Introduction
Cancer is one of the most important challenges of
human health today. Breast cancer (BrC) is the most common
malignancy in women with more than 500,000 deaths globally per year
(1). Despite advances in diagnosis
and treatment, most cases are still diagnosed at advanced stages,
particularly in developing countries in which more than 60% of the
fatalities occur (2). Genetic
modifications in tumor suppressor genes and proto-oncogenes,
together with epigenetic changes in DNA methylation, histone
modifications and altered miRNA expression drive cancer evolution
from a pre-malignant cell to a highly aggressive cancer cell
(3,4).
In aggressive tumors, cells acquire the ability to
detach from neighboring cells, migrate and invade the surrounding
tissue, access the blood or lymphatic vessels and colonize distant
organs (5–7). These processes of invasion and
metastasis are facilitated by epithelial to mesenchymal transition
(EMT). During EMT tumor cells undergo a transcriptional
re-programing, losing expression of epithelial genes, such as
adhesion molecules that sustain cell to cell contacts, and instead
express mesenchymal genes (8).
Invasive cells are characterized by lack of expression of
E-cadherin and gain of expression of vimentin and N-cadherin, which
are commonly used to identify EMT cells. EMT cells also switch from
a cobblestone- to spindle (fibroblast-like) morphology. Expression
of EMT markers often correlates with expression of a stem-like
phenotype (for instance CD44+CD24−/low)
(9–11). Therefore, it is thought that EMT is
a mechanism that also facilitates stemness (12–14).
miRNAs are evolutionarily conserved noncoding RNA
molecules that regulate gene expression. Today, more than 1800
miRNAs have been shown to regulate cellular processes such as cell
differentiation, cell cycle and cell death, and thus their abnormal
function also impacts human diseases. In cancer, different miRNAs
have been classified as tumor suppressors when they regulate the
expression of cellular oncogenes and as oncomiRs when they target
tumor suppressor genes (15). For
instance, recent studies have shown that miRNAs are important
regulators of EMT (16). In the
mammary gland the miR-200 family maintains the epithelial program
by downregulating expression of ZEB1 and ZEB2, the master
transcriptional activators of the EMT program (17). ZEB1 and ZEB2 are also
transcriptional repressors of CDH1, the gene that encodes
E-cadherin (18,19). On the other hand, the miR-221
family is frequently expressed in poorly differentiated aggressive
breast tumors with EMT characteristics (20). miR-650 is a novel oncomiR also
implicated in EMT. miR-650 is often expressed in prostate,
colorectal, hepatocellular, skin and gastric cancers (21–25),
with evidence that it targets tumor suppressors ING4 and NDRG2
(23,26–28).
ING4 and NDRG2 expression is often lost in cancer and different
lines of evidence support their participation in EMT (29–33).
In this study, we examined the chromosomal
abnormalities of three primary cultures isolated from Mexican
patients with BrC using the microarray CytoScan HD and Chromosome
Analysis Suite 3.0 software (Affymetrix, Santa Clara, CA, USA). We
found a common genetic lesion in the three samples, an
amplification of band q11.2 in chromosome 22, in the region that
encodes miR-650. We also found that the three primary isolates were
negative to E-cadherin, positive to vimentin and to the stem cell
marker CD44, and were invasive in Transwell migration assays,
supporting their acquisition of EMT-related properties.
Materials and methods
Patient description and ethics
statement
Patient samples were obtained from the tissue bank
of the Unidad de Investigación En virología y Cáncer, Hospital
Infantil de México Federico Gómez. All patients included in the
study signed an informed consent to participate. The study was
approved by the Scientific, Ethics and Biosafety Institutional
Review Boards of the participating hospitals. Patients included
were diagnosed with invasive ductal carcinoma, histological grade 2
and clinical stage II, with no previous neoadjuvant therapy before
tissue resection. Patients were all female aged 42, 64 and 55
years.
Tissue processing for primary cell
culture isolation
Tumor tissues were rinsed with sterile PBS and
mechanically disaggregated with a scalpel in 1–2-mm fragments,
which were subsequently digested for 2 h at room temperature (RT)
with a mixture of 1 mg/ml collagenase type I (C0130-100 MG, Sigma,
St. Louis, MO, USA) and 100 U/ml hyaluronidase (H3506-100MG, Sigma)
in DMEM/F12 (Dulbecco's modified Eagle's medium/Nutrient Mixture
F-12) (11039-047, Gibco-Invitrogen Cell Culture, Carlsbad, CA, USA)
supplemented with 100 U/ml penicillin and 100 µg/ml
streptomycin (15140-122, Gibco-Invitrogen Cell Culture) in constant
stirring. The resulting suspension was filtered through a wide pore
membrane and a 100 µm membrane. The cells were pelleted and
washed twice with sterile PBS and plated in an enriched medium for
epithelial cells [DMEM/F12 supplemented with 5% horse serum
(26050088, Gibco, Auckland, New Zealand), 10 ng/ml cholera toxin
(C8052), 0.5 µg/ml hydrocortisone (H0888), 5 µg/ml
insulin (91077C), all from Sigma Chemical Co. (St. Louis, MO, USA),
5 ng/ml epidermal growth factor (EGF) (AF-100-15, PeproTech, Rocky
Hill, NJ, USA)] and 1% penicillin/streptomycin 100X, and then
incubated at 37°C in a 5% CO2 atmosphere. To assure the
epithelial origin of the isolated cells, three epithelial markers
were tested by immunocytochemistry: mouse monoclonal anti-human
anti-PanCytokeratin (CM011A, clone: AE1/AE3, Biocare Medical,
Concord, CA, USA), mouse monoclonal anti-human anti-mucin 1 (MUC-1)
[Epithelial Membrane Antigen (EMA) (559774, clone: Mc-5, Biocare
Medical)], and mouse monoclonal anti-human anti-epithelial cell
adhesion molecule (EpCAM) (ab20160, clone: AUA1, Biocare Medical)
all in a working dilution 1:50. The epithelial cell isolates were
named as UIVC-IDC-6, -9 and -10.
Analysis of the primary cultures using
the microarray CytoScan HD and Chromosome Analysis Suite 3.0
software
Total DNA from an early passage (<6) of the three
primary BrC cultures was isolated using the QIAamp® DNA
Micro kit (56304, Qiagen, Gaithersburg, MD, USA) according to the
manufacturer's protocol. To verify the quality of DNA, the
endogenous β-actin gene was amplified by PCR. The Affymetrix
CytoScan HD array (901835, Affymetrix) was used to evaluate copy
number and loss of heterozygosity (LOH). This array contains more
than 2.6 million copy number markers, of which 750,000 are
'genotype-able' single nucleotide polymorphisms (SNPs) and 1.9
million are non-polymorphic probes. Chromosome Analysis Suite
(ChAS) software v3.0 (Affymetrix) was used for data analysis; for
gains and losses we considered a minimum length of 100,000 base
pairs (bp), and for LOH a minimum length of 3 Mbp. We compared all
resulting alterations in the three BrC primary samples against
female and male non-cancer control array data. Data from 1038
phenotypically healthy individuals (Affymetrix) and from Database
of Genomic Variants were used as reference. Alterations found only
in <1% (rare) or never described (new rare) of the reference
population were considered.
Bioinformatic analysis
The data were first classified as gains, losses and
LOH for each individual sample, and then as shared by more than one
sample. An amplification in the region 22q11.2 was found common to
all the primary cultures, harboring the gene for miR-650. We
searched for interaction networks between miR-650 and cancer, and
more specifically with BrC, using the Cytoscape®
Analysis platform (http://www.cytoscape.org). The target genes of miR-650
were confirmed with three databases: Target Scan®
(http://www.targetscan.org/), miRDB
(http://mirdb.org/miRDB/) and microRNA.org (http://www.microrna.org/). We searched for the
proteins encoded by the target genes identified. The online
platform Pathway Linker® (http://pathwaylinker.org) was used to determine
protein-protein interaction networks; this platform uses three
different databases: KEGG (Kyoto Encyclopedia of Genes and
Genomes), Reactome and SignaLink database.
Immunofluorescence assay
Cells (3×104) of each of the primary
isolates were seeded on coverslips for 24 h, fixed with
paraformaldehyde 4% for 10 min, and permeabilized with 0.2% Triton
X-100 in PBS for 20 min (both from Sigma-Aldrich Co., St. Louis,
MO, USA, ref. P6148-500G and T8787-100 ML, respectively). Cells
were blocked with blocking buffer [10% goat serum (Sigma, ref.
G9023-10ML), 0.2% Triton X-100 and 1% BSA (bovine serum albumin,
Sigma, ref. A1933), in PBS 1X] for 1 h, after which, cells were
stained with mouse monoclonal anti-E-cadherin antibody (1:100,
610181, BD Biosciences, San Jose, CA, USA) and with rabbit
monoclonal anti-vimentin antibody-Alexa Fluor-594 (1:2000,
ab154207, Abcam, Cambridge, UK) overnight at 4°C. Cells were then
incubated with a goat anti-mouse-IgG-FITC antibody for 30 min
(1:50, F0257, Sigma-Aldrich Co.). Finally, nuclei were stained with
DAPI (H1200, Vector Laboratories, Youngstown, OH, USA) for 25 min.
Cells were observed using a fluorescence microscope Olympus BX51
and images were acquired with a digital camera (Camedia C4040,
Olympus, Segrate, Milan, Italy).
Invasion assay
Cells (2×104) were resuspended in 200
µl DMEM/F12 medium and placed in the upper chamber of a
Transwell [6.5-mm diameter, 8-µm pore size (3422, Corning
Inc., Corning, NY, USA)] covered with Matrigel (356237, Corning
Inc.). Then, the Transwell was placed in a 24-well culture dish
containing 1 ml of DMEM/F12 medium supplemented with 10% fetal
bovine serum (FBS) (16000-044, Gibco-Invitrogen). After 24 h of
incubation at 37°C, invasive cells were fixed with 4%
paraformaldehyde and stained with Crystal-violet (Hycel Mexico,
S.A. de C.V., ref. 541) and were observed using a microscope Motic
AE31, images were acquired with a digital camera (Moticam 5.0 MP)
both from Motic (China).
Flow cytometry
Cells (3×105) were blocked with PBS 1×
supplemented with 50% FBS and incubated for 30 min with mouse anti
human CD44-Phycoerythrin (555479) and CD24-PE-Cy7 (561646) both
from BD Biosciences. Cells were then incubated with 7AAD (559925,
BD Biosciences). All acquisitions were performed in a FACS Canto II
flow cytometer (Becton Dickinson, Frankin Lakes, NJ, USA). Finally,
the analysis of flow cytometry data was performed on 7-AAD negative
cells using FlowJo V10 software (Tree Star Inc., Ashland, OR,
USA).
Results
Confirmation of the isolation of
epithelial cells of breast tumors
We obtained three primary cultures of Mexican BrC
patients and identified them as UIVC-IDC-6, -9 and -10. Cultures
were chosen in early passages (<6) to reduce the risk of genetic
changes induced during culture. All primary cultures were positive
for epithelial markers PAN-CK, EpCAM and MUC-1 (data not shown).
Expression of these markers is known to be maintained even after
EMT.
Gain of miR-650 in the three BrC primary
cultures
Using the microarray CytoScan HD and the software
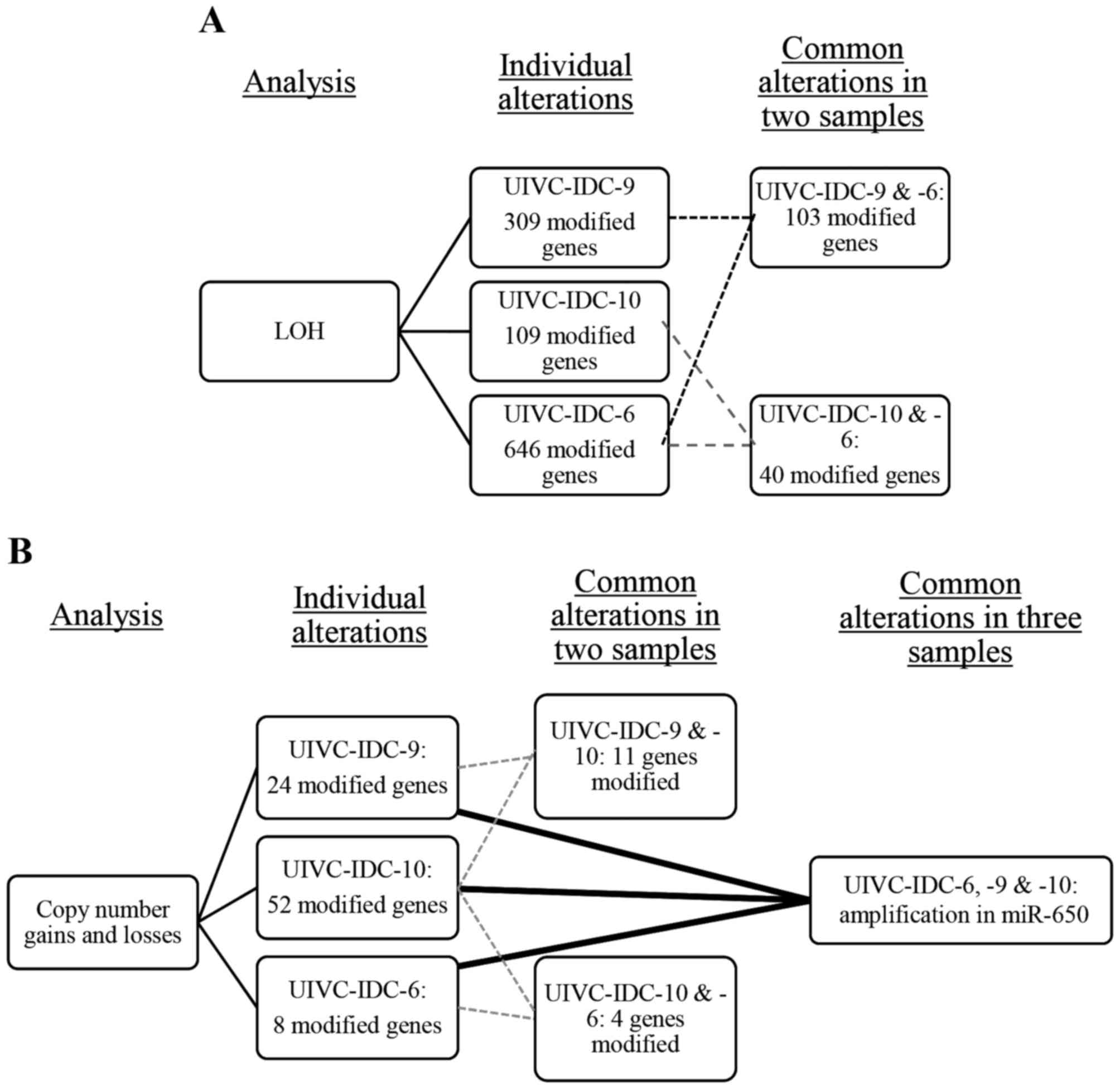
ChAS 3.0, we detected many chromosomal alterations (gains, losses
and LOH) per sample and in common in at least two samples. There
were 103 LOH in common between the sample UIVC-IDC-9 and
UIVC-IDC-6, and 40 between the samples UIVC-IDC-10 and UIVC-IDC-6
(Fig. 1A). There were no LOH
alterations in common in the three BrC samples. For gains and
losses, UIVC-IDC-9 and UIVC-IDC-10 shared 11 alterations, and
samples UIVC-IDC-10 and UIVC-IDC-6 presented 4 common alterations.
We found an amplification on the region 22q11.2 common to all
samples (Fig. 1B), harboring the
gene encoding miR-650 present in all the amplifications.
ING4 and NDRG2, target genes of
miR-650
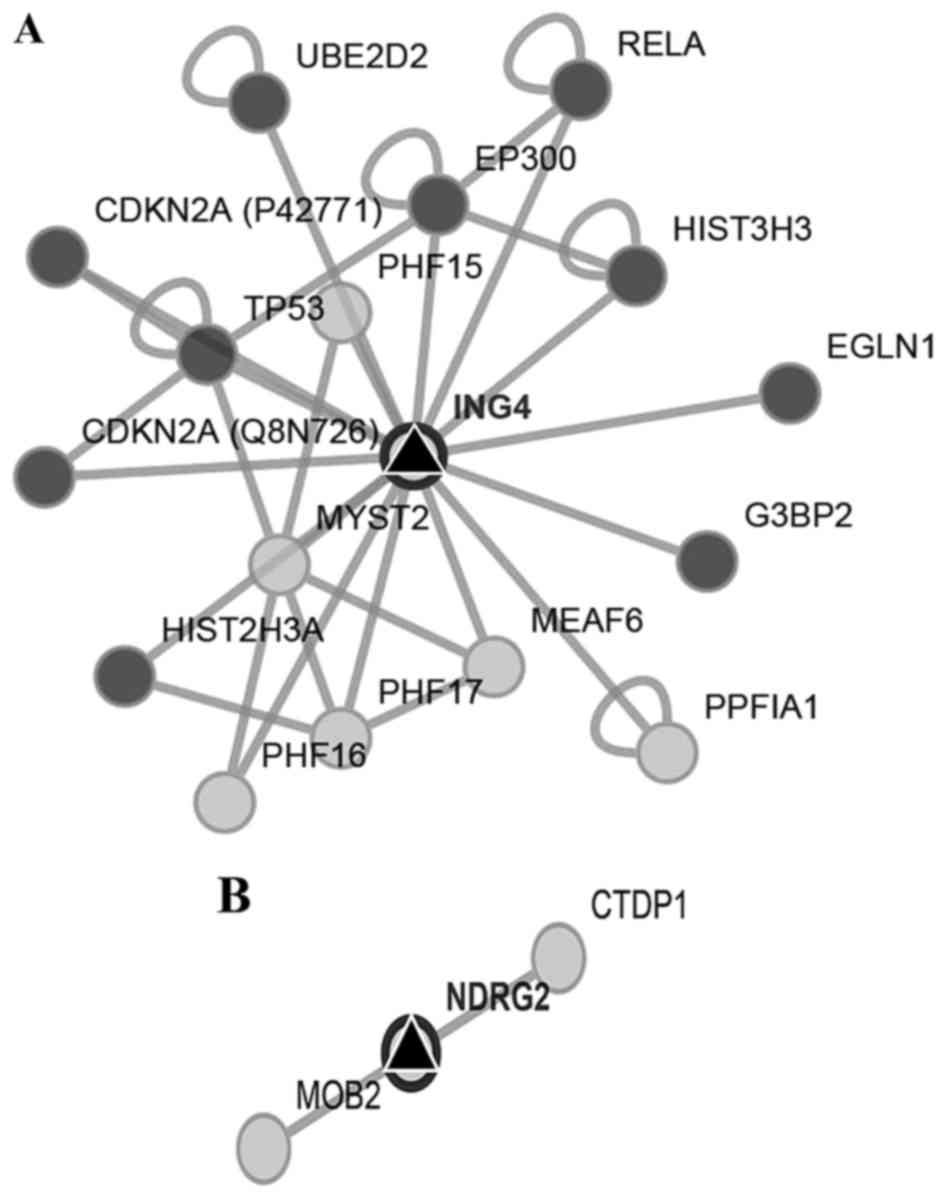
We first searched for the interaction networks of
miR-650 using the platform CytoScape, no relation with other miRNAs
was found. We found that ING4 and NDRG2 genes were
two potential miR-650 targets by reviewing preview reports and
using the platforms Target Scan, miRDB, and MicroRNA.org (Fig. 2A and
B). Then we used Pathway Linker platform to find interaction
networks of these proteins observing an association with cell cycle
and motility regulators (Fig. 3A and
B). However, only for ING4 the platform found significant
associations with signaling pathways and cellular processes
(Table I).
 | Table IING4 significant associations. |
Table I
ING4 significant associations.
| Cellular process or
signaling pathway | P-value |
|---|
| Cancer | 2.5e-10 |
| Pancreatic
cancer | 4.2e-09 |
| Chronic myeloid
leukemia | 4.4e-09 |
| Cell cycle | 2.5e-08 |
| Bladder cancer | 1.7e-07 |
| Non-small cell
carcinoma | 3.9e-07 |
| P53 | 6.2e-07 |
| Melanoma | 6.6e-07 |
| Glioma | 7.0e-07 |
| Prostate
cancer | 1.6e-06 |
| Renal cancer | 0.00015 |
| Systemic lupus
erythematosus | 0.00017 |
| Small cell lung
cancer | 0.00024 |
| Apoptosis | 0.00027 |
| Neurotrophin
signaling pathway | 0.00052 |
| Huntington | 0.00066 |
| Wnt signaling | 0.00077 |
| MAPK signaling | 0.0022 |
Characterization of the EMT profile in
BrC primary cultures
Several studies have shown that ING4 and NDRG2
suppress EMT (29–33). To determinate if the gain of the
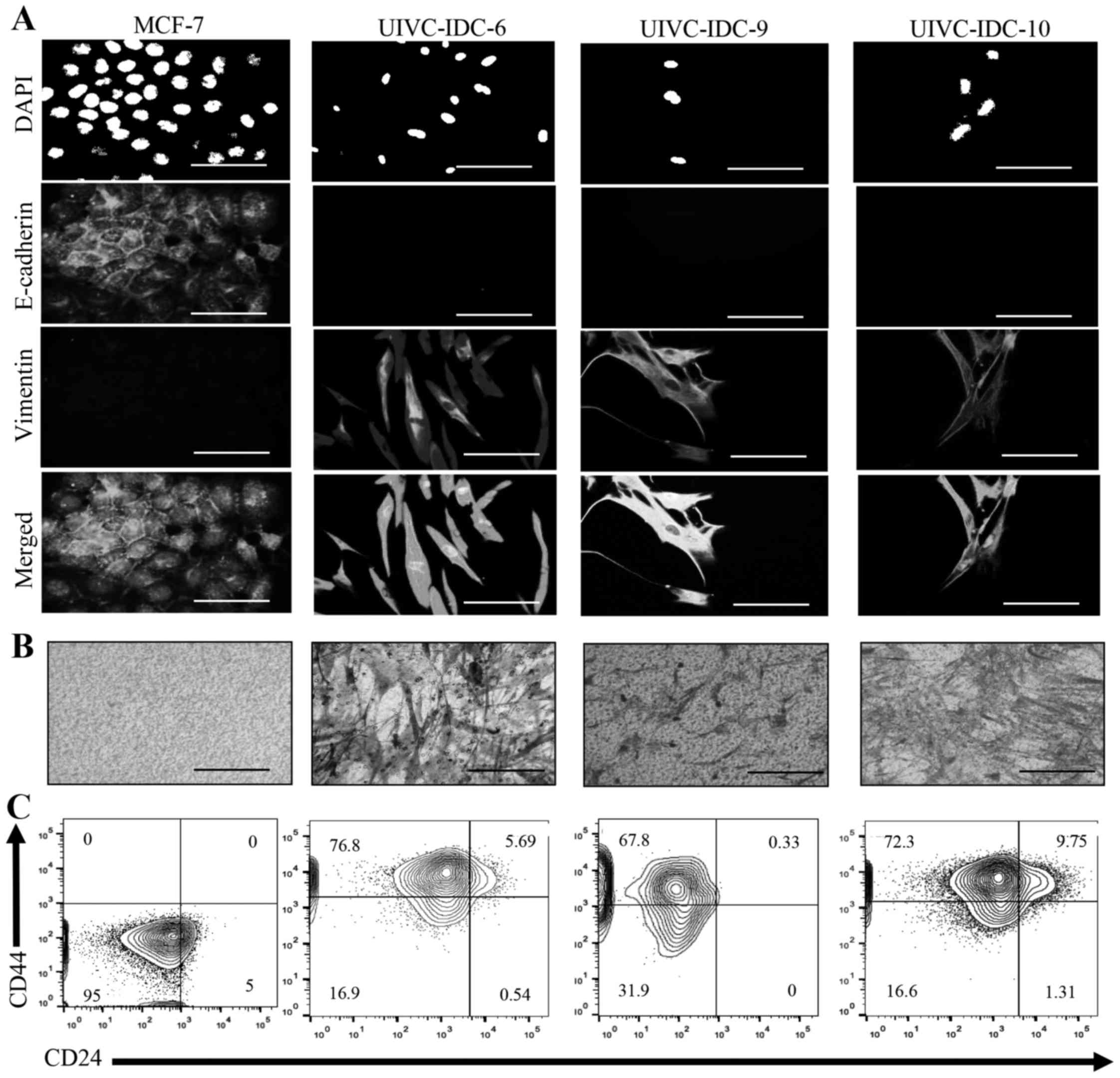
region 22q11.2 was associated with EMT, we analyzed cell morphology
and the presence of EMT markers E-cadherin and vimentin by
immunofluorescence. Noteworthy, all three primary cultures
displayed a very homogeneous profile, the cells shared mesenchymal
characteristics such as spindle-like morphology, negative
expression of E-cadherin and high expression of vimentin (Fig. 4A). EMT identifies aggressive tumors
because of the capacity of invasion of tumor cells. Transwell
invasion assays confirmed that all samples were highly invasive
(Fig. 4B). Moreover, the three
primary isolates contained a high proportion of a population with a
BrC stem cell-like phenotype CD44high
CD24low, UIVC-IDC-6 = 76.8%, UIVC-IDC-9 = 67.8% and
UIVC-IDC-10 = 72.3% (Fig. 4C). The
non-invasive BrC cell line MCF-7 was used as a negative control,
and the highly aggressive BrC cell line MDA-MB-231 was used as
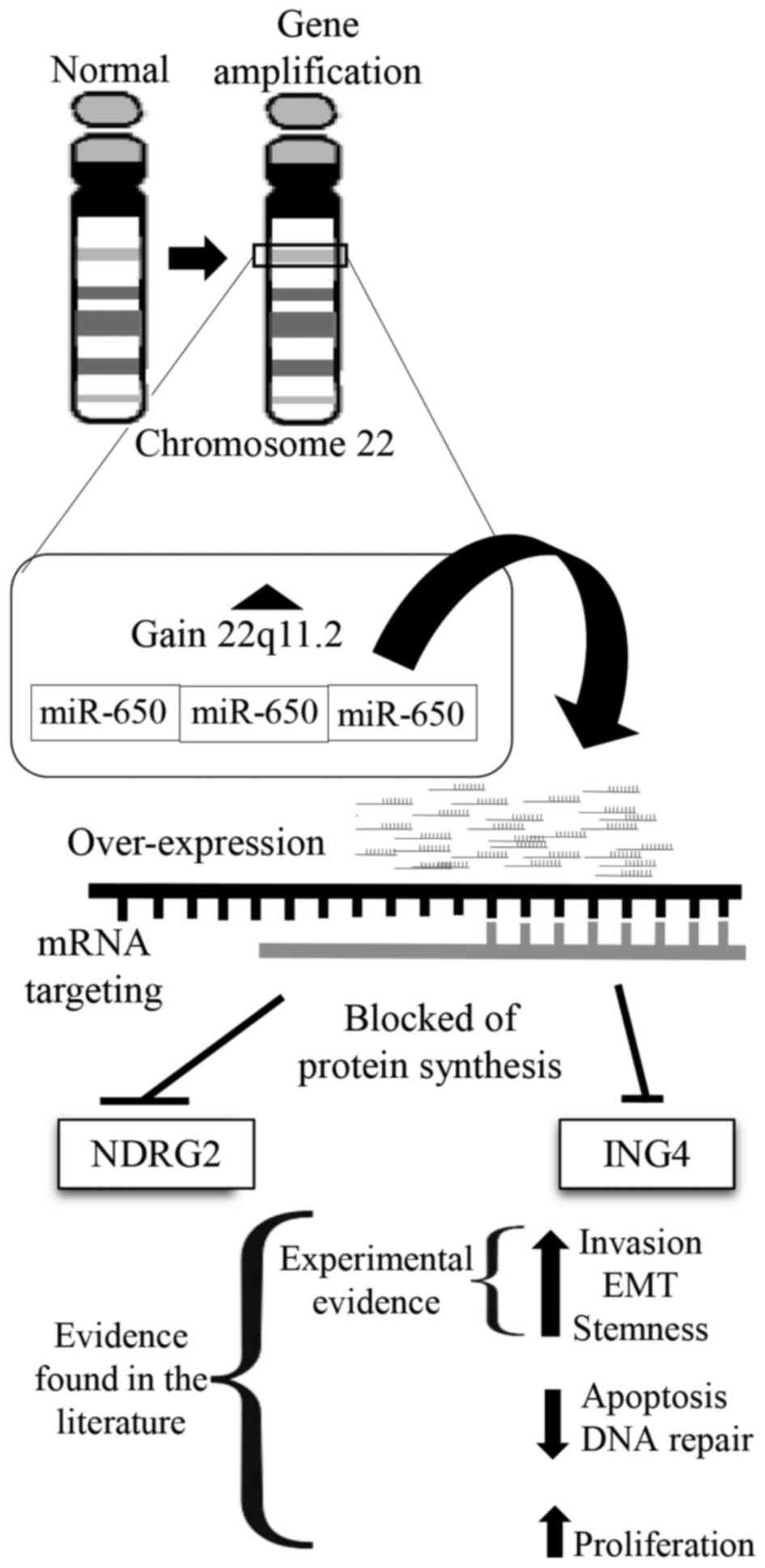
positive control (data not shown). These results are in agreement
with a model in which miR-650 amplification results in
downregulation of ING4 and NDGR4 tumor suppressors and EMT
(Fig. 5).
Discussion
We performed a cytogenetic analysis of three BrC
primary cell cultures, using the CytoScan HD platform and ChAS 3.0
software of Affymetrix in order to search for genetic abnormalities
associated with breast cancer (BrC). We found a large number of
gene copy number alterations, and LOH in each individual sample,
but interestingly, we also observed a previously undescribed
alteration in BrC studies that was common in the three primary
isolates, pointing out a possible new highly represented feature in
Mexican BrC patients.
Although, in vitro and in vivo
evidence supports the association of miR-650 with cancer, to the
best of our knowledge miR-650 has not been previously documented to
be involved in BrC. Furthermore, increased expression of miR-650
often denotes cancers with aggressive features, which is also in
support of our observations. Levels of miR-650 expression in glioma
and hepatocellular carcinoma correlated with the grade of the tumor
(23,34). In gastric cancer the expression
level of miR-650 was significantly associated with metastasis
(25). Overexpression of miR-650
in gastric cancer cell lines increased the size and number of
tumors in xenografted nude mice, and inhibition of miR-650
neutralized this effect (25).
Other human cancers in which miR-650 has been implicated are
prostate cancer, chronic lymphocytic leukemia, osteosarcoma and
lung adenocarcinoma (21,35–37).
miR-650 has been associated with downregulation of tumor
suppressors ING4 and NDRG2 (22,23,25,26).
Because of these associations, it has been proposed that miR-650 is
an oncomiR that regulates, apoptosis, cell cycle, DNA repair, EMT
and metastasis (22,23,34,35).
Thus, miR-650 has also been proposed as a potential target for
cancer treatment.
EMT is a normal de-differentiation process during
embryogenesis. In the inner mass of the blastocyst, the pluripotent
embryonic stem cells have epithelial characteristics and during
gastrulation, the pluripotent epithelial epiblast ingress to form
the primary mesoderm trough EMT (38,39).
In adults EMT has been associated with wound healing, tissue
fibrosis and tissue regeneration, also named as Type 2 EMT
(40). It is believed that in
cancer, cellular changes associated with EMT facilitate cell
invasion and metastasis (41).
Tumor suppressors are important regulators of EMT, and there is
evidence that ING4 and NDRG2 regulate EMT. Wang and collaborators
found that ING4 inhibits migration of a thyroid cancer cell line by
reversing the Wnt/β-catenin pathway induced-EMT (30). Downregulation of ING4 has also been
associated with progression of lung cancer and head and neck
carcinomas (42,43), brain tumors and in some cases, ING4
expression also correlated with tumor grade (44). Using comparative genomic
hybridization (CGH), Kim and colleagues found a deletion of the
ING4 loci in 10–20% of primary breast tumors and BrC cell lines
(26). In a more mechanistic
study, Bayron and collaborators found that ING4 inhibited NF-κB
activity in the BrC cell line T47D (45). On the contrary, upregulation of
ING4 in osteosarcoma cells induced cell apoptosis and suppressed
cell invasion through downregulation of MMP-2, which also
correlated with inhibition of NF-κB activity (46). Other studies support that ING4
negatively regulates NF-κB, and that ING4 downregulation associates
with tumor progression and poor patient outcome (27,45–48).
There is also evidence that NDRG2 supports
EMT-induced aggressive cancer features of different types of cancer
cells, such as invasion and migration. Hong and collaborators found
that epigenetic silencing of NDRG2 induced proliferation and
invasion of primary colorectal cancer cells, and that this could be
associated with advanced stages of the disease (49). Similarly, Lee and collaborators
found that in patients with gallbladder carcinoma loss of NDRG2
expression was an independent predictor of decreased patient
survival and was significantly associated with a more advanced
tumor stage (32). In addition,
loss of NDRG2 expression in gallbladder carcinoma cells resulted in
enhanced proliferation, migration and invasiveness in vitro,
and enhanced tumor growth and metastasis in mice (32). Using immunohistochemistry of
tissues from patients with colon carcinoma, Kim and collaborators
found that NDRG2 and E-cadherin were highly expressed in normal
mucosa and in areas with well differentiated tumor cells, while
areas of poorly differentiated carcinoma there was low to no
expression of both proteins (50).
In the above study, NDRG2 knockdown induced
downregulation of CDH1 (encodes E-cadherin) promoter
activity and upregulation of Snail-1, a master transcription
factor regulator of EMT. In the aggressive BrC cell line
MDA-MB-231, overexpression of NDRG2 results in downregulation of
Snail-1 and upregulation of CDH1 (33). TGF-β is perhaps the most used
inducer of EMT in in vitro studies, and Shen and
collaborators showed that NDRG2 abrogates the TGF-β-induced EMT
(31).
In another study in gallbladder carcinoma cells,
loss of NDRG2 created an EMT positive feedback loop by increasing
the expression of the tyrosine kinase receptor Axl, that regulated
the expression of Slug, another master regulator of EMT
(32). MDA-MB-231 and MCF-7 are
highly metastatic and no-metastatic BrC cell lines, respectively,
in which the metastatic potential correlates with the levels of
NDRG2 (33). NDRG2 also inhibited
NF-κB signaling in MDA-MB-231 cells, together with the
PMA-induction of the tumor-promoting enzyme cyclooxygenase 2
(COX-2) and one of the COX-2 most relevant pro-tumoral products,
prostaglandin E2 (PGE2). NDRG2 strongly repressed phorbol
12-myristate 13-acetate (PMA)-stimulated migration and invasion of
MDA-MB-231 cells. On the other hand, siRNA-mediated knockdown of
NDRG2 in MCF-7 cells resulted in increased COX-2 and NF-κB activity
after PMA activation, correlating with an increased capacity of
cell migration and invasion (51).
There is also evidence that NDRG2 overexpression reduces
proliferation, migration and invasion of lung, bladder, and
colorectal cancer cells (28,52,53).
In solid tumors, a scarce population with capacity
of self-renew has been described, this population seeds new tumors
upon serial passages in mice and reconstitutes all types of tumor
cells both in vivo and in culture. These cells are thought
to be cancer stem cells (CSC) and are considered the most clinical
relevant population because this may be the population that is
responsible for metastases, resistance to chemotherapy and disease
relapse (54–57). There is experimental evidence of a
direct molecular link between EMT and stemness. In 2003, Mani et
al demonstrated that the induction of EMT resulted in
acquisition of both mesenchymal and stem characteristics, for
instance a fibroblastoid morphology together with a BrC stem cell
(BrCSC) phenotype CD44high CD24low (9). Since EMT is a prerequisite for the
invasion-metastasis cascade (58),
evidence shows that metastatic CD44high
CD24low BrCSCs strongly express the TGF-β-induced
signature associated with EMT (59). Of clinical significance,
Santisteban et al demonstrated that induction of EMT
increased the density of BrCSCs, which correlated with high
tumorigenicity in mice, and resistance to pharmacological and
radiation treatment (60). Thus,
there is an important association between EMT, CSCs, relapse and
treatment failure (61–63). Although, there is no direct
evidence linking expression of miR-650, ING4 and NDRG2 with CSCs,
various evidence supports that high miR-650 and/or low ING4 and
NDRG2 correlates with advanced cancers and poor patient outcome
(22,25,33,43,48,53,64).
Furthermore, NDRG2 has been found to downregulate CD24 expression
in hepatocellular carcinoma cell lines and tissues and miR-650 has
been found in microvesicles released by CD105 positive renal
carcinoma stem cells (65,66).
In this study, we found a common amplification in
the 22q11.2 band in primary isolates of three Mexican patients with
BrC. In all the cases this amplification included the region
encoding miR-650, an oncomiRNA previously associated with invasive
cancers, although never before described in BrC. Two of the most
studied targets of miR-650 are tumor suppressors NDGR2 and ING4,
which have been documented to negatively regulate EMT. Our data
support a model in which miR-650 amplification results in NDGR2 and
ING4 downregulation that then triggers EMT, acquisition of cancer
stem cell markers and invasion. A limitation of this study is the
small number of samples analyzed to have a better idea of how
common this genetic lesion is in Mexican patients with BrC. Of
note, our data also suggest that these patients harbor aggressive
tumors. However, these tumors were classified with invasive ductal
carcinoma, histological grade 2 and clinical stage II, considered
of intermediate prognosis. In future studies, it is critical to
follow up BrC patients with the 22q11.2 amplification.
Acknowledgments
This work was supported by CONACyT FONSEC
SSA/IMSS/ISSSTE project no. 233061 to Ezequiel M. Fuentes-Pananá
and by Fondo de Apoyo a la Investigación, Hospital Infantil de
México Federico Gómez (project number HIM-2014-053). N.A.E.-S. is a
doctoral student from Programa de Doctorado en Ciencias Biomédicas,
Universidad Nacional Autónoma de México (UNAM) and received
fellowship 231663 from CONACYT. N.A.E.-S., also acknowledged the
financial support provided by the Mexican Institute of Social
Security (IMSS). M.C.S.-A. acknowledged the scholarship and
financial support provided by Consejo Nacional de Ciencia y
Tecnología (CONACyT #576518). The authors are grateful to A.C.
Velázquez-Wong and C. Jiménez Ramírez for technical assistance in
arrays processing.
References
|
1
|
International Agency for Research on
Cancer (IARC): Latest world cancer statistics, Global cancer burden
rises to 14.1 million new cases in 2012: Marked increase in breast
cancers must be addressed. Press Release 223. 2013, https://www.iarc.fr/en/media-centre/pr/2013/pdfs/pr223_E.pdf.
Accessed December 12, 2013.
|
|
2
|
International Agency for Research on
Cancer (IARC): GLOBOCAN: Estimated Cancer Incidence, Mortality and
Prevalence Worldwide in 2012. 2012, http://globocan.iarc.fr/Default.aspx.
|
|
3
|
Greaves M and Maley CC: Clonal evolution
in cancer. Nature. 481:306–313. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Suzuki HI, Katsura A, Matsuyama H and
Miyazono K: MicroRNA regulons in tumor microenvironment. Oncogene.
34:3085–3094. 2015. View Article : Google Scholar
|
|
5
|
Scheel C, Onder T, Karnoub A, Weinberg RA
and Talmadge JE: Adaptation versus selection: The origins of
metastatic behavior. Cancer Res. 67:11476–11479; discussion
11479–11480. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Thiery JP and Sleeman JP: Complex networks
orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell
Biol. 7:131–142. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kang Y and Massagué J:
Epithelial-mesenchymal transitions: Twist in development and
metastasis. Cell. 118:277–279. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Voulgari A and Pintzas A:
Epithelial-mesenchymal transition in cancer metastasis: Mechanisms,
markers and strategies to overcome drug resistance in the clinic.
Biochim Biophys Acta. 1796:75–90. 2009.PubMed/NCBI
|
|
9
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Morel AP, Lièvre M, Thomas C, Hinkal G,
Ansieau S and Puisieux A: Generation of breast cancer stem cells
through epithelial-mesenchymal transition. PLoS One. 3:e28882008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Scheel C, Eaton EN, Li SH, Chaffer CL,
Reinhardt F, Kah KJ, Bell G, Guo W, Rubin J, Richardson AL, et al:
Paracrine and autocrine signals induce and maintain mesenchymal and
stem cell states in the breast. Cell. 145:926–940. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Beck B, Lapouge G, Rorive S, Drogat B,
Desaedelaere K, Delafaille S, Dubois C, Salmon I, Willekens K,
Marine JC, et al: Different levels of Twist1 regulate skin tumor
initiation, stemness, and progression. Cell Stem Cell. 16:67–79.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Luo M, Brooks M and Wicha MS:
Epithelial-mesenchymal plasticity of breast cancer stem cells:
Implications for metastasis and therapeutic resistance. Curr Pharm
Des. 21:1301–1310. 2015. View Article : Google Scholar :
|
|
14
|
Medema JP: Cancer stem cells: The
challenges ahead. Nat Cell Biol. 15:338–344. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar
|
|
16
|
Mongroo PS and Rustgi AK: The role of the
miR-200 family in epithelial-mesenchymal transition. Cancer Biol
Ther. 10:219–222. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Park SM, Gaur AB, Lengyel E and Peter ME:
The miR-200 family determines the epithelial phenotype of cancer
cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes
Dev. 22:894–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Comijn J, Berx G, Vermassen P, Verschueren
K, van Grunsven L, Bruyneel E, Mareel M, Huylebroeck D and van Roy
F: The two-handed E box binding zinc finger protein SIP1
downregulates E-cadherin and induces invasion. Mol Cell.
7:1267–1278. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Eger A, Aigner K, Sonderegger S, Dampier
B, Oehler S, Schreiber M, Berx G, Cano A, Beug H and Foisner R:
DeltaEF1 is a transcriptional repressor of E-cadherin and regulates
epithelial plasticity in breast cancer cells. Oncogene.
24:2375–2385. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Howe EN, Cochrane DR and Richer JK: The
miR-200 and miR-221/222 microRNA families: Opposing effects on
epithelial identity. J Mammary Gland Biol Neoplasia. 17:65–77.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zuo ZH, Yu YP, Ding Y, Liu S, Martin A,
Tseng G and Luo JH: Oncogenic activity of miR-650 in prostate
cancer is mediated by suppression of CSR1 expression. Am J Pathol.
185:1991–1999. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Feng L, Xie Y, Zhang H and Wu Y:
Down-regulation of NDRG2 gene expression in human colorectal cancer
involves promoter methylation and microRNA-650. Biochem Biophys Res
Commun. 406:534–538. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zeng ZL, Li FJ, Gao F, Sun DS and Yao L:
Upregulation of miR-650 is correlated with down-regulation of ING4
and progression of hepatocellular carcinoma. J Surg Oncol.
107:105–110. 2013. View Article : Google Scholar
|
|
24
|
Chan E, Patel R, Nallur S, Ratner E,
Bacchiocchi A, Hoyt K, Szpakowski S, Godshalk S, Ariyan S, Sznol M,
et al: MicroRNA signatures differentiate melanoma subtypes. Cell
Cycle. 10:1845–1852. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang X, Zhu W, Zhang J, Huo S, Zhou L, Gu
Z and Zhang M: MicroRNA-650 targets ING4 to promote gastric cancer
tumorigenicity. Biochem Biophys Res Commun. 395:275–280. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kim S, Chin K, Gray JW and Bishop JM: A
screen for genes that suppress loss of contact inhibition:
Identification of ING4 as a candidate tumor suppressor gene in
human cancer. Proc Natl Acad Sci USA. 101:16251–16256. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fang F, Luo LB, Tao YM, Wu F and Yang LY:
Decreased expression of inhibitor of growth 4 correlated with poor
prognosis of hepatocellular carcinoma. Cancer Epidemiol Biomarkers
Prev. 18:409–416. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li R, Yu C, Jiang F, Gao L, Li J, Wang Y,
Beckwith N, Yao L, Zhang J and Wu G: Overexpression of N-Myc
downstream-regulated gene 2 (NDRG2) regulates the proliferation and
invasion of bladder cancer cells in vitro and in vivo. PLoS One.
8:e766892013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Qu H, Yin H, Yan S, Tao M, Xie Y and Chen
W: Inhibitor of growth 4 suppresses colorectal cancer growth and
invasion by inducing G1 arrest, inhibiting tumor angiogenesis and
reversing epithelial-mesenchymal transition. Oncol Rep.
35:2927–2935. 2016.PubMed/NCBI
|
|
30
|
Wang CJ, Yang D and Luo YW: Recombinant
ING4 suppresses the migration of SW579 thyroid cancer cells via
epithelial to mesenchymal transition. Exp Ther Med. 10:603–607.
2015.PubMed/NCBI
|
|
31
|
Shen L, Qu X, Ma Y, Zheng J, Chu D, Liu B,
Li X, Wang M, Xu C, Liu N, et al: Tumor suppressor NDRG2 tips the
balance of oncogenic TGF-β via EMT inhibition in colorectal cancer.
Oncogenesis. 3:e862014. View Article : Google Scholar
|
|
32
|
Lee DG, Lee SH, Kim JS, Park J, Cho YL,
Kim KS, Jo DY, Song IC, Kim N, Yun HJ, et al: Loss of NDRG2
promotes epithelial-mesenchymal transition of gallbladder carcinoma
cells through MMP-19-mediated Slug expression. J Hepatol.
63:1429–1439. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim MJ, Lim J, Yang Y, Lee MS and Lim JS:
N-myc downstream-regulated gene 2 (NDRG2) suppresses the
epithelial-mesenchymal transition (EMT) in breast cancer cells via
STAT3/Snail signaling. Cancer Lett. 354:33–42. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sun B, Pu B, Chu D, Chu X, Li W and Wei D:
MicroRNA-650 expression in glioma is associated with prognosis of
patients. J Neurooncol. 115:375–380. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mraz M, Dolezalova D, Plevova K, Stano
Kozubik K, Mayerova V, Cerna K, Musilova K, Tichy B, Pavlova S,
Borsky M, et al: MicroRNA-650 expression is influenced by
immunoglobulin gene rearrangement and affects the biology of
chronic lymphocytic leukemia. Blood. 119:2110–2113. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yun JH, Moon S, Lee HS, Hwang MY, Kim YJ,
Yu HY, Kim Y, Han BG, Kim BJ and Kim JM: MicroRNA-650 in a copy
number-variable region regulates the production of interleukin 6 in
human osteosarcoma cells. Oncol Lett. 10:2603–2609. 2015.PubMed/NCBI
|
|
37
|
Huang JY, Cui SY, Chen YT, Song HZ, Huang
GC, Feng B, Sun M, De W, Wang R and Chen LB: MicroRNA-650 was a
prognostic factor in human lung adenocarcinoma and confers the
docetaxel chemoresistance of lung adenocarcinoma cells via
regulating Bcl-2/Bax expression. PLoS One. 8:e726152013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Acloque H, Adams MS, Fishwick K,
Bronner-Fraser M and Nieto MA: Epithelial-mesenchymal transitions:
The importance of changing cell state in development and disease. J
Clin Invest. 119:1438–1449. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Eastham AM, Spencer H, Soncin F, Ritson S,
Merry CL, Stern PL and Ward CM: Epithelial-mesenchymal transition
events during human embryonic stem cell differentiation. Cancer
Res. 67:11254–11262. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Steinestel K, Eder S, Schrader AJ and
Steinestel J: Clinical significance of epithelial-mesenchymal
transition. Clin Transl Med. 3:172014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gunduz M, Nagatsuka H, Demircan K, Gunduz
E, Cengiz B, Ouchida M, Tsujigiwa H, Yamachika E, Fukushima K,
Beder L, et al: Frequent deletion and down-regulation of ING4, a
candidate tumor suppressor gene at 12p13, in head and neck squamous
cell carcinomas. Gene. 356:109–117. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang QS, Li M, Zhang LY, Jin Y, Tong DD,
Yu Y, Bai J, Huang Q, Liu FL, Liu A, et al: Down-regulation of ING4
is associated with initiation and progression of lung cancer.
Histopathology. 57:271–281. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Garkavtsev I, Kozin SV, Chernova O, Xu L,
Winkler F, Brown E, Barnett GH and Jain RK: The candidate tumour
suppressor protein ING4 regulates brain tumour growth and
angiogenesis. Nature. 428:328–332. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Byron SA, Min E, Thal TS, Hostetter G,
Watanabe AT, Azorsa DO, Little TH, Tapia C and Kim S: Negative
regulation of NF-κB by the ING4 tumor suppressor in breast cancer.
PLoS One. 7:e468232012. View Article : Google Scholar
|
|
46
|
Li M, Zhu Y, Zhang H, Li L, He P, Xia H,
Zhang Y and Mao C: Delivery of inhibitor of growth 4 (ING4) gene
significantly inhibits proliferation and invasion and promotes
apoptosis of human osteosarcoma cells. Sci Rep. 4:73802014.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Shen JC, Unoki M, Ythier D, Duperray A,
Varticovski L, Kumamoto K, Pedeux R and Harris CC: Inhibitor of
growth 4 suppresses cell spreading and cell migration by
interacting with a novel binding partner, liprin alpha1. Cancer
Res. 67:2552–2558. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li J and Li G: Cell cycle regulator ING4
is a suppressor of melanoma angiogenesis that is regulated by the
metastasis suppressor BRMS1. Cancer Res. 70:10445–10453. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hong SN, Kim SJ, Kim ER, Chang DK and Kim
YH: Epigenetic silencing of NDRG2 promotes colorectal cancer
proliferation and invasion. J Gastroenterol Hepatol. 31:164–171.
2016. View Article : Google Scholar
|
|
50
|
Kim YJ, Kang HB, Yim HS, Kim JH and Kim
JW: NDRG2 positively regulates E-cadherin expression and prolongs
overall survival in colon cancer patients. Oncol Rep. 30:1890–1898.
2013.PubMed/NCBI
|
|
51
|
Kim MJ, Kim HS, Lee SH, Yang Y, Lee MS and
Lim JS: NDRG2 controls COX-2/PGE(2)-mediated breast cancer cell
migration and invasion. Mol Cells. 37:759–765. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Faraji SN, Mojtahedi Z, Ghalamfarsa G and
Takhshid MA: N-myc downstream regulated gene 2 overexpression
reduces matrix metalloproteinase-2 and -9 activities and cell
invasion of A549 lung cancer cell line in vitro. Iran J Basic Med
Sci. 18:773–779. 2015.PubMed/NCBI
|
|
53
|
Golestan A, Mojtahedi Z, Ghalamfarsa G,
Hamidinia M and Takhshid MAP: The effects of NDRG2 overexpression
on cell proliferation and invasiveness of SW48 colorectal cancer
cell line. Iran J Med Sci. 40:430–439. 2015.
|
|
54
|
Kreso A and Dick JE: Evolution of the
cancer stem cell model. Cell Stem Cell. 14:275–291. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
O'Brien CA, Pollett A, Gallinger S and
Dick JE: A human colon cancer cell capable of initiating tumour
growth in immunodeficient mice. Nature. 445:106–110. 2007.
View Article : Google Scholar
|
|
57
|
Singh SK, Hawkins C, Clarke ID, Squire JA,
Bayani J, Hide T, Henkelman RM, Cusimano MD and Dirks PB:
Identification of human brain tumour initiating cells. Nature.
432:396–401. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Nieto MA: Epithelial-mesenchymal
transitions in development and disease: Old views and new
perspectives. Int J Dev Biol. 53:1541–1547. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Shipitsin M, Campbell LL, Argani P,
Weremowicz S, Bloushtain-Qimron N, Yao J, Nikolskaya T,
Serebryiskaya T, Beroukhim R, Hu M, et al: Molecular definition of
breast tumor heterogeneity. Cancer Cell. 11:259–273. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Santisteban M, Reiman JM, Asiedu MK,
Behrens MD, Nassar A, Kalli KR, Haluska P, Ingle JN, Hartmann LC,
Manjili MH, et al: Immune-induced epithelial to mesenchymal
transition in vivo generates breast cancer stem cells. Cancer Res.
69:2887–2895. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Dave B, Mittal V, Tan NM and Chang JC:
Epithelial-mesenchymal transition, cancer stem cells and treatment
resistance. Breast Cancer Res. 14:2022012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Davis FM, Stewart TA, Thompson EW and
Monteith GR: Targeting EMT in cancer: Opportunities for
pharmacological intervention. Trends Pharmacol Sci. 35:479–488.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Krause M, Dubrovska A, Linge A and Baumann
M: Cancer stem cells: Radioresistance, prediction of radiotherapy
outcome and specific targets for combined treatments. Adv Drug
Deliv Rev. Feb 12–2016.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Kloten V, Schlensog M, Eschenbruch J,
Gasthaus J, Tiedemann J, Mijnes J, Heide T, Braunschweig T, Knüchel
R and Dahl E: Abundant NDRG2 expression is associated with
aggressiveness and unfavorable patients' outcome in basal-like
breast cancer. PLoS One. 11:e01590732016. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zheng J, Li Y, Yang J, Liu Q, Shi M, Zhang
R, Shi H, Ren Q, Ma J, Guo H, et al: NDRG2 inhibits hepatocellular
carcinoma adhesion, migration and invasion by regulating CD24
expression. BMC Cancer. 11:251–259. 1–9. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Grange C, Tapparo M, Collino F, Vitillo L,
Damasco C, Deregibus MC, Tetta C, Bussolati B and Camussi G:
Microvesicles released from human renal cancer stem cells stimulate
angiogenesis and formation of lung premetastatic niche. Cancer Res.
71:5346–5356. 2011. View Article : Google Scholar : PubMed/NCBI
|