Introduction
Cardiovascular disease is considered to be one of
the most important non-cancer long-term effects of ionizing
radiation, as evidenced by the epidemiological data of atomic bomb
survivors exposed to doses of 0.5 to 2 Gy (1). In the context of space exploration,
high linear energy transfer (LET) radiation found in space produces
high values of relative biological effectiveness (RBE), as compared
to low LET radiation, such as X-rays or gamma-rays, which can
increase the health risks to astronauts (2). Indeed, during long-term missions,
such as a journey to Mars, astronauts are bound to be exposed to
cumulative doses between 0.3 and 4 Sv, depending on the spacecraft
shielding and on the intensity of solar particle events (3).
Heavy ion irradiation is also used for terrestrial
applications, such as non-conventional radiotherapy (hadron
therapy), which takes advantage of the depth distribution of the
dose, which is maximal at the Bragg peak, and of the increased RBE,
allowing the enhanced killing effect on tumor cells while sparing
the healthy tissue (4,5). However, little is known of the
molecular mechanisms involved in the enhanced killing properties of
heavy ion irradiation. Improving our understanding of the effects
of heavy ion radiation, particularly on the cardiovascular system
that may be irradiated during treatment, is therefore of utmost
importance for both long-term space missions and hadron
therapy.
Endothelial cells are critical targets in
radiation-induced cardiovascular damage (1,6,7).
While high doses of low LET radiation induce pro-inflammatory
responses in endothelial cells, the opposite has been observed upon
exposure to low doses (8–10). The mechanisms involved are not yet
fully understood; however, they appear to be at least partly linked
to the transcription factor, nuclear factor (NF)-κB, and the nitric
oxide signaling pathway, which in turn mediates various cellular
responses, including the secretion of cytokines [such as
transforming growth factor (TGF)-β1, interleukin (IL)-6, interferon
(IFN)-γ, IFN-β and tumor necrosis factor (TNF)-α] and chemokines
(9–11). Another possible mechanism of
radiation-induced cardiovascular alteration, as shown upon low LET
radiation (12–16), is the endothelial retraction and
the impairment of cellular adhesion. Matrix metalloproteinases
(MMPs), Rho GTPases, calcium signaling and reactive oxygen species
seem to be important factors that stimulate modifications in cell
junctions and the cytoskeleton through adhesion molecules and actin
(12–16). Although high LET radiation has
been shown to reduce the length of a 3D human endothelial vessel
model, both developing and mature (17), only a few studies have been
conducted to identify the mechanisms involved in the endothelial
response to high LET radiation (18,19).
Thus, the aim of this study was to investigate the
effects of moderate and high doses of high LET nickel ion (Ni)
irradiation on gene expression in endothelial cells in order to
elucidate the molecular mechanisms responsible for
radiation-induced cardiovascular damage. For this purpose, the
EA.hy926 cell line, which originates from human umbilical vein
endothelial cells, was irradiated with nickel ions (LET, 183
keV/μm) at moderate (0.5 Gy) and high (2 and 5 Gy) doses after
which gene expression was determined by whole-genome microarray
analysis.
Materials and methods
Cell culture
The human EA.hy926 endothelial cells were obtained
from the American Type Culture Collection (ATCC; Manassas, VA,
USA). They were cultured (37°C-5% CO2) in Dulbecco’s
modified Eagle’s medium supplemented with 10% fetal bovine serum
and 1% penicillin/streptomycin (all from N.V. Invitrogen S.A.,
Merelbeke, Belgium). The cells were regularly examined for the
absence of mycoplasma using the LookOut® Mycoplasma PCR
Detection kit (Sigma-Aldrich, St. Louis, MO, USA).
Nickel irradiation
The cells were seeded at a density of 105
cells in 12.5 cm2 flasks. Twenty-four hours after
plating, the flasks were placed in a transportable incubator (37°C)
and moved from the resident laboratory (Mol, Belgium) to the GSI
Helmholtzzentrum für Schwerionenforschung GmbH (Darmstadt,
Germany). Forty-eight hours after plating, the subconfluent cells
were irradiated in flasks completely filled with culture medium
with a 1 GeV/u Ni beam at the SIS facility at GSI with the
intensity controlled raster scanning technique as described by
Haberer et al (20). The
ion energy at the sample position was approximately 930 MeV/u with
a LET of 183 keV/μm (calculated with the program code ATIMA). The
culture flasks were placed vertically and exposed perpendicularly
to the nickel ion beam at the following doses: 0.5, 2 and 5 Gy.
Non-irradiated control samples were treated similarly to the
irradiated samples, but placed out of the beam. Following
irradiation, the cells were incubated (37°C, 5% CO2) in
2 ml of conditioned medium until fixation time points (2, 8 and 24
h).
DNA double-strand break detection
(detection of γ-H2AX foci)
The cells were fixed in 4% paraformaldehyde (Merck
KGaA, Darmstadt, Germany) 2 and 24 h after irradiation. They were
then treated with 0.25% Triton X for 5 min, blocked with 3% bovine
serum albumin (both from Sigma-Aldrich) for 30 min and incubated
overnight with mouse anti-γ-H2AX antibody (Abcam, Cambridge, MA,
USA) at 4°C. After a second blocking of 10 min, the cells were
incubated for 1 h with anti-mouse secondary antibody coupled to
FITC (Sigma-Aldrich) at 37°C and then mounted in Vectashield
mounting medium (Vector Laboratories, Burlingame, CA, USA) with
DAPI. Between each of the previous steps, the slides were washed
with phosphate-buffered saline (PBS).
An automated inverted fluorescence microscope
(TE2000-E; Nikon, Tokyo, Japan), equipped with a motorized XYZ
stage, emission and excitation filter wheels, shutters and a triple
dichroic mirror (436/514/604) was used for the image acquisition of
the immunostained slides. Images were acquired with a 40X Plan
Fluor oil objective (NA 1.3) and an Andor iXon EMCCD camera (Andor
Technology, South Windsor, CT, USA). For each sample, at least 12
fields were acquired on 5 z-stack focusses (1 μm). The γ-H2AX spot
number and spot occupancy were analyzed with the INSCYDE plugin for
ImageJ as previously described (21). Spot occupancy was defined for each
nucleus as the sum of the spot areas divided by the nucleus area
(spot_occupancy = sum (spot_area)/nuclear_area). A minimum number
of 100 cells was analyzed in 2 biological replicates per condition.
For statistical analyses, the data were analyzed using the
Mann-Whitney U test with SPSS version 17.0 software (IBM Corp.,
Chicago, IL, USA) and box plots were generated using the same
software. P-values <0.05 were considered to indicate
statistically significant differences.
RNA extraction
At 2 time points after irradiation (8 and 24 h), the
adherent cells were washed in PBS, lysed in 350 ml of AllPrep
DNA/RNA/Protein Mini kit lysis buffer (Qiagen, Hilden, Germany) and
frozen at −80°C. RNA was extracted using the same kit and its
concentration was measured using a NanoDrop spectrophotometer
(Thermo Fisher Scientific, Waltham, MA, USA), while its quality
(RNA integrity number, RIN) was determined using Agilent’s
lab-on-chip Bioanalyzer 2100 (Agilent Technologies, Santa Clara,
CA, USA). All RNA samples had a RIN value >9.0.
Affymetrix microarrays and data
analysis
RNA was processed using the GeneChip WT cDNA
Synthesis and Amplification kit (Affymetrix, Santa Clara, CA, USA)
according to the manufacturer’s instructions. The resulting RNA was
hybridized to Affymetrix Human Gene 1.0 ST arrays which contain an
estimated number of 28,869 genes based on the March 2006 [UCSC Hg
18; National Center for Biotechnology Information (NCBI) build 36]
human genome assembly. Biological triplicates were collected for
each condition.
Raw data (.cel-files) were imported at exon level in
Partek Genomics Suite version 6.5 (Partek, Inc., St. Louis, MO,
USA). Briefly, robust multi-array average (RMA) background
correction was applied, data were normalized by quantile
normalization and probeset summarization was performed by the
median polish method. Gene summarization was performed using
one-step Tukey’s biweight method. The obtained data were analyzed
with Partek Genomics Suite for single gene analysis. One- or
two-way ANOVA, taking into consideration the scan date (where
applicable) and the dose as factors, were performed for each time
point. In order to determine statistical significance, thresholds
were set on the p-value <0.001 and on the fold-change
>1.5.
The enrichment of the transcription factor binding
motifs was analyzed using Pscan Ver. 1.2 (22) and the JASPAR database, scanning in
a region from −950 to +50 base pairs from the transcription start
site.
Results
DNA damage
To assess DNA damage induction by nickel ion
irradiation and evaluate the cell capacity required to repair this
damage, we performed a high content cytometric assay of γ-H2AX, 2
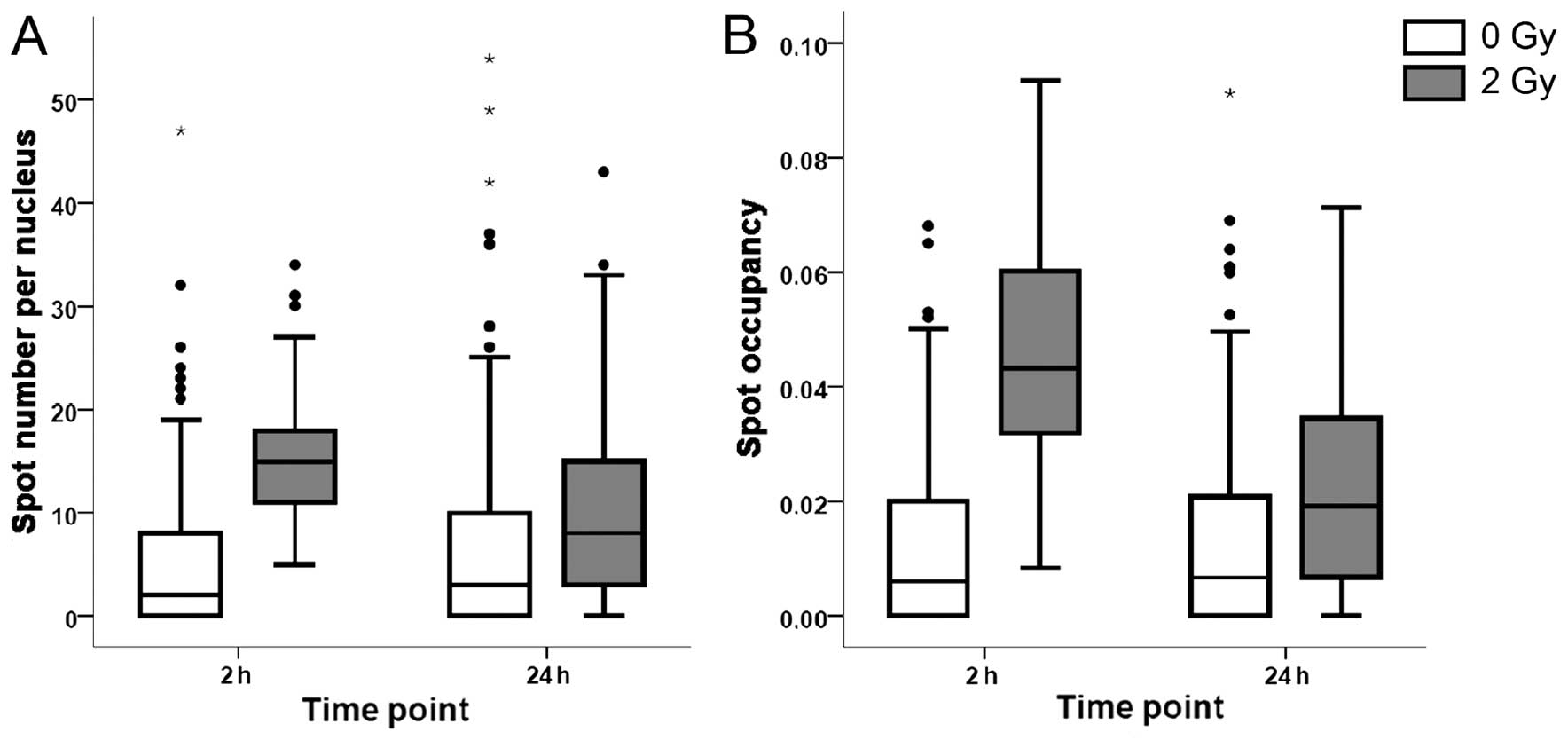
and 24 h after exposure. As measured by the number of γ-H2AX foci,
DNA damage was significantly increased 2 h after nickel ion
irradiation (2 Gy), with an average number of 15 foci per nucleus
vs. 2 foci per nucleus in the control samples (Fig. 1A). Twenty-four hours after
irradiation, the number of foci decreased to 9 per nucleus, which
was significantly higher than the values of controls, indicating
that part of the DNA damage persisted for at least 24 h. Similar
trends were observed for the spot occupancy, which is the fraction
of the projected area of the nucleus occupied by the signal from
the γ-H2AX foci (Fig. 1B).
Effects of nickel irradiation on gene
expression
In order to evaluate gene expression, we performed
microarrays 8 and 24 h after irradiation. A 0.5 Gy irradiation,
both after 8 and 24 h, elicited a subtle effect on gene expression
in the EA.hy926 cells. Six annotated genes were differentially
regulated with fold changes (FC) between 1.5 and 1.8 after 8 h, and
18 genes were differentially regulated with an FC between 1.5 and
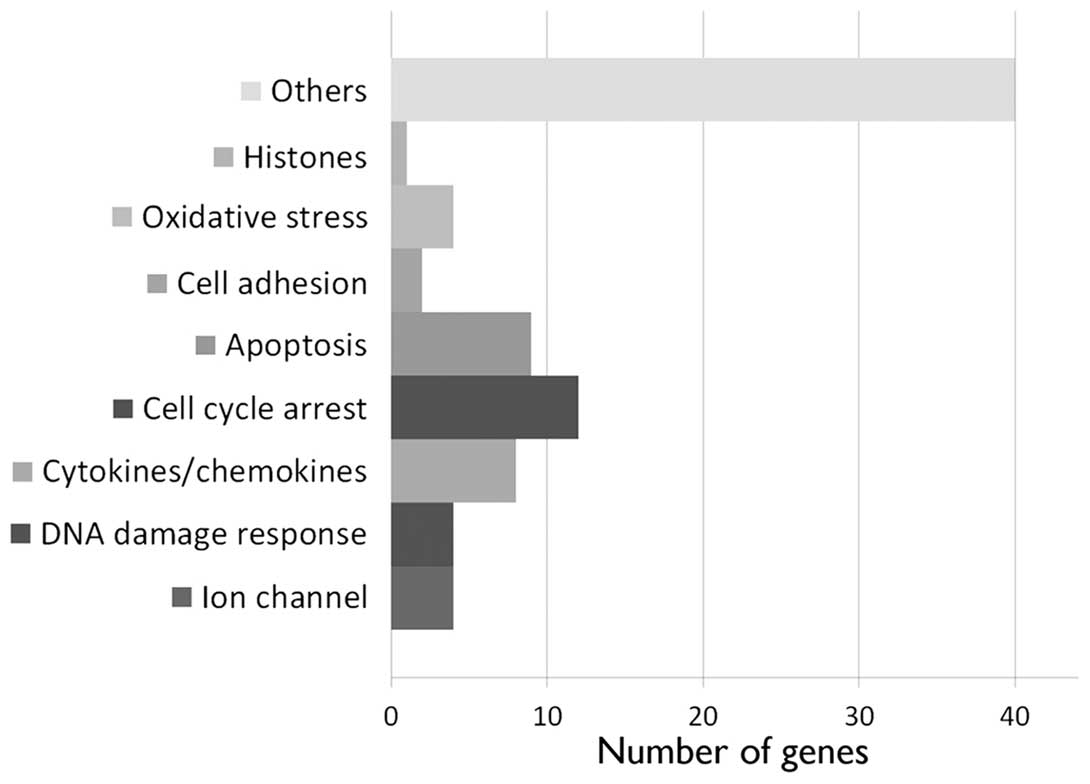
2.3 after 24 h. A more drastic effect was observed at 5 Gy, 24 h
after irradiation. At this time point, we detected the upregulation
of 77 annotated genes (Fig. 2 and
Table I; maximum FC, 3.4). Among
these genes, cytokines and chemokines (CXCL5, TGFA, TRIM22, TNFSF9,
EBI3, IL-6, IL-11 and CD70) were identified, as well as genes
involved in DNA damage response (SPATA18, POLL, APOBEC3H and
SESN1), cell cycle arrest (ZMAT3, MXD4, TP53INP1, HSPB8, TGFA,
SESN2, BTG2, DTX3, TOB1, HBP1, CDKN1A and PLK3) and apoptosis
(TP53INP1, HSPB8, TGFA, TP53I3, MOAP1, CYFIP2, TRADD, DTX3 and
FAS). In addition, we observed the upregulation of genes coding for
ion channels (SLC22A4, KCNJ2, ORAI3 and CLIC3), cell adhesion
(CEACAM1 and NEU1) and oxidative stress response proteins (FMO4,
FDXR, SIRT2 and SESN1).
 | Table IList of the differentially expressed
genes at 8 and 24 h after 0.5 and 5 Gy of nickel ion
irradiation. |
Table I
List of the differentially expressed
genes at 8 and 24 h after 0.5 and 5 Gy of nickel ion
irradiation.
| Downregulated
genes | Upregulated
genes |
|---|
|
|
|---|
| Gene symbol | GenBank | p-value | FC | Gene symbol | GenBank | p-value | FC |
|---|
| List of
differentially expressed genes 8 h after 0.5 Gy nickel ion
irradiation |
| CRYBB2 | NM_000496 | 1,02E-03 | −1,800 | HSP90AA6P | NR_036751 | 6,55E-03 | 1,630 |
| GNAT1 | NM_144499 | 5,54E-03 | −1,601 | RFT1 | NM_052859 | 6,83E-03 | 1,572 |
| DNAJB13 | NM_153614 | 3,79E-03 | −1,521 | DEFB123 | NM_153324 | 8,50E-03 | 1,518 |
| List of
differentially expressed genes 24 h after 0.5 Gy nickel-ion
irradiation |
| UPF3A | NM_023011 | 1,53E-03 | −1,932 | C1orf113 |
ENST00000312808 | 8,96E-03 | 2,224 |
|
E2F8a | NM_024680 | 5,38E-03 | −1,716 | SNORD53 | NR_002741 | 6,78E-03 | 2,182 |
| LIMA1 | NM_001113546 | 9,63E-03 | −1,670 | SLC45A4 | BC033223 | 8,10E-03 | 2,049 |
| C16orf55 | AK303024 | 7,00E-04 | −1,600 | HIST1H2BD | NM_021063 | 9,01E-03 | 2,028 |
| HLA-DRB4 | AK293020 | 3,18E-03 | −1,583 | ZNF16 | NM_001029976 | 3,65E-03 | 1,952 |
|
MCM10a | NM_182751 | 9,77E-03 | −1,530 | RNF207 | NM_207396 | 7,07E-03 | 1,677 |
| PROK2 | NM_001126128 | 8,29E-03 | −1,518 |
LCE1Eb | NM_178353 | 4,89E-03 | 1,619 |
| | | | C10orf72 | NM_001031746 | 6,04E-04 | 1,592 |
| | | | PMCH | NM_002674 | 7,49E-03 | 1,591 |
| | | | RUNDC3B | NM_138290 | 1,98E-03 | 1,571 |
| | | |
ORAI3b | NM_152288 | 7,01E-03 | 1,515 |
| List of
differentially expressed genes 24 h after 5 Gy nickel-ion
irradiation |
|
FAM111Ba | NM_198947 | 5,65E-03 | −5,894 |
ACTA2b | NM_001141945 | 2,01E-05 | 3,424 |
|
PCBP1a | NM_006196 | 6,05E-04 | −3,611 | TP53INP1 | NM_033285 | 1,02E-04 | 2,976 |
| DHRS2 | NM_182908 | 6,48E-05 | −3,090 |
CD70b | NM_001252 | 9,56E-03 | 2,702 |
|
MCM6a | NM_005915 | 1,81E-05 | −3,069 | PHOSPHO1 | NM_001143804 | 9,47E-04 | 2,629 |
| HIST1H1T | NM_005323 | 6,99E-03 | −2,762 | CDKN1A | NR_037151 | 8,27E-03 | 2,221 |
|
ZNF367a | NM_153695 | 1,61E-04 | −2,669 | CEACAM1 | NM_001712 | 5,56E-04 | 2,184 |
| KIF20A | NM_005733 | 4,17E-04 | −2,595 |
BTG2b | NM_006763 | 1,08E-03 | 2,170 |
| LMNB1 | NM_005573 | 9,94E-04 | −2,561 |
APOBEC3Hb | NM_001166003 | 4,32E-03 | 2,070 |
| HIST1H1D | NM_005320 | 4,45E-03 | −2,478 |
TRIM22b | NM_006074 | 4,27E-04 | 2,063 |
|
E2F8a | NM_024680 | 3,88E-04 | −2,469 |
KCNJ2b | NM_000891 | 1,88E-03 | 2,055 |
| HAUS8 | NM_033417 | 1,71E-05 | −2,444 |
SPATA18b | NM_145263 | 1,16E-06 | 2,026 |
| MYBL2 | NM_002466 | 7,56E-04 | −2,369 |
ZNF223b | NM_013361 | 4,56E-03 | 1,997 |
|
UHRF1a | NM_001048201 | 5,93E-03 | −2,363 |
LCE1Eb | NM_178353 | 8,26E-04 | 1,986 |
| SRP19 |
ENST00000512790 | 6,66E-03 | −2,337 | FDXR | NM_024417 | 1,72E-03 | 1,972 |
| UPF3A | NM_023011 | 4,54E-04 | −2,292 | TUBA4A | NM_006000 | 1,36E-03 | 1,932 |
|
DLGAP5a | NM_014750 | 9,85E-03 | −2,264 | PSTPIP2 | NM_024430 | 5,51E-04 | 1,924 |
|
ATAD2a | NM_014109 | 7,92E-03 | −2,239 |
ACY3b | NM_080658 | 8,81E-03 | 1,922 |
|
UNGa | NM_003362 | 2,21E-05 | −2,220 |
SLC40A1b | NM_014585 | 2,55E-03 | 1,921 |
|
HELLSa | NM_018063 | 8,16E-03 | −2,194 | TMEM150A | NM_001031738 | 5,23E-04 | 1,904 |
|
MCM3a | NM_002388 | 1,81E-03 | −2,189 | MXD4 | NM_006454 | 1,09E-04 | 1,903 |
| FIGNL1 | NM_001042762 | 6,39E-03 | −2,184 |
IL-6b | NM_000600 | 2,89E-03 | 1,883 |
| KIF11 | NM_004523 | 9,41E-03 | −2,164 | NCRNA00219 | NR_015370 | 8,85E-03 | 1,864 |
| FBXO5 | NM_012177 | 1,38E-05 | −2,150 |
KBTBD8b | NM_032505 | 8,57E-03 | 1,833 |
|
E2F2a | NM_004091 | 3,11E-04 | −2,126 |
NIPAL3b | NM_020448 | 1,16E-06 | 1,819 |
| WDR76 | NM_024908 | 4,11E-04 | −2,108 | RAB4B | NM_016154 | 3,30E-03 | 1,811 |
|
HIST1H2BGa | NM_003518 | 1,90E-03 | −2,106 |
SAT1b | NR_027783 | 7,52E-03 | 1,810 |
| LOC1720 | NR_033423 | 1,54E-03 | −2,101 | SLC22A4 | NM_003059 | 7,67E-04 | 1,796 |
| CAMK2N1 | NM_018584 | 1,24E-03 | −2,091 | NEU1 | NM_000434 | 2,68E-03 | 1,778 |
|
DLEU2a | NR_002612 | 6,87E-03 | −2,088 |
CYFIP2b | NM_001037332 | 5,48E-03 | 1,770 |
|
MCM5a | NM_006739 | 2,95E-04 | −2,084 | TNFSF9 | NM_003811 | 7,09E-04 | 1,736 |
| ANKRD36B | NM_025190 | 3,63E-03 | −2,082 |
TMEM217b | NM_145316 | 7,60E-03 | 1,730 |
|
POLA1a | NM_016937 | 2,54E-03 | −2,073 | IL-11 | NM_000641 | 3,38E-03 | 1,721 |
| BUB1B | NM_001211 | 1,55E-03 | −1,995 | ATP6V0A4 | NM_020632 | 7,57E-03 | 1,712 |
| GPSM2 | NM_013296 | 2,43E-04 | −1,988 | FMO4 | NM_002022 | 1,05E-03 | 1,707 |
| HMGB2 | NM_001130688 | 3,20E-03 | −1,979 | WIPI1 | NM_017983 | 7,41E-04 | 1,705 |
| DHCR24 | NM_014762 | 3,24E-04 | −1,970 | HBP1 | NM_012257 | 7,81E-03 | 1,684 |
|
MCM7a | NM_005916 | 2,60E-05 | −1,956 | UCN2 | NM_033199 | 4,81E-03 | 1,683 |
| ANLN | NM_018685 | 7,33E-03 | −1,953 | bEBI3 | NM_005755 | 1,99E-03 | 1,676 |
| NDC80 | NM_006101 | 2,04E-04 | −1,953 | bFAS | NM_000043 | 9,18E-03 | 1,673 |
|
MCM2a | NM_004526 | 2,05E-04 | −1,927 |
CLIC3b | NM_004669 | 8,25E-03 | 1,666 |
| CDKN3 | NM_005192 | 9,25E-03 | −1,917 | TGFA | NM_003236 | 1,70E-04 | 1,662 |
| EMP2 | NM_001424 | 1,87E-03 | −1,915 |
NCF2b | NM_000433 | 6,46E-03 | 1,656 |
| TACC3 | NM_006342 | 2,72E-03 | −1,906 | NADSYN1 | NM_018161 | 4,99E-04 | 1,651 |
| LHX2 | NM_004789 | 8,33E-03 | −1,883 |
CXCL5b | NM_002994 | 2,65E-05 | 1,643 |
| NCAPG2 | NM_017760 | 1,59E-03 | −1,881 |
SESN2b | NM_031459 | 8,22E-04 | 1,642 |
| DHFR | NM_000791 | 1,91E-05 | −1,878 | HSPB8 | NM_014365 | 1,53E-04 | 1,634 |
|
PER3a | NM_016831 | 8,02E-03 | −1,867 |
FAM84Ab | NM_145175 | 3,91E-04 | 1,623 |
| SEMA3D | NM_152754 | 7,14E-03 | −1,867 |
ORAI3b | NM_152288 | 3,54E-03 | 1,616 |
| KIFC1 | NM_002263 | 1,96E-04 | −1,860 |
C9orf150b | NM_203403 | 2,10E-03 | 1,614 |
| DEPDC1B | NM_018369 | 5,71E-03 | −1,853 | C2orf80 | NM_001099334 | 5,67E-04 | 1,613 |
| USP1 | NM_003368 | 1,74E-03 | −1,848 | PLK3 | NM_004073 | 8,79E-03 | 1,611 |
| CCNE2 | NM_057749 | 1,34E-03 | −1,841 |
MAGED4b | NM_001098800 | 1,62E-03 | 1,611 |
| PRC1 | NM_003981 | 1,06E-03 | −1,838 | LRRC29 | NM_012163 | 2,25E-06 | 1,589 |
| DEPDC1 | NM_001114120 | 1,89E-04 | −1,819 | POLL | NM_001174084 | 2,96E-03 | 1,583 |
| ORC1 | NM_004153 | 2,50E-03 | −1,811 | DFNA5 | NM_004403 | 6,35E-04 | 1,578 |
|
CDCA7a | NM_031942 | 1,42E-04 | −1,807 | CRYAB | NM_001885 | 3,49E-04 | 1,577 |
|
MCM10a | NM_182751 | 2,11E-03 | −1,801 | WBP5 | NM_016303 | 2,50E-04 | 1,569 |
|
CDT1a | NM_030928 | 5,55E-03 | −1,800 | SESN1 | NM_014454 | 6,34E-03 | 1,565 |
|
FAM111Aa | NM_022074 | 5,61E-04 | −1,798 |
RDH10b | NM_172037 | 8,25E-03 | 1,556 |
|
STX11a | NM_003764 | 1,61E-03 | −1,795 |
BHLHE40b | NM_003670 | 9,87E-03 | 1,553 |
|
MSH2a | NM_000251 | 7,76E-04 | −1,794 | FAM113A | AK293638 | 7,81E-04 | 1,550 |
| MKKS | NM_018848 | 6,55E-04 | −1,794 | LOC100130581 | NR_027413 | 6,57E-03 | 1,546 |
| CEP78 | NM_001098802 | 9,07E-03 | −1,790 | MOAP1 | NM_022151 | 2,89E-04 | 1,543 |
| RFC4 | NM_002916 | 3,92E-03 | −1,789 |
TP53I3b | NM_004881 | 1,71E-04 | 1,543 |
| KIF23 | NM_138555 | 3,79E-03 | −1,789 | OR51B6 | NM_001004750 | 5,49E-03 | 1,542 |
|
MLF1IPa | NM_024629 | 2,81E-04 | −1,785 |
NIPSNAP1b | NM_003634 | 3,88E-04 | 1,541 |
| CEP55 | NM_018131 | 4,87E-04 | −1,782 |
HHATb | NM_001170580 | 2,41E-03 | 1,536 |
|
TCF19a | NM_007109 | 6,78E-04 | −1,781 | ARR3 | NM_004312 | 7,01E-04 | 1,535 |
| BUB1 | NM_004336 | 6,08E-04 | −1,780 |
SIRT2b | NM_012237 | 4,30E-03 | 1,533 |
| CHAF1B | NM_005441 | 1,22E-03 | −1,773 |
C15orf33b | NM_152647 | 1,43E-03 | 1,526 |
|
EZH2a | NM_004456 | 5,38E-04 | −1,772 |
RBKSb | NM_022128 | 5,82E-03 | 1,523 |
| PLK4 | NM_014264 | 2,16E-04 | −1,757 |
DTX3b | NM_178502 | 4,95E-03 | 1,519 |
|
E2F1a | NM_005225 | 1,61E-03 | −1,755 | TOB1 | NM_005749 | 6,18E-03 | 1,517 |
|
H1F0a | NM_005318 | 1,51E-04 | −1,740 |
ADCY4b | NM_001198592 | 5,24E-03 | 1,514 |
| CEP57L1 | NM_001083535 | 5,12E-03 | −1,739 | ARL15 | NM_019087 | 1,98E-03 | 1,512 |
|
NUSAP1a | NM_016359 | 8,15E-05 | −1,730 |
TRADDb | NM_003789 | 2,41E-03 | 1,509 |
| ESPL1 | NM_012291 | 9,12E-03 | −1,724 |
ZMAT3b | NM_022470 | 5,71E-05 | 1,502 |
| KIF2C | NM_006845 | 7,54E-04 | −1,718 | | | | |
| DEPDC4 | NM_152317 | 3,25E-03 | −1,714 | | | | |
| MSH6 | NM_000179 | 6,14E-03 | −1,712 | | | | |
|
CDC6a | NM_001254 | 2,07E-03 | −1,710 | | | | |
|
PM20D2a | NM_001010853 | 7,65E-04 | −1,706 | | | | |
| PVRL1 | NM_002855 | 9,24E-03 | −1,693 | | | | |
| C16orf55 | AK303024 | 3,86E-04 | −1,691 | | | | |
|
RRM2a | NM_001165931 | 7,98E-03 | −1,687 | | | | |
|
HIST1H3Fa | NM_021018 | 8,20E-03 | −1,682 | DDX12 | NR_033399 | 3,55E-03 | −1,570 |
| ZNF716 | NM_001159279 | 2,70E-04 | −1,682 | FLJ30064 | AK054626 | 2,40E-03 | −1,562 |
| KIAA1524 | NM_020890 | 1,99E-04 | −1,680 |
USP37a | NM_020935 | 3,15E-04 | −1,566 |
|
FANCAa | NM_000135 | 6,54E-06 | −1,680 | PLS1 | NM_001172312 | 9,23E-04 | −1,559 |
| KLHL23 | NM_144711 | 8,07E-03 | −1,679 | MT4 | NM_032935 | 7,81E-03 | −1,558 |
| CDCA2 | NM_152562 | 3,22E-03 | −1,677 | GTSE1 | NM_016426 | 6,65E-03 | −1,556 |
| WHSC1 | NM_133330 | 7,07E-04 | −1,677 |
KCTD12a | NM_138444 | 7,18E-03 | −1,555 |
| MMS22L | NM_198468 | 2,44E-04 | −1,675 |
ZNF749a | NM_001023561 | 3,89E-03 | −1,553 |
| FAM72D | AB096683 | 2,28E-03 | −1,674 |
CENPHa | NM_022909 | 1,88E-03 | −1,547 |
|
KIAA0101a | NM_014736 | 2,64E-03 | −1,663 | DDX11 | NM_030653 | 7,35E-04 | −1,545 |
| AREG | NM_001657 | 8,49E-03 | −1,658 | SNX5 | NM_152227 | 4,96E-03 | −1,543 |
|
GINS2a | NM_016095 | 7,86E-03 | −1,657 |
MTBPa | NM_022045 | 4,20E-03 | −1,539 |
|
ARHGAP11Ba | NM_001039841 | 1,80E-03 | −1,657 | GAR1 | NM_018983 | 8,34E-03 | −1,539 |
| LYAR | NM_017816 | 9,45E-03 | −1,656 | NUF2 | NM_145697 | 2,74E-04 | −1,531 |
| YAP1 | NM_001130145 | 1,60E-03 | −1,655 |
CCNFa | NM_001761 | 8,64E-04 | −1,529 |
| PKP4 | NM_003628 | 4,89E-03 | −1,653 |
PBKa | NM_018492 | 3,92E-03 | −1,528 |
| FGF12 | NM_021032 | 2,85E-03 | −1,649 | NCAPH | NM_015341 | 3,03E-05 | −1,521 |
| NFKBIL2 | NM_013432 | 2,44E-03 | −1,632 |
EXO1a | NM_130398 | 3,35E-03 | −1,521 |
|
FOXD4L3a | NM_199135 | 3,14E-03 | −1,631 | NOS1AP | NM_014697 | 7,34E-03 | −1,520 |
| CALML4 | NM_033429 | 6,12E-03 | −1,609 | RACGAP1 | NM_013277 | 6,33E-03 | −1,517 |
| DSCC1 | NM_024094 | 2,26E-03 | −1,602 | CLCNKA | NM_004070 | 2,68E-03 | −1,517 |
| PRIM1 | NM_000946 | 2,41E-05 | −1,593 | FAM133B | NM_001040057 | 7,16E-03 | −1,515 |
|
DTLa | NM_016448 | 2,55E-04 | −1,591 |
DUX4L4a | NM_001177376 | 8,97E-03 | −1,514 |
| WDHD1 | NM_007086 | 5,97E-04 | −1,590 | GABRA6 | NM_000811 | 3,58E-03 | −1,513 |
| SUN2 | NM_015374 | 2,49E-03 | −1,586 | L2HGDH | NM_024884 | 3,44E-03 | −1,512 |
|
PHF10a | NM_018288 | 2,15E-03 | −1,583 | CDKN2C | NM_001262 | 1,50E-03 | −1,511 |
| SKA1 | NM_001039535 | 4,42E-04 | −1,576 | ARHGAP19 | NM_032900 | 1,78E-03 | −1,510 |
|
CNTNAP3a | NM_033655 | 2,76E-04 | −1,576 | SLFN11 | NM_001104587 | 5,13E-03 | −1,508 |
|
RAD51a | NM_002875 | 2,80E-03 | −1,575 | C14orf80 | NM_001134875 | 1,09E-03 | −1,506 |
| CDCA8 | NM_018101 | 4,13E-04 | −1,573 | NCAPG | NM_022346 | 3,17E-03 | −1,501 |
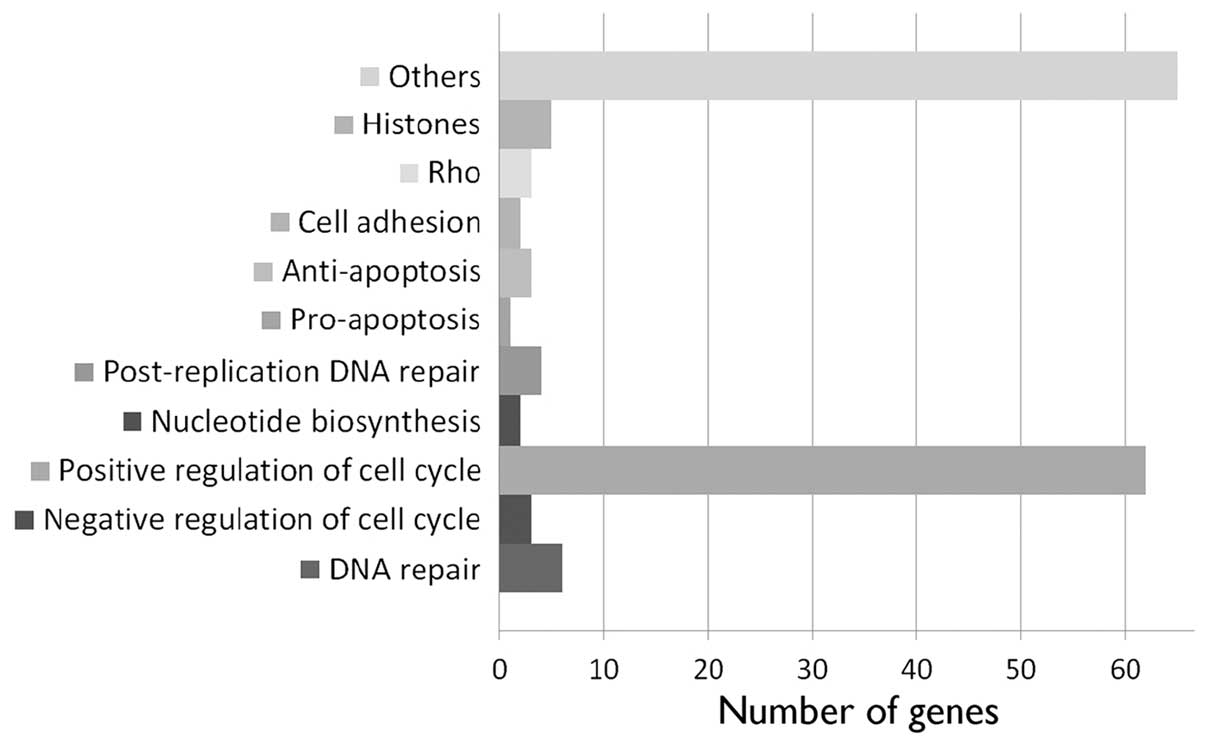
A total of 145 annotated genes was downregulated 24
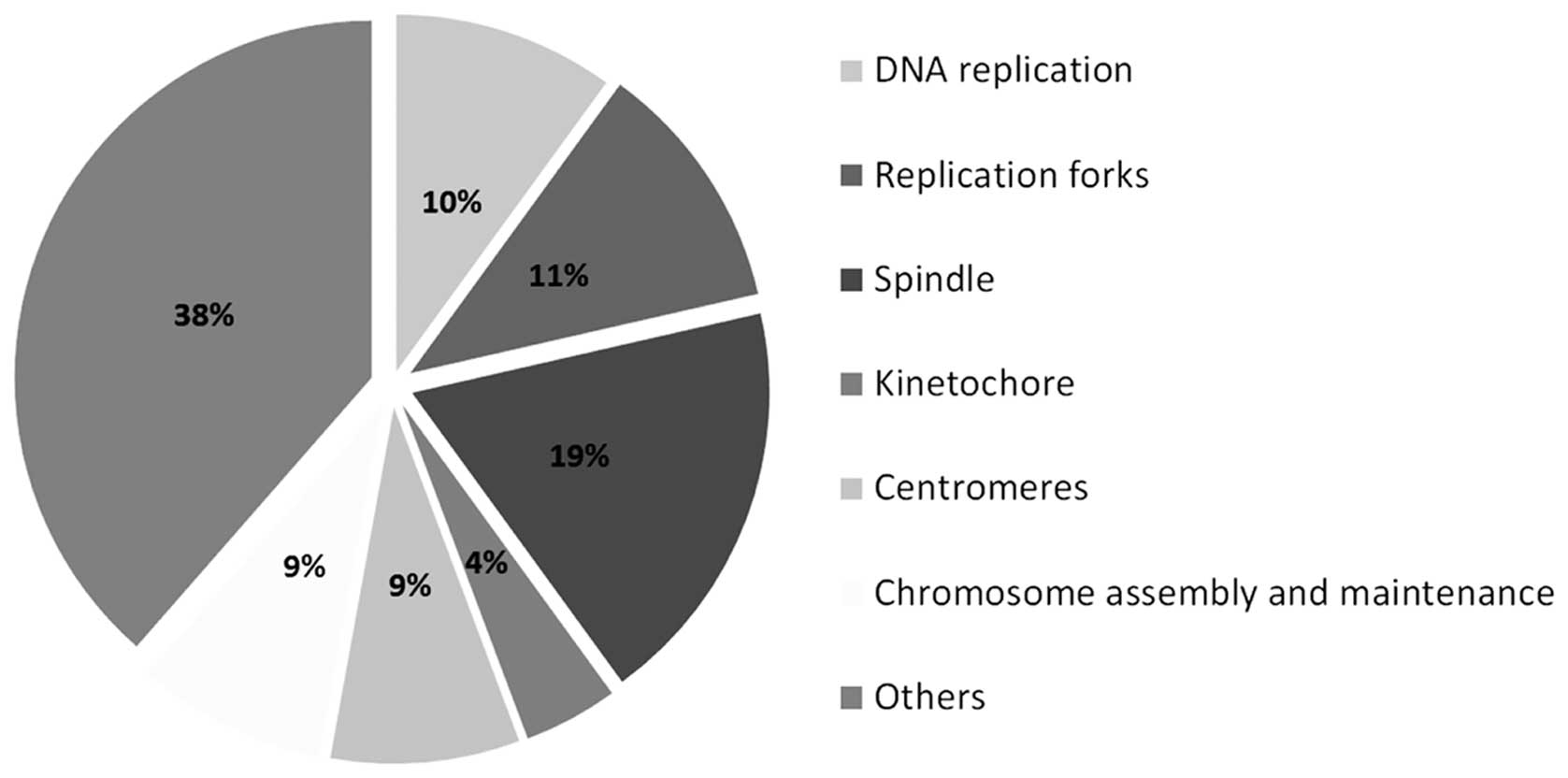
h after nickel ion irradiation (5 Gy) (Fig. 3 and Table I). The majority (62 genes) is
known to be involved in various aspects of cell division, such as
DNA replication, replication forks and chromosome assembly and
segregation (Table II and
Fig. 4). Other downregulated
genes found have been implicated in post-replicative DNA repair
(UNG, UPF3A, MSH2 and MSH6), nucleotide biosynthesis (DHFR and
RRM2), DNA repair (FANCA, MMS22L, NFKBIL2, RAD51, EXO1 and HMGB2),
positive (YAP1) and negative regulation of apoptosis (DHRS2, DHCR24
and MTBP), Rho signaling (ARHGAP19, ARHGAP11B and RACGAP1) and cell
adhesion (PVRL1 and DLGAP5).
 | Table IIList of the downregulated genes
involved in cell cycle progression 24 h after 5 Gy of nickel ion
irradiation. |
Table II
List of the downregulated genes
involved in cell cycle progression 24 h after 5 Gy of nickel ion
irradiation.
| DNA
replication | Replication
forks | Spindle | Kinetochore | Centromeres | Chromosome
formation/stability |
|---|
| PRIM1 |
MCM6a | HAUS8 | NDC80 | PLK4 | NCAPH |
|
CDC6a |
MCM7a | KIFC1 | NUF2 | MLF1IP | DDX11 |
| DSCC1 |
MCM2a |
NUSAP1a | SKA1 | CEP55 |
PHF10a |
| ORC1 |
MCM5a | CDCA8 | |
CENPHa | NCAPG2 |
|
POLA1a |
MCM3a | KIF20A | | CEP57L1 | NCAPG |
|
GINS2a |
MCM10a | SKA1 | | CEP78 | CDCA2 |
|
CDT1a | NFKBIL2 | BUB1 | | | |
| RFC4 | KIF2C | | | |
| | PRC1 | | | |
| | BUB1B | | | |
| | KIF23 | | | |
| | ESPL1 | | | |
| | KIF11 | | | |
Enrichment of transcription factor
binding motifs
In order to identify the transcription factors
potentially responsible for the differential gene expression upon
irradiation, we scanned sequences close to the transcription start
sites of these genes using Pscan (22). We found motifs for E2F1 among the
transcription factor binding motifs enriched in the downregulated
gene list, (p-value <10–19). On the other hand, we found two
members of the REL family (RelA and NF-κB) with significantly
enriched binding motifs in the list of upregulated genes (p-values
<0.05).
Discussion
DNA damage persists 24 h after
irradiation
We measured a significant increase in the number of
γ-H2AX foci 2 h following nickel ion irradiation. This number was
lower than the 30 spots per nucleus that we measured on average
upon X-irradiation with the same dose (data not shown). However, it
is not so surprising since high LET irradiation deposits high
amounts of energy along well-separated tracks. For nickel ions with
a LET of 183 keV/μm and at a dose of 2 Gy, we calculated an average
of 6.8 direct particle hits per nucleus (100 μm2), which
follows a Poisson distribution. However, we observed an average of
15 spots per nucleus. This may be due to the secondary radiation
from the ion track and the basal level of endogenous γ-H2AX foci as
observed in the controls.
Considering that the imaging of γ-H2AX foci was
performed at the same angle as ion tracks produced by the
irradiation beam, the complexity of the damage along these tracks
could not be taken into account. However, the DNA damage complexity
is known to be important in high LET irradiation (23–26). Although significantly increased,
the γ-H2AX spot occupancy did not seem to be able to account for
the complexity of DNA damage and showed similar results to the spot
number measurement. This complex DNA damage is associated with
slower repair (27) and therefore
leads to a more pronounced delayed cellular damage (26). Our results revealed a significant
level of γ-H2AX foci 24 h following nickel ion irradiation, as
compared to controls; this suggests the presence of complex DNA
damage.
Effects of high LET irradiation on the
cell cycle
Nickel ion irradiation at a dose of 0.5 Gy elicited
a lower gene expression response as compared to a dose of 5 Gy, in
terms of the number of regulated genes and FC. At 24 h
post-irradiation (5 Gy), we observed an upregulation of 12 genes
involved in cell cycle arrest and a downregulation of 62 genes
involved in cell cycle progression, among which were 3 members of
the E2F transcription factor family (E2F1, E2F2 and E2F8).
Moreover, the transcription factor binding motifs for E2F1 were
found to be highly enriched in the list of downregulated genes. E2F
is a family of transcription factors known to control G1- to
S-phase transition (28), and to
regulate the expression of a large variety of genes involved in DNA
replication, DNA repair and apoptosis (29). Among the E2F transcription
factors, E2F1 is known to be stabilized upon DNA damage through its
phosphorylation by ataxia telangiectasia-mutated (ATM) kinase, ATM
and Rad3-related (ATR) kinase and checkpoint kinase 2 (CHK2), as
well as through its acetylation (29). Our results suggest a major role of
E2F transcription factors in the response of EA.hy926 cells to high
LET irradiation.
Six components of the minichromosome maintenance
(MCM) complex, a heterohexamer helicase essential for the
initiation and elongation step of DNA replication (30), were downregulated. This helicase
may be a target for replication checkpoints (31), and is thought to be regulated
mostly through post-transcriptional modifications (32). However, our results indicate a
possible transcriptional regulation of several members of the MCM
complex. Apart from MCM, many of the observed downregulated genes
are involved in DNA replication and in chromosome formation,
maintenance and segregation, indicating their key role in cell
cycle regulation in response to high LET radiation. Of note, we
also reported the downregulation of 4 genes involved in
post-replication DNA repair (UNG, UPF3A, MSH2 and MSH6), which may
be silenced in the absence of active replication.
During this study, irradiation was performed on
proliferating endothelial cells. The results gathered on cell cycle
gene expression are therefore of moderate interest for mature blood
vessels where proliferation is limited. However, as far as hadron
therapy is concerned, our data indicate that high LET radiation may
have a significant impact on the cellular proliferation of newly
formed vascular vessels in the vicinity of the targeted tumor.
DNA damage response, oxidative stress and
apoptosis
The expression of several genes involved in the DNA
damage response, oxidative stress response and apoptosis was
induced 24 h after 5 Gy of nickel ion irradiation, with a
concomitant reduction of genes involved in DNA repair. However,
these effects were not significant at a dose of 0.5 Gy, at either
time points (8 and 24 h). These results suggest that a high dose of
nickel ion irradiation induces a global DNA damage response,
accompanied by cell cycle arrest and an increase in pro-apoptotic
gene expression 24 h after irradiation.
Impact of radiation on genes related to
cell adhesion
The impermeability of the endothelium is essential
for the vasculature integrity and is determined by the cooperation
of cell junctions and the cytoskeleton (33,34). In turn, adhesion molecules
regulate cell homeostasis, growth and apoptosis (33). A number of cellular pathways are
known to regulate cell adhesion in endothelial cells. These include
growth factors, Rho GTPases, protein kinases and calcium signaling
(34,35). The alteration of these pathways or
of adhesion molecules may trigger the radiation-induced retraction
observed by others in endothelial cells (13,14). Our study identified the
differential expression of a number of genes known to be involved
in cell adhesion (CEACAM1 and NEU1), cytoskeleton architecture
(TUBA4A, LIMA1 and PLS1), Rho signaling (ARHGAP19, ARHGAP11B and
RACGAP1) and calcium metabolism (ORAI3, CAMK2N1 and CALML4) 24 h
after 5 Gy of nickel ion irradiation, which are potentially
involved in endothelial cell retraction.
Expression of cytokines and
chemokines
Inflammatory responses mediated by endothelial cells
are believed to be involved in radiation-induced cardiovascular
disease (7). Our study revealed
the upregulation of 8 cytokines or chemokines that may be linked to
inflammation (CXCL5, TGFA, TRIM22, TNFSF9, EBI3, IL-6, IL-11 and
CD70). Of note, a search for transcription factor binding motifs
that are significantly enriched in the list of upregulated genes
upon 5 Gy of irradiation, revealed 2 members of the REL family
(RelA and NF-κB). This family of transcription factors induces the
expression of a multitude of genes, such as cytokines,
proliferation, pro-survival and anti-apoptotic genes (36). For instance, we found IL-6 to be
upregulated after 5 Gy of nickel ion irradiation. IL-6 expression
was also shown to be upregulated by low-dose radiation therapy
(10). IL-6 is known to be
activated by NF-κB (36,37) and is thought to play a role in
radiation-induced cardiovascular disease (1,7).
The secretion of cytokines may also affect non-irradiated cells by
a bystander effect. Indeed, in human fibroblasts, the external
addition of IL-6 has been shown to increase γH2AX spot occupancy
(38). The activation of NF-κB
may be linked to the transcription factor, signal transducer and
activator of transcription 3 (STAT3) (37), of which we also found significant
binding motif enrichment.
In conclusion, we observed a downregulation of
multiple genes involved in cell division, particularly at 24 h
after nickel ion irradiation. Our results suggest an important role
for E2F transcription factors in this process. The endothelial
function being based on a plethora of intercellular interactions
within a dynamic structure involving cell movements and turnover,
cell cycle arrest may play a role in the radiation-induced
cardiovascular disease. On the other hand, we observed an
upregulation of various cytokines which may be induced by NF-κB.
Other studies have also suggested that these cytokines may be
linked to radiation-induced cardiovascular disease (10). The effects on gene expression were
observed upon high doses of acute irradiation and are less relevant
to space exploration. However, during hadron therapy, healthy
tissues surrounding tumors, such as endothelial cells, may be
subjected to high doses, which may lead to complications. In this
study, we identified a multitude of potential molecular targets for
further mechanistic studies out of which the gene expression
changes upon high doses of nickel ion irradiation may be important
for patients treated with hadron-therapy.
Acknowledgements
This study was supported by 4 PRODEX/BELSPO/ESA
contracts (C90-303, C90-380, C90-391 and 42-000-90-380) and the ESA
IBER-2 program. The authors wish to thank Professor Marco Durante
for providing access to the GSI irradiation facilities.
References
|
1
|
Schultz-Hector S and Trott KR:
Radiation-induced cardiovascular diseases: is the epidemiologic
evidence compatible with the radiobiologic data? Int J Radiat Oncol
Biol Phys. 67:10–18. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Blaber E, Marçal H and Burns BP:
Bioastronautics: the influence of microgravity on astronaut health.
Astrobiology. 10:463–473. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brinckmann E: Biology in Space and Life on
Earth: Effects of Spaceflight on Biological Systems. WILEY-VCH
Verlag GmbH & Co. KGaA; Weinheim: 2007, View Article : Google Scholar
|
|
4
|
Rong Y and Welsh J: Basics of particle
therapy II biologic and dosimetric aspects of clinical hadron
therapy. Am J Clin Oncol. 33:646–649. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Blakely EA and Chang PY: Biology of
charged particles. Cancer J. 15:271–284. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Halle M, Hall P and Tornvall P:
Cardiovascular disease associated with radiotherapy: Activation of
nuclear factor kappa-B. J Intern Med. 269:469–477. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hildebrandt G: Non-cancer diseases and
non-targeted effects. Mutat Res. 687:73–77. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rödel F, Frey B, Capalbo G, et al:
Discontinuous induction of x-linked inhibitor of apoptosis in ea.
Hy926 endothelial cells is linked to NF-κB activation and mediates
the anti-inflammatory properties of low-dose ionising-radiation.
Radiother Oncol. 97:346–351. 2011.PubMed/NCBI
|
|
9
|
Rödel F, Hofmann D, Auer J, et al: The
anti-inflammatory effect of low-dose radiation therapy involves a
diminished CCL20 chemokine expression and granulocyte/endothelial
cell adhesion. Strahlenther Onkol. 184:41–47. 2008.
|
|
10
|
Rödel F, Keilholz L, Herrmann M, Sauer R
and Hildebrandt G: Radiobiological mechanisms in inflammatory
diseases of low-dose radiation therapy. Int J Radiat Biol.
83:357–366. 2007.PubMed/NCBI
|
|
11
|
Rödel F, Schaller U, Schultze-Mosgau S, et
al: The induction of TGF-beta(1) and NF-kappaB parallels a biphasic
time course of leukocyte/endothelial cell adhesion following
low-dose X-irradiation. Strahlenther Onkol. 180:194–200.
2004.PubMed/NCBI
|
|
12
|
Ando K, Ishibashi T, Ohkawara H, et al:
Crucial role of membrane type 1 matrix metalloproteinase (MT1-MMP)
in Rhoa/Rac1-dependent signaling pathways in thrombin-stimulated
endothelial cells. J Atheroscler Thromb. 18:762–773. 2011.
View Article : Google Scholar
|
|
13
|
Kantak SS, Diglio CA and Onoda JM: Low
dose radiation-induced endothelial cell retraction. Int J Radiat
Biol. 64:319–328. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Onoda JM, Kantak SS and Diglio CA:
Radiation induced endothelial cell retraction in vitro: correlation
with acute pulmonary edema. Pathol Oncol Res. 5:49–55. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gabrys D, Greco O, Patel G, Prise KM,
Tozer GM and Kanthou C: Radiation effects on the cytoskeleton of
endothelial cells and endothelial monolayer permeability. Int J
Radiat Oncol Biol Phys. 69:1553–1562. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pluder F, Barjaktarovic Z, Azimzadeh O, et
al: Low-dose irradiation causes rapid alterations to the proteome
of the human endothelial cell line EA. hy926. Radiat Environ
Biophys. 50:155–166. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Grabham P, Hu B, Sharma P and Geard C:
Effects of ionizing radiation on three-dimensional human vessel
models: differential effects according to radiation quality and
cellular development. Radiat Res. 175:21–28. 2011. View Article : Google Scholar
|
|
18
|
Takahashi Y, Teshima T, Kawaguchi N,
Hamada Y, Mori S, Madachi A, Ikeda S, Mizuno H, Ogata T, Nojima K,
Furusawa Y and Matsuura N: Heavy ion irradiation inhibits in vitro
angiogenesis even at sublethal dose. Cancer Res. 63:4253–4257.
2003.PubMed/NCBI
|
|
19
|
Kiyohara H, Ishizaki Y, Suzuki Y, Katoh H,
Hamada N, Ohno T, Takahashi T, Kobayashi Y and Nakano T:
Radiation-induced ICAM-1 expression via TGF-β1 pathway on human
umbilical vein endothelial cells; comparison between X-ray and
carbon-ion beam irradiation. J Radiat Res. 52:287–292.
2011.PubMed/NCBI
|
|
20
|
Haberer T, Becher W, Schardt D and Kraft
G: Magnetic scanning system for heavy ion therapy. Nucl Instrum
Methods Phys Res A. 330:296–305. 1993. View Article : Google Scholar
|
|
21
|
De Vos WH, Van Neste L, Dieriks B, Joss GH
and Van Oostveldt P: High content image cytometry in the context of
subnuclear organization. Cytometry A. 77:64–75. 2010.PubMed/NCBI
|
|
22
|
Zambelli F, Pesole G and Pavesi G: Pscan:
finding over-represented transcription factor binding site motifs
in sequences from co-regulated or co-expressed genes. Nucleic Acids
Res. 37:W247–W252. 2009. View Article : Google Scholar
|
|
23
|
Fokas E, Kraft G, An H and
Engenhart-Cabillic R: Ion beam radiobiology and cancer: time to
update ourselves. Biochim Biophys Acta. 1796:216–229.
2009.PubMed/NCBI
|
|
24
|
Held KD: Effects of low fluences of
radiations found in space on cellular systems. Int J Radiat Biol.
85:379–390. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Costes SV, Boissière A, Ravani S, Romano
R, Parvin B and Barcellos-Hoff MH: Imaging features that
discriminate between foci induced by high- and low-let radiation in
human fibroblasts. Radiat Res. 165:505–515. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Blakely EA and Kronenberg A: Heavy-ion
radiobiology: new approaches to delineate mechanisms underlying
enhanced biological effectiveness. Radiat Res. 150:S126–S145. 1998.
View Article : Google Scholar
|
|
27
|
Chappell LJ, Whalen MK, Gurai S, Ponomarev
A, Cucinotta FA and Pluth JM: Analysis of flow cytometry DNA damage
response protein activation kinetics after exposure to x rays and
high-energy iron nuclei. Radiat Res. 174:691–702. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dyson N: The regulation of E2F by
pRB-family proteins. Genes Dev. 12:2245–2262. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Biswas AK and Johnson DG: Transcriptional
and nontranscriptional functions of E2F1 in response to DNA damage.
Cancer Res. 72:13–17. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Costa A and Onesti S: The MCM complex:
(just) a replicative helicase? Biochem Soc Trans. 36:136–140. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Forsburg SL: The MCM helicase: Linking
checkpoints to the replication fork. Biochem Soc Trans. 36:114–119.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chuang CH, Yang D, Bai G, Freeland A,
Pruitt SC and Schimenti JC: Post-transcriptional homeostasis and
regulation of MCM2-7 in mammalian cells. Nucleic Acids Res.
40:4914–4924. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dejana E, Orsenigo F, Molendini C, Baluk P
and McDonald DM: Organization and signaling of endothelial
cell-to-cell junctions in various regions of the blood and
lymphatic vascular trees. Cell Tissue Res. 335:17–25. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Prasain N and Stevens T: The actin
cytoskeleton in endothelial cell phenotypes. Microvasc Res.
77:53–63. 2009. View Article : Google Scholar
|
|
35
|
Bogatcheva NV and Verin AD: The role of
cytoskeleton in the regulation of vascular endothelial barrier
function. Microvasc Res. 76:202–207. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Oeckinghaus A and Ghosh S: The NF-kappaB
family of transcription factors and its regulation. Cold Spring
Harb Perspect Biol. 1:a0000342009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Karin M: NF-kappaB as a critical link
between inflammation and cancer. Cold Spring Harb Perspect Biol.
1:a0001412009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dieriks B, De Vos WH, Derradji H, Baatout
S and Van Oostveldt P: Medium-mediated DNA repair response after
ionizing radiation is correlated with the increase of specific
cytokines in human fibroblasts. Mutat Res. 687:40–48. 2010.
View Article : Google Scholar
|