Introduction
Prostate cancer (PCa) is the most common non-skin
malignancy in males worldwide (1). Approximately 80% of PCa patients are
diagnosed at an early stage, and the disease is confined to the
prostate, and therefore, radical prostatectomy (RP) is the
treatment of choice for organ-confined prostate tumors (2). However, ~30% of patients who
received the surgical treatment developed tumor recurrence within
10 years postoperation (2).
Presently, prognostic markers, such as the level of
prostate-specific antigen, clinical stage and the grade of tumor
(Gleason score), have been used to predict recurrence; however,
they do not explain the interindividual variation in the clinical
outcomes among the patients who received RP. Therefore, new
biomarkers are required to identify the patients who are at a
greater risk of developing recurrence following surgery.
MicroRNAs (miRNAs or miRs), ~22 nucleotides in
length, have emerged as one of the key factors of the regulatory
network that regulates a wide spectrum of cellular activities, such
as proliferation, apoptosis, migration and differentiation, by
modulating the expression of target genes via binding to the seed
sequence at the 3′ untranslated region (3′UTR) of target mRNAs,
resulting in translational repression or mRNA degradation (3). In recent years, the function of miRs
in human cancer has attracted increasing attention (4–6).
Numerous studies have revealed the role of miRs in the development
and progression in tumors by microarray assays (7,8).
The aberrant expression of miRs could be categorized into two
classes: Upregulated and downregulated miRs. Certain miRs, known as
tumor-suppressor miRs, in cancer have been shown to suppress the
expression of oncogenes, leading to the inhibition of cancer cell
proliferation (9,10). While other highly expressed miRs,
known as oncogenic miRs, in cancer have been shown to inhibit
tumor-suppressor genes and promote tumor proliferation and
metastasis (11,12). Although specific miRs are
overexpressed in cancer cells, the majority of cancer-related miRs
are downregulated in tumors, indicating the fact that there are
more tumor-suppressor compared to oncogenic miRs (13,14). Microarray-based expression
profiling analysis highlighted the critical role of miRNAs in the
pathogenesis of PCa (14).
However, the association between the miRNA expression and PCa
recurrence remains largely unknown. To date, only a few studies
have investigated miRNA expression and PCa recurrence following RP
(15–20), showing that miRNAs, including
miR-21, miR-221 and miR-222, are potential prognostic markers for
recurrence.
Cancer stem cell (CSC) theory was initially
introduced by Mackillop et al (21) and was validated in acute myeloid
leukemia for the first time in 1997 (22). In this theory, the population of
cancer cells retains a hierarchical system, and CSCs constitute a
small part of tumor cells, which are characterized by their ability
to seed new tumors. The CSC theory has been subsequently validated
in a wide spectrum of human cancers, such as breast (23), brain (24), pancreatic (25) and liver cancer (26), and PCa (27). In PCa, CSCs are believed to be
involved in regulating metastasis, relapse and therapy resistance
(28–31).
Based on the above-mentioned evidence, we
hypothesize that the differentially expressed miRNAs in the PCa
stem cells may be responsible for the pathogenesis of the disease
recurrence. To test the hypothesis, the expression levels of 6
candidate miRNAs that are differentially expressed in prostate CSCs
were evaluated and compared based on microarray analysis in a
previous study (32), and let-7a
was substantially downregulated in recurrent compared with
non-current PCa. let-7a was established as an effective biomarker
to predict recurrence of the cancer.
Materials and methods
Patients
A total of 68 patients with histologically confirmed
PCa by RP, including 32 recurrent and 36 non-recurrent cases, were
recruited in the First Affiliated Hospital of Dalian Medical
University, Department of Urology (Liaoning, China). The
clinicopathological characteristics of the recurrent and
non-recurrent patients are presented in Table I. Patients were followed up for ≥4
years (defined as non-recurrent cases) or until prostate-specific
antigen (PSA) recurrence (recurrence was defined as two consecutive
serum PSAs >0.2 ng/ml). The study was approved by an internal
institutional review board at Dalian Medical University. Patients
were included into the study upon giving their written informed
consent.
 | Table IClinicopathological characteristics
of the subjects recruited in the study. |
Table I
Clinicopathological characteristics
of the subjects recruited in the study.
| Clinicopathological
parameters | Non-recurrent cases
(n=36) | Recurrent cases
(n=32) |
|---|
| Age, years
(range) | 66 (49–77) | 65 (45–75) |
| Pre-operative
PSA |
| Mean ng/ml
(range) | 22 (1–86) | 45 (18–148) |
| Gleason score, n
(%) |
| ≤6 | 2 (5.55) | 2 (6.25) |
| 7 | 16 (44.44) | 7 (21.88) |
| 8 | 9 (25.00) | 11 (34.38) |
| 9 | 5 (13.89) | 9 (28.13) |
| 10 | 4 (11.11) | 3 (9.38) |
| Pathological tumor
stage, n (%) |
| pT2 | 4 (11.11) | 3 (9.38) |
| pT3a | 14 (38.89) | 11 (34.38) |
| pT3b | 10 (27.78) | 15 (46.88) |
| pT4 | 8 (22.22) | 4 (12.50) |
| Lymph node
metastasis, n (%) |
| Positive | 8 (22.22) | 11 (34.38) |
| Negative | 28 (77.78) | 22 (68.75) |
Total RNA isolation
Total RNA was isolated from 32 recurrent and 36
non-recurrent tissue samples using TRIzol reagent (Invitrogen Life
Technologies, San Diego, CA, USA) according to the manufacturer's
instructions. The purities and concentrations of RNA samples were
determined spectrophotometrically using NanoDrop ND-2000c (Thermo
Fisher Scientific, Inc., Wilmington, DE, USA). RNA integrity was
examined using gel electrophoresis.
cDNA synthesis and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
To validate the differential expression of
miR-143/145, let-7a, miR-200a, miR-10 and miR-17, RNA samples were
isolated from a different set of 32 recurrent and 36 non-recurrent
patients. For miRNA RT-qPCR experiments, equal amounts of total RNA
from each sample were used for first-strand DNA (cDNA) synthesis
using the TaqMan MicroRNA Reverse Transcription kit according to
the manufacturer's instructions (Applied Biosystems, Foster City,
CA, USA). The TaqMan Universal Master mix (Applied Biosystems) was
used for the specific chromosome segment amplification, including
TaqMan miR-143 (assay ID: 002249), miR-145 (assay ID:
Hs03303169_pri), miR-200a (assay ID: 000502), miR-10 (assay ID:
000387), miR-17 (assay ID: 002308) and let-7a (assay ID: 000377)
amplification kits that were obtained from Applied Biosystems.
miRNA expression analysis by RT-qPCR was carried out using a Roche
LightCycler 480 II real-time thermal cycler (Roche Diagnostics,
Basel, Switzerland). miRNA expression data were normalized to
RNU43. Relative miRNA expression was calculated with the
comparative ΔCt-method (ΔCt sample = Ct sample − Ct RNU6B). The
2−ΔΔCt method was used to assess fold changes in miR
expression between samples and controls. Mean Ct was determined
from triplicate PCR experiments.
Cell culture
LNCaP cells, originally purchased from American Type
Culture Collection (Manassas, VA, USA), were maintained in
RPMI-1640 supplemented with 10% fetal bovine serum, 10 mmol/l
Hepes, 50 U/ml penicillin and 50 mg/ml streptomycin (all from
Invitrogen Life Technologies, Carlsbad, CA, USA). The cells were
cultured in a 5% CO2 humidified atmosphere at 37°C, and
genotypically characterized to support the authenticity of these
cells, which was consistent with their origin.
miRNA mimics/inhibitors and
transfection
Cells were transfected with 50 nmol/l of let-7a
mimics or anti-let-7a inhibitors or Negative Control #1 (Ambion,
Austin, TX, USA) using DharmaFECT 3 Transfection reagent
(Dharmacon, Inc., Lafayette, CO, USA). After 24, 48 and 72 h
transfection, the cells were harvested and subjected to the
3-(4,5-dimethylthi-azol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay to evaluate the survivals, and sections of the cells
harvested 48 h after transfection were also used for RT-qPCR,
western blotting and the apoptosis assay.
Western blotting
Cells were lysed following preparation and
determination of equal amounts of proteins. The proteins were
separated on SDS-PAGE and later transferred to a PVDF membrane
(Millipore, Billerica, MA, USA). For protein expression of
insulin-like growth factor 1 receptor (IGF1R), 1 mg/ml goat
polyclonal antibodies (Cat. no. ab4065; Abcam, Cambridge, MA, USA)
was used and β-actin (Cat. no. ab6276; Abcam) served as a
reference. For visualization, horseradish peroxidase-coupled
secondary antibodies (Cat. no. ab6728; Abcam) and the ECL Plus kit
(Pierce Biotechnology, Inc., Rockford, IL, USA) were used to
develop the signals. For quantification of band intensity, ImageJ
(http://imagej.nih.gov/ij/) (NIH,
Baltimore, MD, USA) was used.
Luciferase assay
LNCaP3 cells were seeded at a density of
6×103 cells/well in a 96-well plate and incubated for 24
h. The cells were co-transfected with wild-type or mutant IGF1R
3′UTR luciferase plasmid or Renilla luciferase plasmid and control
miRNA, and the let-7a mimics using DharmaFECT DUO Transfection
reagent (Dharmacon, Inc.). After 48 h of incubation, luciferase
activity was assayed using the Steady-Glo Luciferase Assay System
(Promega Corp., Madison, WI, USA). The Renilla luciferase activity
was used as a control for transfection efficiency.
Apoptosis assay
Flow cytometry-based apoptosis was analyzed. At 48 h
post-transfection, the LNCaP cells were harvested and resuspended
in phosphate-buffered saline (PBS) and subsequently fixed in
ethanol at room temperature overnight. The cells were washed with
PBS and resuspended in staining solution (50 mg/ml propidium
iodide, 1 mg/ml RNase A and 0.1% Triton X-100 in PBS; all purchased
from Invitrogen Life Technologies). The stained cells were
subsequently analyzed for apoptosis with the Becton Dickinson Flow
Cytometer (PT. Madagasi Brosa, Inc., Sumatera Utara,
Indonesia).
MTT assay
LNCaP cells transfected with either NC or let-7a
mimics, or let-7a inhibitor were plated on 96-well plates at
1×104 cells/well. Viable cells were measured 24, 48 and
72 h after transfection. Following incubation with MTT, the cells
were lysed in 150 ml of 100% dimethyl sulfoxide (both from
Sigma-Aldrich, St. Louis, MO, USA) and UV-visible absorbance was
read at 490 nm using the 96-well plate reader. Each sample was run
in triplicate.
Statistical analysis
Differences between each group were determined by
the t-test or Mann-Whitney U test using Statistical SPSS software
package 19.00 (IBM, Corp., Armonk, NY, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
Patient characteristics
A total of 32 recurrent and 36 non-recurrent tumors
from RP were included in the study. The average age of the patients
with recurrent PCa was 65 years, whereas those without recurrence
had an average age of 66 years. All the participants recruited in
the study were of Han ethnicity. The PSA level ranged from 18 to
148 ng/ml and 1 to 86 ng/ml in patients with and without
recurrence, respectively. As expected, the mean pre-operative PSA
level of recurrent patients was almost twice that of the
non-recurrent patients (22 vs. 45 ng/ml). The clinicopathological
features, such as Gleason score, tumor stages and lymph node
metastasis, are described in Table
I.
Evaluation of miRNAs expressed in
recurrent PCa
To evaluate the differentially expressed miRNAs in
recurrent PCa, 6 candidate miRNAs were selected that have been
reported to be differentially expressed in PCa stem cells based on
the miRNA microarray analysis, considering the significant role of
CSCs in the pathogenesis of recurrent PCa. RT-qPCR was performed to
examine the expression levels of the 6 candidate miRNAs in 32
recurrent and 36 non-recurrent case samples, and only 1 miRNA,
let-7a, was significantly downregulated in the recurrent groups
with all the other 5 miRNAs similarly expressed in the two groups,
as shown in Fig. 1. Therefore,
the following functional analysis was focused on let-7a.
Identification of the target gene of
let-7a in PCa
To identify the potential target gene of let-7a in
PCa, the online miRNA database (www.mirdb.org)
was searched and IGF1R was found to be a potential target gene
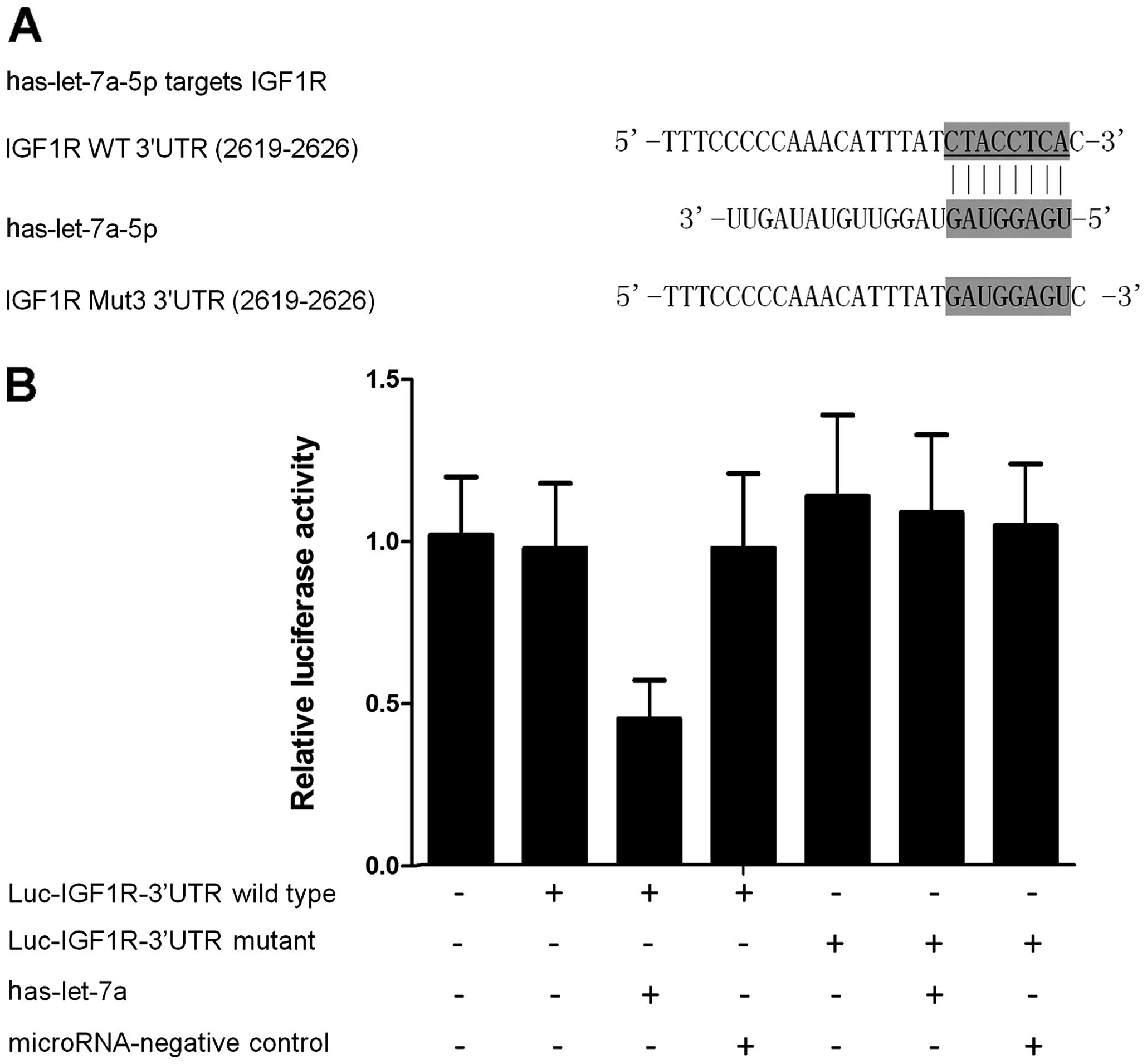
(Fig. 2A). Subsequently, the
wild-type of 3′UTR of IGF1R was subcloned and inserted into the
vector that contained the luciferase gene, and the 'seed sequence'
in the 3′UTR of IGF1R was replaced using site-directed mutagenesis
(Fig. 2A). The results of the
luciferase assay showed that the relative luciferase activities in
the let-7a-overexpressing PCa cells transfected with wild-type
3′UTR of IGF1R were substantially lower than those cells
transfected with mutant 3′UTR of IGF1R (Fig. 2B), suggesting that IGF1R was an
effective target of let-7a with the 'seed sequence' in the 3′UTR
acting as the binding site of the miRNA.
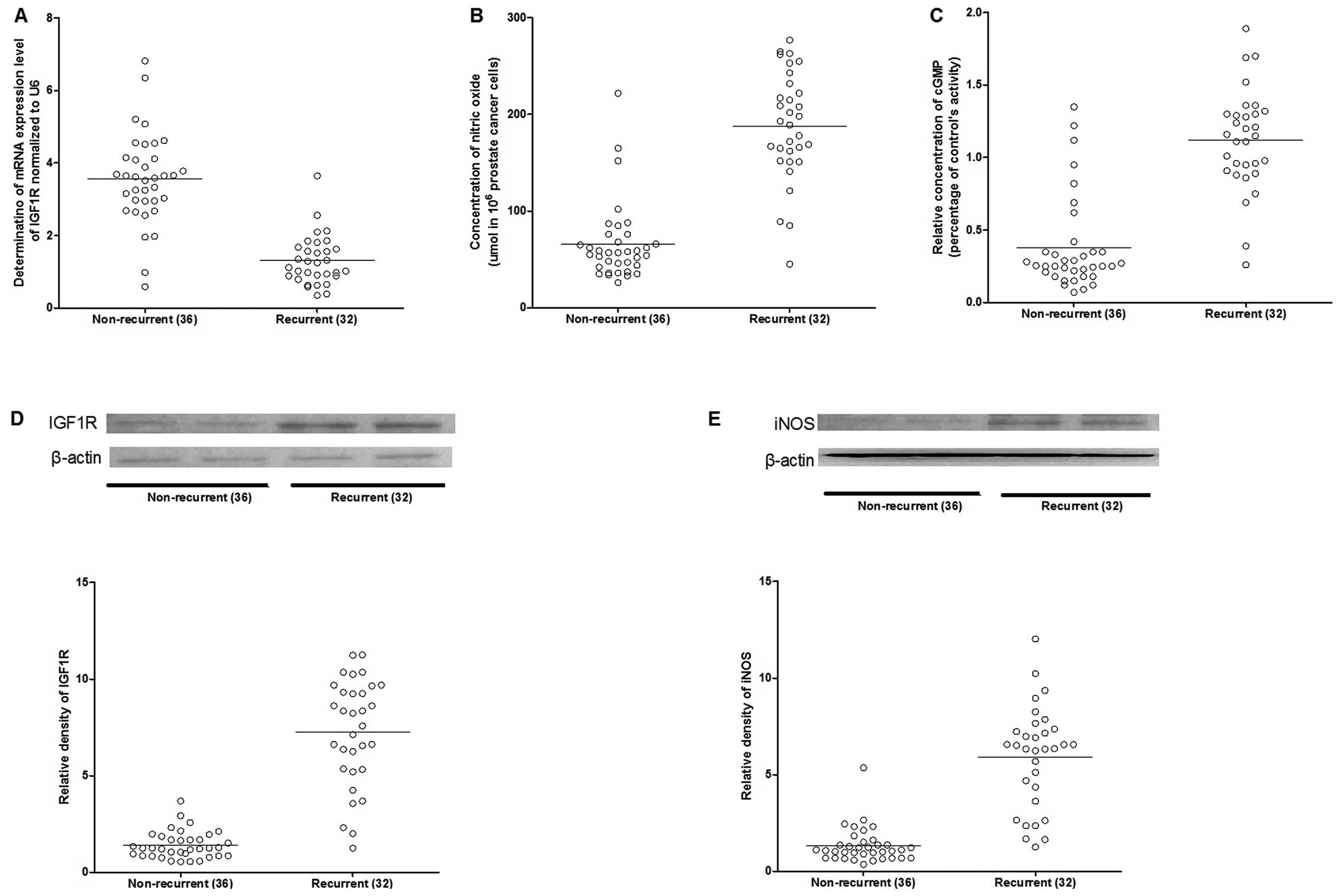
Assessment of protein and mRNA expression
level patterns
To identify the miRNA-gene association in the
recurrence of the disease, the mRNA and protein expression patterns
in the recurrent and non-recurrent cases were assessed using
RT-qPCR and western blotting. The mRNA and protein expression
levels of IGF1R were significantly upregulated in the recurrent
cases (Fig. 3). As a downstream
effector of IGF1R, the expression level of inducible nitric oxide
synthase (iNOS) was also increased in the recurrent cases (Fig. 3), and consistently, as the
catalytic products of iNOS, the concentration of nitric oxide and
cGMP were markedly higher in the recurrent compared to
non-recurrent groups (Fig.
3).
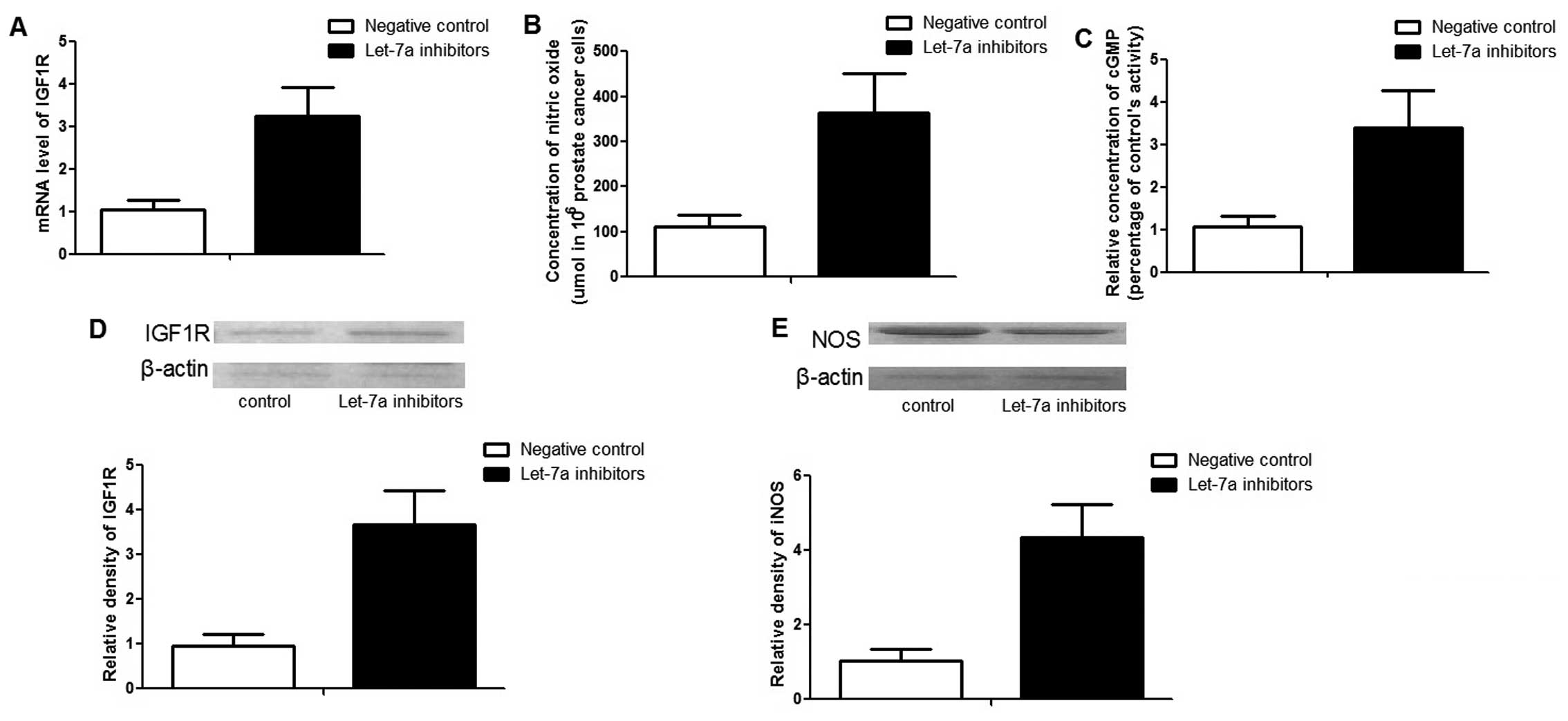
Function of let-7a in PCa cells
To further characterize the role of let-7a,
'gain-of-function' analysis was performed by transfecting let-7a
mimics. After 48 h transfection, the expression level of let-7a was
~30 times higher than those cells transfected with the negative
controls (data not shown). As expected, exogenous overexpression of
let-7a significantly downregulated the expression of IGF1R, as well
as its downstream effector, iNOS, as shown in Fig. 4. The NO and cyclic guanosine
mono-phosphate (cGMP) concentrations were much lower in the cells
transfected with let-7a mimics compared to the controls (Fig. 4).
To confirm the function of let-7a in PCa cells,
'loss-of-function' analysis was performed by transfecting
anti-let-7a inhibitors. After 48 h transfection, the expression
level of let-7a was ~12 times lower than those cells transfected
with negative controls (data not shown). In line with the
'gain-of-function' result, the downregulation of let-7a
significantly promoted the expression of IGF1R and iNOS. In
addition, the concentrations of NO and cGMP were also enhanced by
the inhibitors (Fig. 5).
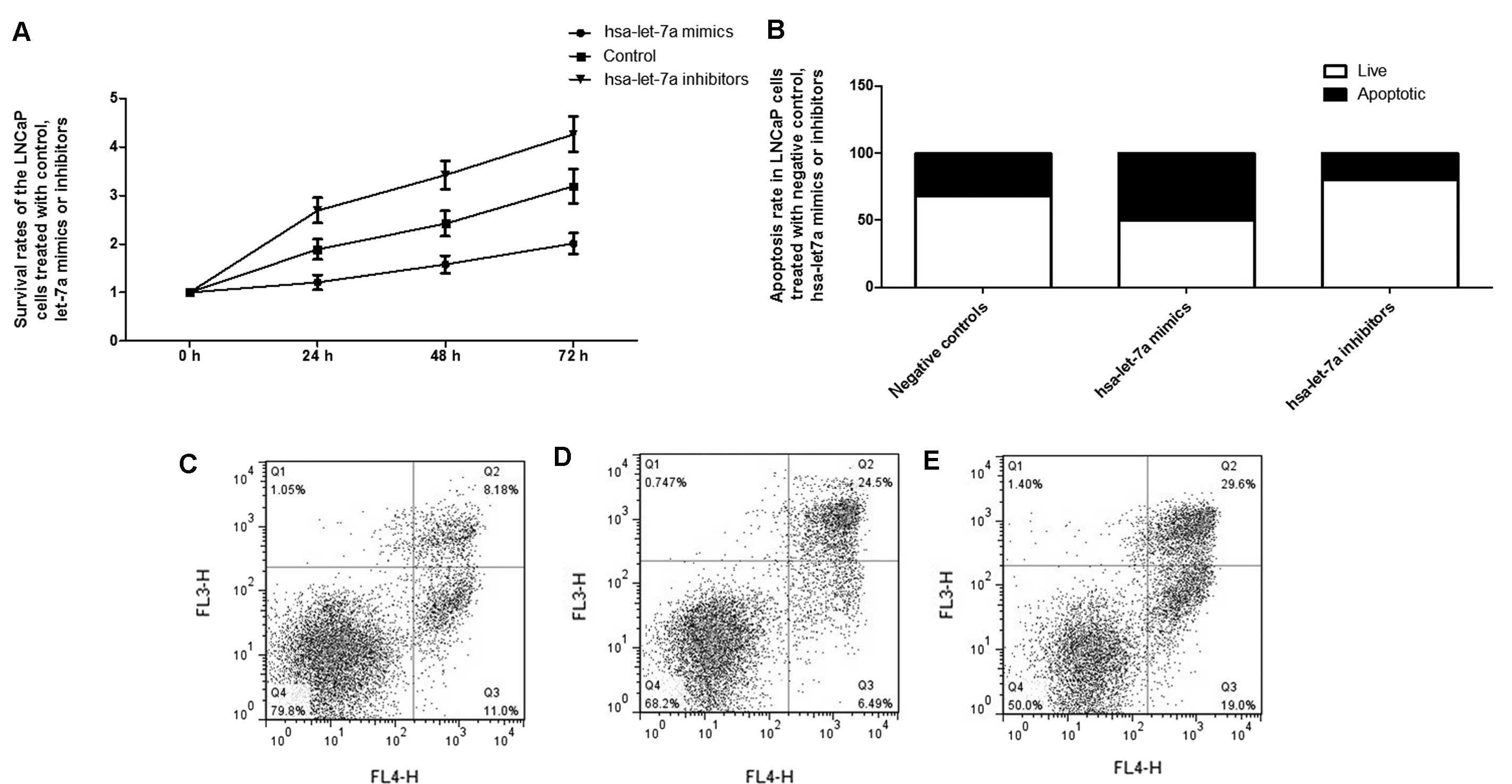
Considering the significant role of the
let-7a-IGF1R-iNOS-NO-cGMP axis in the control of cellular
proliferation, the effect of let-7a expression alternation on the
growth of PCa cells was further evaluated, identifying that
exogenous overexpression of let-7a substantially suppressed the
proliferation of the cells, while inhibition of let-7a evidently
promoted the proliferation (Fig.
6). Additionally, flow cytometry analysis was used to explore
the molecular mechanism underlying the proliferation-regulating
effect, and exogenous overexpression of let-7a significantly
introduced apoptosis to the LNCaP cells, while transfection with
let-7a inhibitors reduced the apoptosis, as shown in Fig. 6.
Discussion
PCa is characterized by highly heterogeneous disease
courses, and ~30% of the PCa patients develop recurrence even
following a successful surgical intervention or adjuvant therapy
(33,34). The most commonly used biomarkers
to predict the pathological stage of the tumor and the treatment
efficiency include primary tumor stage, serum PSA level and biopsy
Gleason scores; however, none of these indicators are reliable to
predict the clinical outcomes of PCa (35,36). Recurrence is the main cause of
fatality for PCa patients and CSCs are proposed to have important
roles in cancer recurrence (37).
In the present study, confirmatory RT-qPCR was performed to
evaluate the expression levels of 6 candidate miRNAs that have been
shown to be differentially expressed in prostate CSCs based on
microarray analysis in a previous study (32), and identified that let-7a was
substantially down-regulated in recurrent compared with non-current
PCa.
First identified in Caenorhabditis elegans,
let-7 has been intensively studied. The human let-7 family is
composed of 9 members, which are let-7a, let-7b, let-7c, let-7d,
let-7e, let-7f, let-7g, let-7i and miR-98. The let-7 family is gen
erally believed to function as a tumor suppressor by targeting
certain known oncogenes, such as Ras (38), high-mobility group A2 (39) and c-myc (40). Downregulated let-7 expression has
been identified in numerous cancers, including PCa (41), and it has been associated with
poor patient prognosis in lung cancer (42), head and neck squamous cell
carcinoma (43), and ovarian
cancer (44). Furthermore, let-7
family members have been shown to be involved in the control of the
self-renewal capacity of breast cancer cells (45) by regulating the genes, such as
Oct4 and Sox2, which have been functionally associated with the
stem cell (46). Accumulative
evidence showed that let-7 could alter the expression of Lin28 and
In28B, which in turn block the accumulation of mature let-7,
forming a feedback loop and having a critical role in regulating
'stemness' by controlling self-renewal (47–51), and such stemness and self-renewal
are the biological characteristics of CSCs that are associated with
tumor aggressiveness and recurrence.
As an important member of the let-7 family, let-7a
has been shown to fulfill tumor-suppressive functions by
suppressing certain CSC properties in PCa (4), as well as in certain other cancer
types (45,52). In the present study, let-7a was
found to virtually target IGF1R, a gene that has been shown to
promote proliferation of variable types of cells by inhibiting
apoptosis (53), and such an
miR-gene association was confirmed by the luciferase assay, as well
as 'loss-of-function' and 'gain-of-function' analysis by
transfecting let-7a mimics and inhibitors, respectively.
The inability of a cell to regulate its growth and
proliferation is an important characteristic of cancer. Activation
of insulin-like growth factor 1 (IGF1)/IGF1R signaling is
reportedly critical for PCa cell growth and progression. IGF1 is a
universal factor exhibiting pleiotropic effects on a variety of
cell types, and IGF1R is a receptor tyrosine kinase that mediates
IGF1-induced signaling events, including cell survival and
proliferation. The IGF1/IGF1R signaling pathway has an important
role in the development and growth of numerous tissues (1,2).
In the mammary gland, IGF1 is the primary mediator of growth
hormone signaling and controls ductal development and terminal end
bud formation (54). Pacher et
al (55) reported that
activation of the IGF1/IGF1R signaling pathway promoted the
expression of a wide spectrum of genes, contributing to the
stimulatory effects of IGF signaling on the global protein
synthesis rate, cell proliferation and tumor formation.
IGF1, as one of the growth factors and cytokines
that control apoptosis, is a potent survival factor. Introduction
of IGF1 could prevent serum deprivation-induced apoptosis by
binding its ligand, IGF1R, and activating the signaling pathway,
and such an effect was proven to be NO independent (56), indicating a potential role of iNOS
in the control of apoptosis by the IGF1/IGF1R pathway. The
shrinkage of apoptotic cells, resulting from a shortage of
cytosolic ions and water in response to apoptosis inducers
(57), is an initial prerequisite
for apoptosis that antecedes the majority of other morphological
alterations during the apoptotic process. Jin et al
(58) demonstrated that
IGF1/IGF1R have a significant role in the inhibition of cell
shrinkage, as well as the attenuation of SD-induced apoptosis, and
an iNOS-NO-dependent mechanism accounted for a significant role of
the effects of IGF1/IGF1R on the control of cell proliferation.
Results of the present study suggest that
upregulation of IGF1R, caused by downregulation of let-7a, could
promote the proliferation of cancer cells by inducing an increase
in the expression level of iNOS and concentrations of NO and cGMP.
Consistently, downregulation of IGF1R by upregulation of let-7a
could significantly suppress the proliferation of PCa cells by
suppressing the expression of iNOS and reducing the concentration
of NO and cGMP.
Several limitations are recognized in the present
study. Firstly, the sample pool recruited in the present study was
small, which makes the conclusion drawn from the study
statistically limited, and it requires to be interpreted with
caution. Secondly, the participants enrolled were all Chinese, and
the conclusion could be further limited by the lack of diversity of
ethnicity. Future large-scale investigations involving populations
of other ethnicities are therefore warranted to confirm these
findings and to evaluate ethnic differences.
In conclusion, the present findings that let-7a
directly targets IGF1R in PCa cells could further reveal the
mechanism of prostate recurrence. let-7a may partly contribute to
IGF1R overexpression in recurrent PCa, by increasing cell survival
and proliferation. let-7a may be a novel therapeutic candidate to
prevent recurrent PCa given its ability to induce apoptosis and
inhibit cell growth.
Acknowledgments
The project was fully sponsored by the National
Natural Science Foundation of China with grant no. 30371825.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siddiqui SA, Inman BA, Sengupta S, Slezak
JM, Bergstralh EJ, Leibovich BC, Zincke H and Blute ML: Obesity and
survival after radical prostatectomy: A 10-year prospective cohort
study. Cancer. 107:521–529. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nolan T and Cogoni C: The long hand of the
small RNAs reaches into several levels of gene regulation. Biochem
Cell Biol. 82:472–481. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu C, Kelnar K, Vlassov AV, Brown D, Wang
J and Tang DG: Distinct microRNA expression profiles in prostate
cancer stem/progenitor cells and tumor-suppressive functions of
let-7. Cancer Res. 72:3393–3404. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Duan Z, Choy E, Harmon D, Liu X, Susa M,
Mankin H and Hornicek F: MicroRNA-199a-3p is downregulated in human
osteosarcoma and regulates cell proliferation and migration. Mol
Cancer Ther. 10:1337–1345. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Duan Z, Choy E, Nielsen GP, Rosenberg A,
Iafrate J, Yang C, Schwab J, Mankin H, Xavier R and Hornicek FJ:
Differential expression of microRNA (miRNA) in chordoma reveals a
role for miRNA-1 in Met expression. J Orthop Res. 28:746–752.
2010.
|
|
7
|
Ratert N, Meyer HA, Jung M, Mollenkopf HJ,
Wagner I, Miller K, Kilic E, Erbersdobler A, Weikert S and Jung K:
Reference miRNAs for miRNAome analysis of urothelial carcinomas.
PLoS One. 7:e393092012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lang MF, Yang S, Zhao C, Sun G, Murai K,
Wu X, Wang J, Gao H, Brown CE, Liu X, et al: Genome-wide profiling
identified a set of miRNAs that are differentially expressed in
glioblastoma stem cells and normal neural stem cells. PLoS One.
7:e362482012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Akao Y, Nakagawa Y, Hirata I, Iio A, Itoh
T, Kojima K, Nakashima R, Kitade Y and Naoe T: Role of antioncomirs
miR-143 and -145 in human colorectal tumors. Cancer Gene Ther.
17:398–408. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Johnson SM, Grosshans H, Shingara J, Byrom
M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D and Slack
FJ: RAS is regulated by the let-7 microRNA family. Cell.
120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B,
Korzh V, Lodish HF and Lim B: MicroRNA-125b is a novel negative
regulator of p53. Genes Dev. 23:862–876. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Y, Gao JS, Tang X, Tucker LD,
Quesenberry P, Rigoutsos I and Ramratnam B: MicroRNA 125a and its
regulation of the p53 tumor suppressor gene. FEBS Lett.
583:3725–3730. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Garzon R, Marcucci G and Croce CM:
Targeting microRNAs in cancer: Rationale, strategies and
challenges. Nat Rev Drug Discov. 9:775–789. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bader AG, Brown D and Winkler M: The
promise of microRNA replacement therapy. Cancer Res. 70:7027–7030.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fendler A, Jung M, Stephan C, Honey RJ,
Stewart RJ, Pace KT, Erbersdobler A, Samaan S, Jung K and Yousef
GM: miRNAs can predict prostate cancer biochemical relapse and are
involved in tumor progression. Int J Oncol. 39:1183–1192.
2011.PubMed/NCBI
|
|
16
|
Leite KR, Tomiyama A, Reis ST,
Sousa-Canavez JM, Sañudo A, Dall'Oglio MF, Camara-Lopes LH and
Srougi M: MicroRNA-100 expression is independently related to
biochemical recurrence of prostate cancer. J Urol. 185:1118–1122.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Long Q, Johnson BA, Osunkoya AO, Lai YH,
Zhou W, Abramovitz M, Xia M, Bouzyk MB, Nam RK, Sugar L, et al:
Protein-coding and microRNA biomarkers of recurrence of prostate
cancer following radical prostatectomy. Am J Pathol. 179:46–54.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schaefer A, Jung M, Mollenkopf HJ, Wagner
I, Stephan C, Jentzmik F, Miller K, Lein M, Kristiansen G and Jung
K: Diagnostic and prognostic implications of microRNA profiling in
prostate carcinoma. Int J Cancer. 126:1166–1176. 2010.
|
|
19
|
Spahn M, Kneitz S, Scholz CJ, Stenger N,
Rüdiger T, Ströbel P, Riedmiller H and Kneitz B: Expression of
microRNA-221 is progressively reduced in aggressive prostate cancer
and metastasis and predicts clinical recurrence. Int J Cancer.
127:394–403. 2010.
|
|
20
|
Tong AW, Fulgham P, Jay C, Chen P, Khalil
I, Liu S, Senzer N, Eklund AC, Han J and Nemunaitis J: MicroRNA
profile analysis of human prostate cancers. Cancer Gene Ther.
16:206–216. 2009.
|
|
21
|
Mackillop WJ, Ciampi A, Till JE and Buick
RN: A stem cell model of human tumor growth: Implications for tumor
cell clonogenic assays. J Natl Cancer Inst. 70:9–16.
1983.PubMed/NCBI
|
|
22
|
Bonnet D and Dick JE: Human acute myeloid
leukemia is organized as a hierarchy that originates from a
primitive hematopoietic cell. Nat Med. 3:730–737. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Singh SK, Hawkins C, Clarke ID, Squire JA,
Bayani J, Hide T, Henkelman RM, Cusimano MD and Dirks PB:
Identification of human brain tumour initiating cells. Nature.
432:396–401. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li C, Heidt DG, Dalerba P, Burant CF,
Zhang L, Adsay V, Wicha M, Clarke MF and Simeone DM: Identification
of pancreatic cancer stem cells. Cancer Res. 67:1030–1037. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai
P, Chu PW, Lam CT, Poon RT and Fan ST: Significance of
CD90+ cancer stem cells in human liver cancer. Cancer
Cell. 13:153–166. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Collins AT, Berry PA, Hyde C, Stower MJ
and Maitland NJ: Prospective identification of tumorigenic prostate
cancer stem cells. Cancer Res. 65:10946–10951. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sugihara E and Saya H: Complexity of
cancer stem cells. Int J Cancer. 132:1249–1259. 2013. View Article : Google Scholar
|
|
29
|
Freitas DP, Teixeira CA, Santos-Silva F,
Vasconcelos MH and Almeida GM: Therapy-induced enrichment of
putative lung cancer stem-like cells. Int J Cancer. 134:1270–1278.
2014. View Article : Google Scholar
|
|
30
|
Ratajczak M, Tarnowski M, Staniszewska M,
Sroczynski T and Banach B: Mechanisms of cancer metastasis:
Involvement of cancer stem cells? Minerva Med. 101:179–191.
2010.PubMed/NCBI
|
|
31
|
Chen X, Rycaj K, Liu X and Tang DG: New
insights into prostate cancer stem cells. Cell Cycle. 12:579–586.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rane JK, Scaravilli M, Ylipää A, Pellacani
D, Mann VM, Simms MS, Nykter M, Collins AT, Visakorpi T and
Maitland NJ: MicroRNA expression profile of primary prostate cancer
stem cells as a source of biomarkers and therapeutic targets. Eur
Urol. 67:7–10. 2015. View Article : Google Scholar
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
34
|
Han M, Partin AW, Zahurak M, Piantadosi S,
Epstein JI and Walsh PC: Biochemical (prostate specific antigen)
recurrence probability following radical prostatectomy for
clinically localized prostate cancer. J Urol. 169:517–523. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Brookman-Amissah N, Nariculam J, Freeman
A, Willamson M, Kirby RS, Masters JR and Feneley MR: Allelic
imbalance at 13q14.2 approximately q14.3 in localized prostate
cancer is associated with early biochemical relapse. Cancer Genet
Cytogenet. 179:118–126. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Barron N, Keenan J, Gammell P, Martinez
VG, Freeman A, Masters JR and Clynes M: Biochemical relapse
following radical prostatectomy and miR-200a levels in prostate
cancer. Prostate. 72:1193–1199. 2012. View Article : Google Scholar
|
|
37
|
Li X, Liu Y, Chen W, Fang Y, Xu H, Zhu HH,
Chu M, Li W, Zhuang G and Gao WQ: TOP2Ahigh is the phenotype of
recurrence and metastasis whereas TOP2Aneg cells represent cancer
stem cells in prostate cancer. Oncotarget. 5:9498–9513. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kumar MS, Erkeland SJ, Pester RE, Chen CY,
Ebert MS, Sharp PA and Jacks T: Suppression of non-small cell lung
tumor development by the let-7 microRNA family. Proc Natl Acad Sci
USA. 105:3903–3908. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mayr C, Hemann MT and Bartel DP:
Disrupting the pairing between let-7 and Hmga2 enhances oncogenic
transformation. Science. 315:1576–1579. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kim HH, Kuwano Y, Srikantan S, Lee EK,
Martindale JL and Gorospe M: HuR recruits let-7/RISC to repress
c-Myc expression. Genes Dev. 23:1743–1748. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Barh D, Malhotra R, Ravi B and Sindhurani
P: MicroRNA let-7: An emerging next-generation cancer therapeutic.
Curr Oncol. 17:70–80. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Landi MT, Zhao Y, Rotunno M, Koshiol J,
Liu H, Bergen AW, Rubagotti M, Goldstein AM, Linnoila I, Marincola
FM, et al: MicroRNA expression differentiates histology and
predicts survival of lung cancer. Clin Cancer Res. 16:430–441.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Childs G, Fazzari M, Kung G, Kawachi N,
Brandwein-Gensler M, McLemore M, Chen Q, Burk RD, Smith RV,
Prystowsky MB, et al: Low-level expression of microRNAs let-7d and
miR-205 are prognostic markers of head and neck squamous cell
carcinoma. Am J Pathol. 174:736–745. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Helland Å, Anglesio MS, George J, Cowin
PA, Johnstone CN, House CM, Sheppard KE, Etemadmoghadam D, Melnyk
N, Rustgi AK, et al: Australian Ovarian Cancer Study Group:
Deregulation of MYCN, LIN28B and LET7 in a molecular subtype of
aggressive high-grade serous ovarian cancers. PLoS One.
6:e180642011. View Article : Google Scholar
|
|
45
|
Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong
C, Huang Y, Hu X, Su F, Lieberman J, et al: let-7 regulates self
renewal and tumorigenicity of breast cancer cells. Cell.
131:1109–1123. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kong D, Banerjee S, Ahmad A, Li Y, Wang Z,
Sethi S and Sarkar FH: Epithelial to mesenchymal transition is
mechanistically linked with stem cell signatures in prostate cancer
cells. PLoS One. 5:e124452010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Viswanathan SR, Daley GQ and Gregory RI:
Selective blockade of microRNA processing by Lin28. Science.
320:97–100. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yang X, Lin X, Zhong X, Kaur S, Li N,
Liang S, Lassus H, Wang L, Katsaros D, Montone K, et al:
Double-negative feedback loop between reprogramming factor LIN28
and microRNA let-7 regulates aldehyde dehydrogenase 1-positive
cancer stem cells. Cancer Res. 70:9463–9472. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhong X, Li N, Liang S, Huang Q, Coukos G
and Zhang L: Identification of microRNAs regulating reprogramming
factor LIN28 in embryonic stem cells and cancer cells. J Biol Chem.
285:41961–41971. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ji J and Wang XW: A Yin-Yang balancing act
of the lin28/let-7 link in tumorigenesis. J Hepatol. 53:974–975.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
King CE, Cuatrecasas M, Castells A,
Sepulveda AR, Lee JS and Rustgi AK: LIN28B promotes colon cancer
progression and metastasis. Cancer Res. 71:4260–4268. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Peter ME: Let-7 and miR-200 microRNAs:
Guardians against pluripotency and cancer progression. Cell Cycle.
8:843–852. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Esquela-Kerscher A, Trang P, Wiggins JF,
Patrawala L, Cheng A, Ford L, Weidhaas JB, Brown D, Bader AG and
Slack FJ: The let-7 microRNA reduces tumor growth in mouse models
of lung cancer. Cell Cycle. 7:759–764. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kleinberg DL, Feldman M and Ruan W: IGF-I:
An essential factor in terminal end bud formation and ductal
morphogenesis. J Mammary Gland Biol Neoplasia. 5:7–17. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Pacher M, Seewald MJ, Mikula M, Oehler S,
Mogg M, Vinatzer U, Eger A, Schweifer N, Varecka R, Sommergruber W,
et al: Impact of constitutive IGF1/IGF2 stimulation on the
transcriptional program of human breast cancer cells.
Carcinogenesis. 28:49–59. 2007. View Article : Google Scholar
|
|
56
|
Kang BP, Urbonas A, Baddoo A, Baskin S,
Malhotra A and Meggs LG: IGF-1 inhibits the mitochondrial apoptosis
program in mesangial cells exposed to high glucose. Am J Physiol
Renal Physiol. 285:F1013–F1024. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wei L, Xiao AY, Jin C, Yang A, Lu ZY and
Yu SP: Effects of chloride and potassium channel blockers on
apoptotic cell shrinkage and apoptosis in cortical neurons.
Pflugers Arch. 448:325–334. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Jin C and Guo J, Qiu X, Ma K, Xiang M, Zhu
X and Guo J: IGF-1 induces iNOS expression via the p38 MAPK signal
pathway in the anti-apoptotic process in pulmonary artery smooth
muscle cells during PAH. J Recept Signal Transduct Res. 34:325–331.
2014. View Article : Google Scholar : PubMed/NCBI
|