, Sakagami H and Yamamoto M: Evaluation of cytotoxicity
and tumor-specificity of licorice flavonoids based on chemical
structure. Anticancer Res. 33:3061–3068. 2013.PubMed/NCBI
|
|
33
|
Kinghorn AD, Su BN, Jang DS, Chang LC, Lee
D, Gu JQ, Carcache-Blanco EJ, Pawlus AD, Lee SK, Park EJ, et al:
Natural inhibitors of carcinogenesis. Planta Med. 70:691–705. 2004.
View Article : Google Scholar
|
|
34
|
Broxterman HJ and Georgopapadakou NH:
Anticancer therapeutics: ‘Addictive’ targets, multi-targeted drugs,
new drug combinations. Drug Resis Updat. 8:183–197. 2005.
|
|
35
|
Shoshan MC and Linder S: Promiscuous and
specific anti-cancer drugs: combatting biological complexity with
complex therapy. Cancer Ther. 2:297–304. 2004.
|