Introduction
Lung cancer is one of the increasingly potential
causes of cancer deaths in both males and females worldwide
(1). It was estimated that 595,690
Americans will die from cancer in 2016, with >27% being due to
lung cancer (2). Cancer statistics
in China indicated that an estimated 4,292,000 new cancer cases and
2,814,000 cancer deaths would occur in 2015, with lung cancer being
the most common incident cancer and the leading cause of cancer
death (3). The main lung cancer
types include small cell lung cancer and non-small cell lung cancer
(NSCLC), which together account for ~87% of all lung cancer cases
(4). Most patients with NSCLC are
observed to be in advanced stages of cancer at diagnosis, however
even when diagnosed at an early stage and treated surgically,
cancer recurrence has been observed in patients, with spread to the
lymph node, liver and other tissues.
Metastasis is the major cause of death in patients
with NSCLC. The process in which immobile epithelial cells
transform to invasive cell types in what has been termed as
epithelial-mesenchymal transition (EMT), is increasingly being
accepted as having an instrumental role in tumor metastasis
(5). EMT is characterized by the
loss of cell-cell adhesions, and the transition to spindle-like
cells that are capable of invading the extracellular matrix
(6). An increasing number of
studies have demonstrated that cancer stem cell (CSC) molecule
CD133 promotes tumor invasion and metastasis by inducing EMT
processes (7–15).
Human CD133 is a five transmembrane single-chain
glycoprotein that belongs to the prominin family containing two
large extracellular and two small intracellular loops (16). Lee et al examined the
effects of IL-6 on growth, EMT processes and metastatic ability of
CD133+ and CD133− cell subpopulations
isolated from NSCLC cells. Of which they demonstrated the dual
roles of IL-6 in regulating growth of CD133− and
CD133+ subpopulations of lung cancer cells, as well as
the positive regulation of EMT/metastasis increase by IL-6 in
CD133+ cells, but not in CD133− cells
(14). Some researchers have
suggested that CSCs are present in both blood and tumor tissue of
NSCLC patients, and that EMT-inducing transcriptional factors Bmi1
may play an important role in initiation and maintenance of CSCs
and might be involved in vascular dissemination of NSCLC (12). Others have shown that elevated
CD133 expression is the signature marker of EMT and CSC association
in lung adenocarcinoma (17).
Additionally, Bertolini et al showed that
CD133+/CXCR4+/EpCAM− lung
cancer-initiating cells sustain metastasis and correlate with poor
prognosis (18).
CXCR4, a seven-transmembrane G protein-coupled
receptor, is a physiological receptor for stromal-derived-factor-1
(19) which is highly expressed in
various types of human tumors (20–24).
A number of studies have demonstrated the vital role of CXCR4 in
cancer cell survival, proliferation, invasion and metastasis, and
that CXCR4 promotes EMT processes in various cancers (25–29),
with CD133+CXCR4+ cells showing a stronger
invasiveness capacity (18,30–33).
Although researchers have suggested a possible role
for CD133 and CXCR4 in NSCLC metastasis, the precise relationship
of CD133, CXCR4 and EMT processes are unclear and requires further
investigation. Therefore, in this study, we detected the expression
of CD133 and CXCR4 in NSCLC patients and investigated the effects
of CD133 in NSCLC tumorigenesis. We further investigated the
CXCR4/CD133 interaction and explored the possible molecular
mechanisms by which CXCR4/CD133 regulates NSCLC metastasis via the
EMT process.
Materials and methods
GEO databases analysis
Gene expression data were obtained from the publicly
available GEO databases. We chose GSE10072, GSE40275 and GSE63459
to identify the expression signature of CD133 because these sets
included normal (n>30) and tumor samples (n>30). Original
values were normalized by application of log2 method and
comparisons were performed by two-tailed Student's t-test for
GSE10072 and GSE40275, and paired t-test for GSE63459. We chose
GSE30219 because it was the largest lung cancer dataset to identify
the correlation between CD133 and CXCR4 by two-tailed bivariate
analysis.
NSCLC tissue samples
A total of 64 specimens was collected from patients
with primary NSCLC surgical resection at Renmin Hospital of Wuhan
University from May 2012 to April 2015. Patients were followed-up
by telephone until recurrence/metastasis or until May 2016. This
study was carried out in accordance with the principles of the
Helsinki declaration and approved by the ethics committee of the
Renmin Hospital of Wuhan University.
NSCLC cell lines
A549 was obtained from American Type Culture
Collection (ATCC) and maintained in Dulbecco's modified Eagle's
medium (DMEM) with high glucose (Gibco, USA) supplemented with 10%
fetal bovine serum (Hyclone, USA), containing 100 U/ml penicillin
and 100 mg/ml streptomycin. A549 cells were cultured in a 5%
CO2 air incubator at 37°C and passaged using 0.25%
trypsin-EDTA (Gibco) when they reached confluence.
Immunohistochemistry
Paraffin-embedded tissue sections were dewaxed and
rehydrated, and antigen retrieval was performed by microwaving in
10 mM sodium citrate buffer, pH 6.0, for 20 min. Sections were then
incubated with 3% hydrogen peroxide for 30 min at room temperature
to block endogenous peroxidase, then blocked in 10% normal goat
serum for 0.5 h. Immunostaining was performed by incubating with
anti-CXCR4 (1:500, ab124824; Abcam Corp., UK), anti-CD133 (1:200,
orb99113; Biorbyt Ltd., UK), anti-E cadherin (1:500, ab40772; Abcam
Corp.) or anti-vimentin (1:500, ab92547; Abcam Corp.) at 4°C
overnight. Slides were then washed in PBS and incubated with
secondary antibody (anti-rabbit detection system; Boster, China)
for 30 min at 37°C. Staining was visualized with 3,
3-diaminobenzidine and counterstained with hematoxylin. The final
score is the average of the percentage of stained area [scored as 0
(without staining), 1 (<25%), 2 (25–50%) and 3 (>50%)] and
intensity [scored as 0 (without staining), 1 (light yellow), 2
(yellow) and 3 (brown yellow)] of stained cells The expression
levels were divided into low expression (mean score <2).
Magnetic cell sorting
A549 cells were washed and suspended in PBS at a
maximal concentration of 1.0×108 cells. Cells were
incubated at 4°C with 100 µl FcR blocking reagent and 100
µl CD133 Micro Beads (130-097-049, Miltenyi Biotec, Germany)
for 30 min. Cells were isolated into CD133+ and
CD133− cells by using MS columns (130-042-201, Miltenyi
Biotec) and LD columns (130-042-901, Miltenyi Biotec) according to
the manufacturer's instructions.
Immunofluorescence assay
Cells were fixed and permeabi-lized with 2%
paraformaldehyde and 0.5% Triton X-100. After overnight incubation
with anti-CD133 (1:50, 18470-1-AP; Proteintech, USA), the specimens
were rinsed thoroughly and treated with anti-goat antibodies
(1:100, BA1032, Boster), respectively. Nuclei were stained using
0.3 µM DAPI (C1002, Beyotime Biotechnology, China). The
digital images were then captured with a cooled CCD camera and
processed with the help of photoshop (Adobe) software.
Clone formation assay
Cells (500 cells/well) were seeded in 6-well plate
and allowed to grow for 2 weeks in 37°C incubator. The cells were
then washed twice with ice-cold PBS, fixed by methanol for 15 min
and stained with Giemsa for 15 min. The images of staining and
clone spheres of cells were obtained by a charge coupled device
camera.
Cell proliferation assay
Cells were seeded in a 96-well plate at the density
of 1.0×104 cells per well for 48 h, 10 µl CCK8
solution was added to each well at indicated times and incubated
for another 2 h. The absorbance of each well was obtained from
PerkinElmer 2030 Victor X multi-label plate reader (Perkin-Elmer,
Waltham, MA, USA) at 450 nm.
SiRNA transfect assay
Cells were plated 24 h before transfection at 30–50%
confluence, cells were transfected using Lipofectamine™ 2000
(Invitrogen, USA) with siRNA duplexes specific for human CD133
(Ribobio, China) or negative control (NC) siRNA. The following were
the sequences of CD133 siRNA used in the study:
sense-CUGGGAAGCUAUUUAAUAA; antisense-UUAUUAAAUAGCUUCCCAG. In
addition, control was included where cells were treated with
Lipofectamine 2000 alone. The siRNA experiment was carried out for
48 h to analyse the RNA expression level by quantitative RT-PCR and
72 h to analyse the protein expression level by western blot
analysis.
Quantitative RT-PCR
Total RNA was extracted by using TRIzol (Invitrogen)
according to the manufacturer's specifications and quantified by
NanoDrop 2000 (Thermo Scientific, USA). Total 1 µg RNA was
reverse-transcribed to cDNA with random primers using the Promega
reverse transcriptase kit (USA) according to manufacturer's
protocol. To assess gene expression, cDNAs were amplified with the
SYBR® Premix Ex Taq™ II (Tli RNaseH Plus) (Takara,
Japan) using the QuantStudio 6 Flex Real-Time PCR system (Life
Technologies (AB and Invitrogen) USA]. The relative levels were
calculated by the comparative Ct method (2−ΔΔCt). The
sequences of specific primers are shown in Table I.
 | Table IThe sequences of specific primers for
PCR or qPCR in this study. |
Table I
The sequences of specific primers for
PCR or qPCR in this study.
| Gene | Primer
sequences |
|---|
| β-actin | F:
5′-AGCGAGCATCCCCCAAAGTT-3′ |
| R:
5′-GGGCACGAAGGCTCATCATT-3′ |
| CD133 | F:
5′-GCACTCTATACCAAAGCGTCAA-3′ |
| R:
5′-CACGATGCCACTTTCTCACT-3′ |
| CXCR4 | F:
5′-ATCTGTGACCGCTTCTACCC-3′ |
| R:
5′-CGATGCTGATCCCAATGTAG-3′ |
| E-cadherin | F:
5′-CGTAGCAGTGACGAATGTGG-3′ |
| R:
5′-CTGGGCAGTGTAGGATGTGA-3′ |
| Vimentin | F:
5′-CGCCAACTACATCGACAAGG-3′ |
| R:
5′-GGCTTTGTCGTTGGTTAGCT-3′ |
| Snail | F:
5′-TTTACCTTCCAGCAGCCCTACGA-3′ |
| R:
5′-GCCTTTCCCACTGTCCTCATCT-3′ |
| Slug | F:
5′-TCAAGAAGCATTTCAACGCCTC-3′ |
| R:
5′-TGAGCTGAGGATCTCTGGTTGT-3′ |
| Twist | F:
5′-AGCAACAGCGAGGAAGAGCC-3′ |
| R:
5′-CACAGCCCGCAGACTTCTTG-3′ |
Semi-quantitative RT-PCR
To assess gene expression, cDNAs were amplified with
the 2X Ex Taq™ master mix (CWBio, China) using the Mycycler thermal
cycler PCR system (Bio-Rad, USA). The expression of each gene was
captured by Gel imaging system (BiD-1D/VILBER, France). The
sequences of specific primers are shown in Table I.
Western blot analysis
Cells were washed with ice-cold PBS and lysed by
RIPA buffer supplemented with protease inhibitor PMSF on ice. Total
protein concentrations were measured by BCA kit (Thermo Scientific)
and absorbance was measured by the PerkinElmer 2030 Victor X
multi-label plate reader. The extracted proteins were separated by
a 12% SDS-polyacrylamide gel electrophoresis and transferred to
PVDF membranes (Millipore, USA). The membranes were blocked with 5%
non-fat milk TBS-T (0.1% Tween-20, 100 nM Tris-HCl, 0.9% NaCl) and
subsequently incubated with anti-CXCR4 (1:200, sc-6190; Santa Cruz,
USA), anti-CD133 (1:500, orb99113; Biorbyt Ltd.), anti-E-cadherin
(1:10,000, ab40772; Abcam Corp.) or anti-vimentin (1:2,000,
ab92547; Abcam Corp.) respectively, with gentle shaking at 4°C
overnight. After washing three times, the membranes were incubated
with HRP-conjugated secondary antibodies for 2 h at room
temperature. The protein bands were visualized with ECL plus
western blot analysis detection reagents (Thermo Scientific).
Transwell migration assay
A total of 5×104 cells was plated in the
upper chamber, which had a 6.5-mm diameter polycarbonate membrane
with an 8-µm pore size coated filter (Corning, USA). The
lower chamber was filled with 600 µl DMEM solution
containing 10% FBS, which acted as a chemoattractant. After 24 h of
incubation at 37°C, non-migrated cells were removed with cotton
swabs and migrated cells were fixed in methanol and stained with
0.1% crystal violet. Images were obtained by light microscope with
a charge coupled device camera. Cells from four different fields
were counted to obtain the migration rates.
Statistical analysis
All assays were performed in triplicates.
Statistical analysis was performed using SPSS 19.0 for Windows or
GraphPad Prism 5 software. Data are reported as means ± SD.
Statistical differences were analyzed by Student's t-test for data
between control and treated groups, or a one-way analysis of
variance (ANOVA) for data from multiple groups, with the level of
significance set at p<0.05.
Results
CD133 and CXCR4 are highly expressed in
NSCLC patients with metastasis
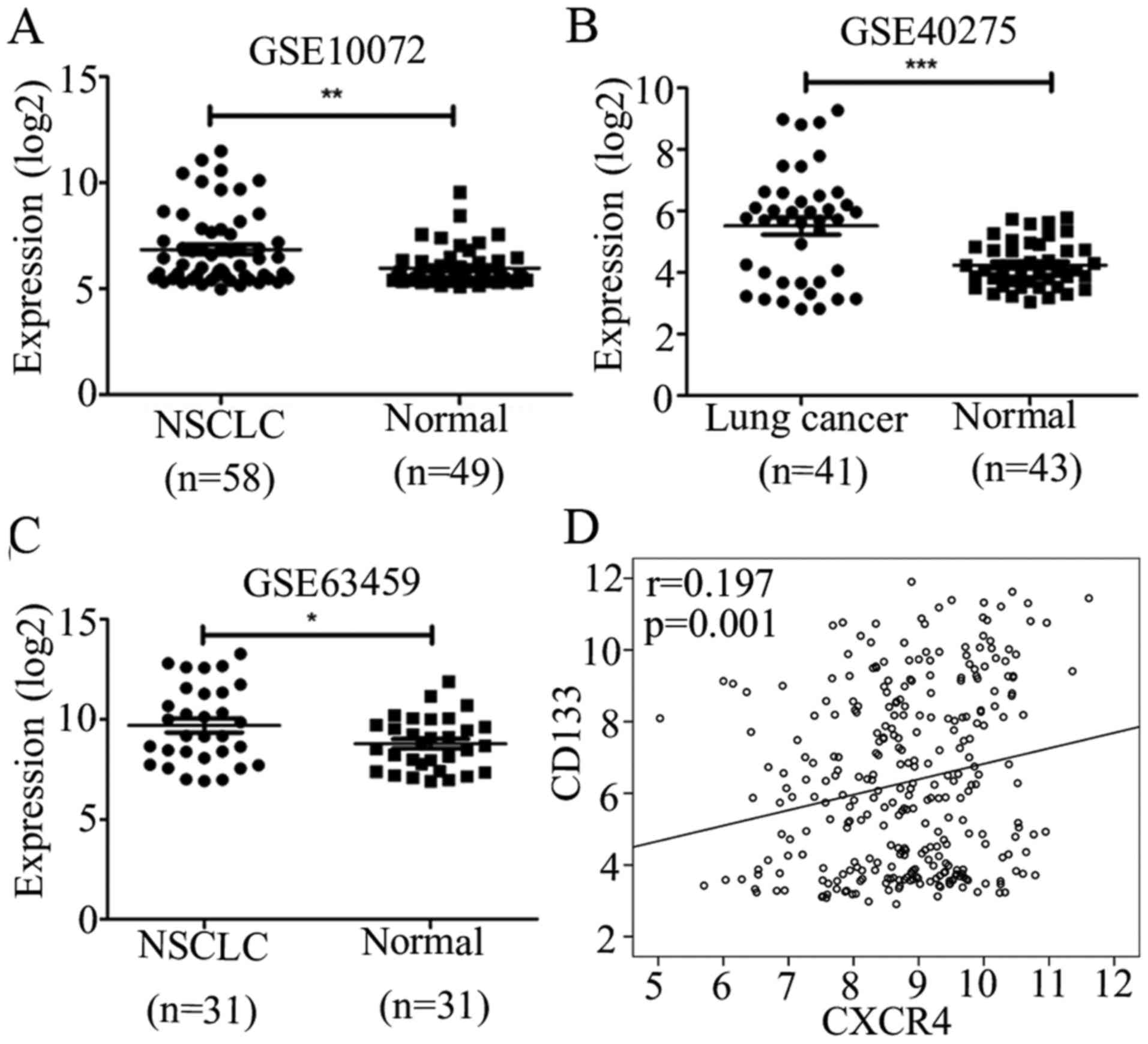
To explore the effect of CD133 in lung cancer, we
downloaded GSE10072, GSE40275 and GSE63459 to identify the
expression of CD133 in lung cancer and normal lung tissues. These
results showed that CD133 was highly expressed in lung cancer
compared to normal lung tissues (Fig.
1A–C), and we chose GSE30219, which is the largest dataset from
GEO datasets to identify the correlation of CD133 and CXCR4, it
showed that CD133 positively correlated with CXCR4 (Fig. 1D). Further, to investigate the role
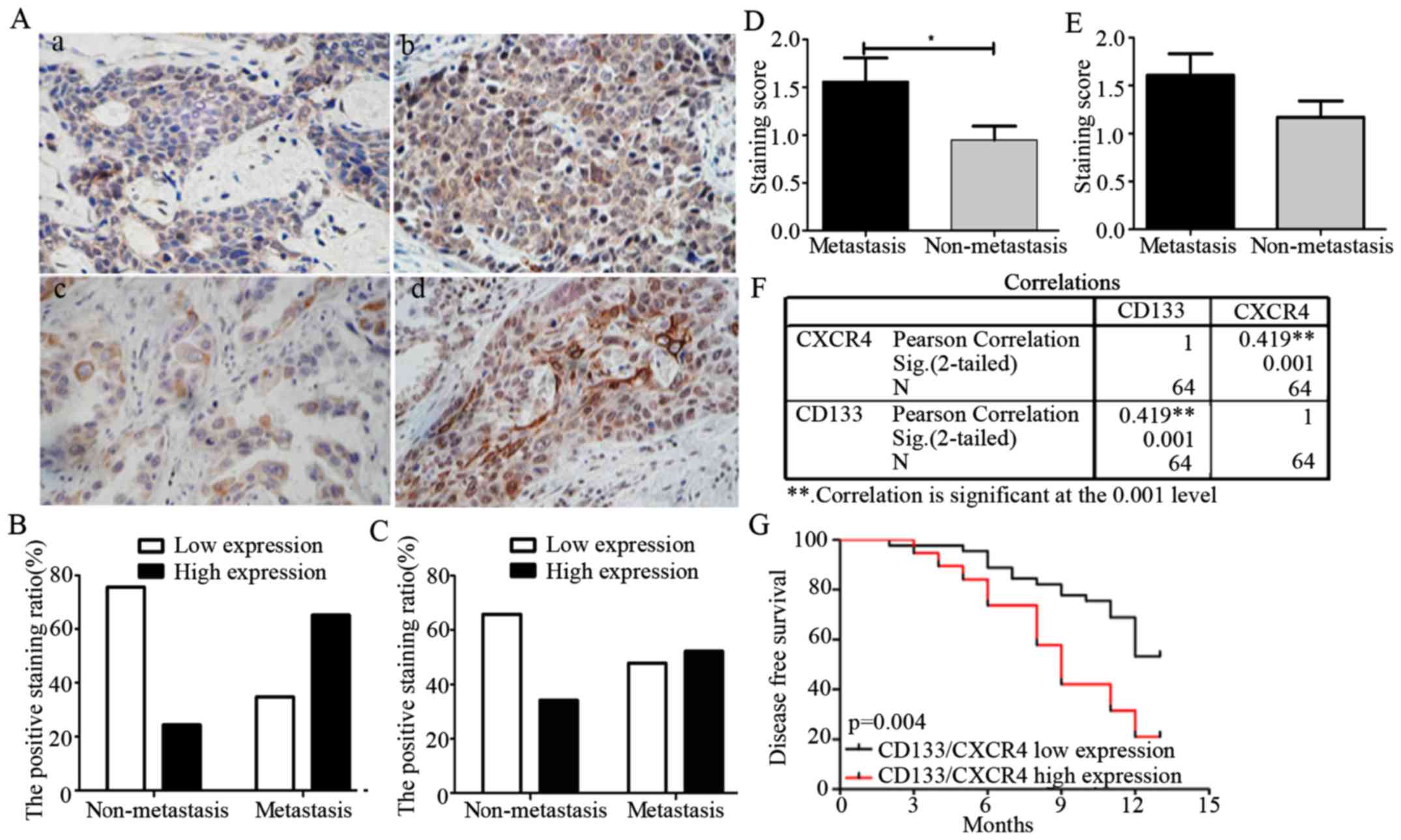
of CD133 and CXCR4 in NSCLC metastasis, we first detected the
expression levels of CD133 and CXCR4 in 64 NSCLC patients by
immunohistochemistry, the clinical and pathological characteristics
of the patients are shown in Table
II. These results showed that CD133 and CXCR4 levels
significantly increased in NSCLC metastatic patients as compared to
those in non-metastatic NSCLC patients (Fig. 2A). We categorized NSCLC patients
with metastasis into low (34.8%) and high group (75.6%), and
patients with non-metastasis into low (65.2%) and high group
(24.4%) for CD133 expression (Fig.
2B), then we categorized NSCLC patients with metastasis into
low (47.8%) and high group (65.9%) and patients with non-metastasis
into low (52.2%) and high group (34.1%) for CXCR4 expression,
respectively (Fig. 2C). The
staining scores were 1.57±0.24 in metastasis group and 0.95±0.14 in
non-metastasis group for CD133 expression (Fig. 2D), were 1.61±0.22 in metastasis
group and 1.17±0.17 in non-metastasis group for CXCR4 expression
(Fig. 2E), respectively.
 | Table IIThe clinical and pathological
characteristics of the patients. |
Table II
The clinical and pathological
characteristics of the patients.
| Features | Patients
(n=64) |
|---|
| Age (years) |
| ≤65 | 15 |
| >65 | 49 |
| Gender |
| Male | 44 |
| Female | 20 |
| T stage |
| T2 | 43 |
| T3 | 14 |
| T4 | 7 |
| N stage |
| N0 | 36 |
| N1 | 7 |
| N2 | 21 |
| M stage |
| M0 | 62 |
| M1 | 2 |
| Histology |
|
Adenocarcinoma | 39 |
| Squamous
carcinoma | 25 |
These results show a significant association between
CD133 and CXCR4 expression levels with NSCLC metastatic state in
patients. A positive correlation between CD133 and CXCR4 expression
was observed as shown in Fig. 2F
(r=0.419, p=0.001).
To further study the association between CD133/CXCR4
expression and cancer patient prognosis, we divided the patients
into two groups which including CD133/CXCR4 high expression (score
≥4) and low expression (score <4) group according to the
immunohistochemistry staining score average (stained area and
intensity) and we analyzed the correlation between CD133/CXCR4
co-expression and cancer patient disease-free survival, and we
observed that patients with high expression of both CD133 and CXCR4
presented a significantly poor survival rate (Fig. 2G). Using univariate and
multivariate Cox regression models, we also observed that N stage
and CD133/CXCR4 co-expression correlated with a shorter
disease-free survival. These results therefore suggest that
CD133/CXCR4 co-expression may be a potential independent predictor
of NSCLC patient survival (Table
III).
 | Table IIIUnivariate and multivariate Cox
regression analyses in NSCLC patients. |
Table III
Univariate and multivariate Cox
regression analyses in NSCLC patients.
| Variables | Univariate model
| Multivariate model
|
|---|
| HR | 95% CI of HR | P-value | HR | 95% CI of HR | P-value |
|---|
| Gender | 0.858 | 0.422–1.745 | 0.672 | 0.924 | 0.385–2.218 | 0.860 |
| Age | 1.007 | 0.969–1.046 | 0.729 | 0.999 | 0.952–1.048 | 0.965 |
| Histology | 0.754 | 0.388–1.463 | 0.404 | 1.363 | 0.532–3.488 | 0.519 |
| T stage | 1.475 | 0.953–2.282 | 0.081 | 1.514 | 0.917–2.499 | 0.105 |
| N stage | 1.659 | 1.182–2.329 | 0.003 | 1.672 | 1.107–2.526 | 0.014 |
| M stage | 2.610 | 0.617–11.047 | 0.192 | 2.291 | 0.412–12.732 | 0.343 |
| CD133/CXCR4
co-expression | 2.722 | 1.400–5.294 | 0.003 | 2.480 | 1.077–5.709 | 0.033 |
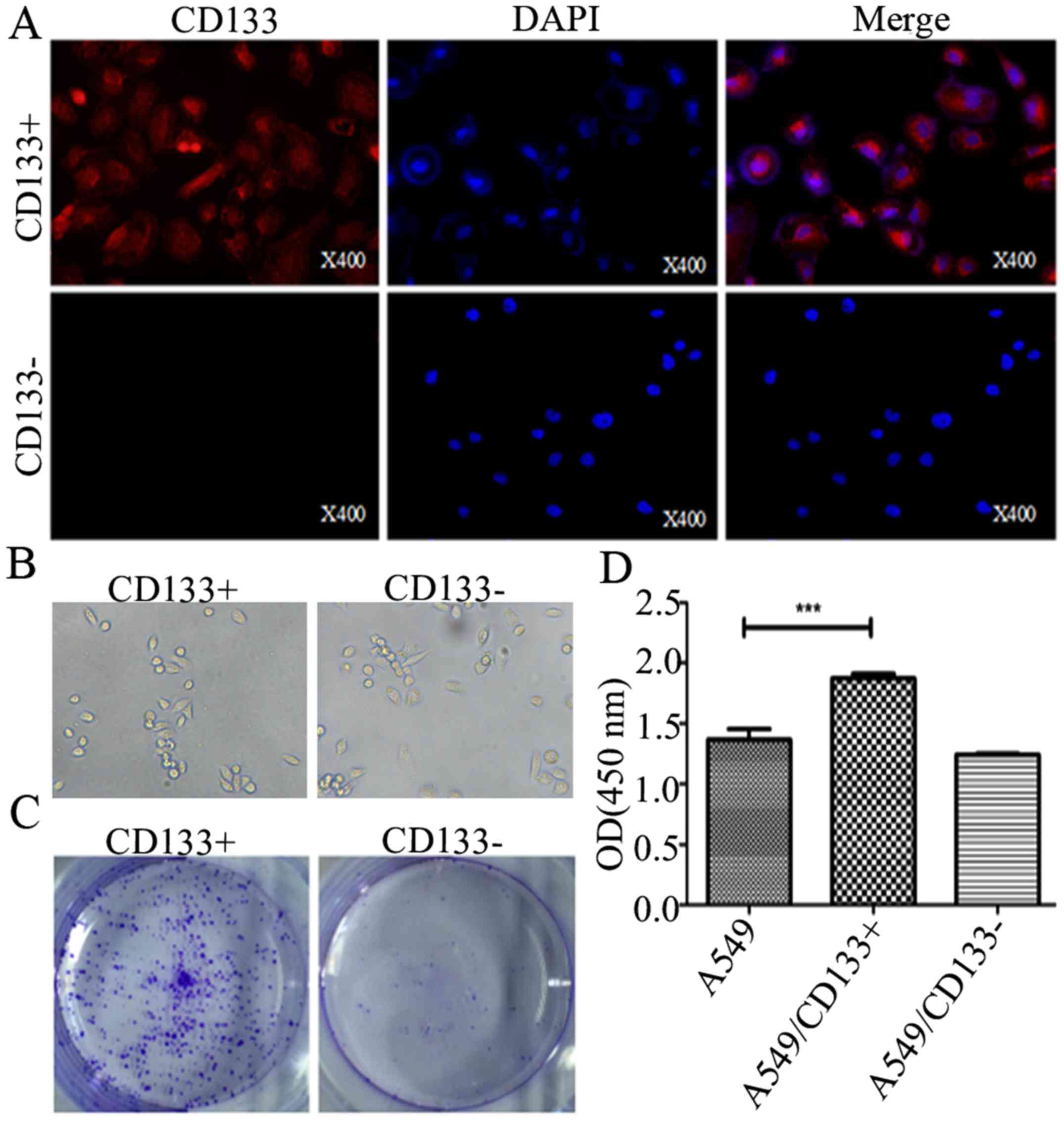
CD133 promotes NSCLC tumorigenesis
To investigate the role of CD133 in NSCLC
tumorigenesis, CD133+ and CD133− A549 cells
were sorted by magnetic cell sorting and verified by
immunofluorescence assay. Our results indicated that CD133
glycoproteins were generally expressed in the CD133+ as
compared to the CD133− groups (Fig. 3A). Furthermore, colony formation
assay and CCK8 assay results revealed that CD133+ cells
had a significant growth advantage over the control cells as shown
in Fig. 3B and C.
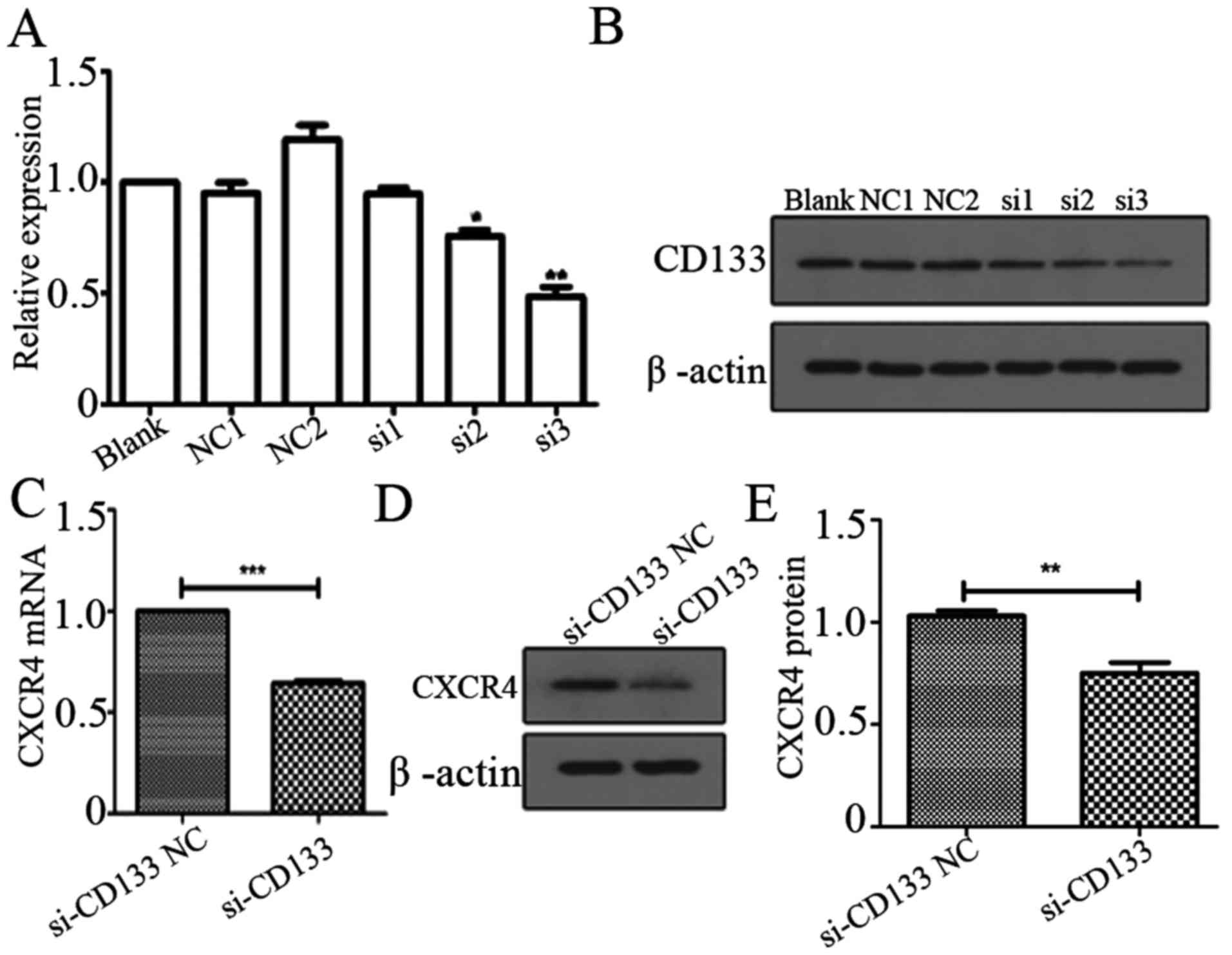
CD133 mediates the expression of CXCR4 in
NSCLC
We previously found that CD133 and CXCR4 were highly
expressed in A549 cells, so here to investigate the effect of CD133
on CXCR4 expression, we first demonstrated that CD133 mRNA and
protein levels were significantly inhibited by CD133 siRNA
(Fig. 4A and B). We then observed
the effect of CD133 on the mRNA and protein expression of CXCR4 by
CD133 silencing using CD133 siRNA (si3). We observed that CXCR4
mRNA and protein levels were significantly downregulated as a
result of CD133 silencing (Fig.
4C–E). Taken together, these results demonstrate that CD133
might play a significant role in the CXCR4 regulation.
CD133+CXCR4+
promotes EMT process in NSCLC
To investigate whether CXCR4 plays a vital role in
CD133-induced EMT processes, we first sorted A549 cells into
CD133-CXCR4+, CD133+CXCR4− and
CD133+CXCR4+ groups. Our results showed
remarkably higher migration rates in
CD133+CXCR4+ group as compared to the
CD133−CXCR4+ group as revealed by Transwell
migration assay (Fig. 5A and B).
We also detected the differential protein expression of E-cadherin
and Vimentin in CD133−CXCR4+,
CD133+CXCR4− and
CD133+CXCR4+ groups by western blot analysis,
and results showed that E-cadherin was decreased, while Vimentin
was increased in CD133+CXCR4+ as compared to
the CD133-CXCR4+ (Fig.
5C).
 | Figure 5CD133+CXCR4+
promotes EMT process in NSCLC cells. (A) Transwell assay for the
invasion of CD133−CXCR4+,
CD133+CXCR4− and
CD133+CXCR4+ cells. (B) Western blot assay
for the expression of E-cadherin and Vimentin in
CD133−CXCR4+,
CD133+CXCR4− and
CD133+CXCR4+ cells. (C) qPCR for the
expression of E-cadherin, Vimentin, Snail, Slug and Twist in
A549/CD133+, A549/CD133+ shRNA-NC,
A549/CD133+ shRNA-CXCR4 and A549/CD133+ with
amd3100. (D and E) RT-PCR for the expression of E-cadherin,
Vimentin, Snail, Slug and Twist in A549/CD133+,
A549/CD133+ shRNA-NC and A549/CD133+
shRNA-CXCR4 cells. |
We then went a step further and detected the
expression of EMT-related molecules (E-cadherin, Vimentin) and
transcriptional factors (Snail, Slug and Twist) in
CD133+A549 cells in the presence of CXCR4 inhibitors
(amd3100) or CXCR4 siRNA. Our findings indicated that E-cadherin
was significantly upregulated, while Vimentin, Snail, Slug and
Twist were significantly downregulated as a result of CXCR4
silencing or presence of CXCR4 inhibitors (Fig. 5D and E). These results demonstrate
that CD133 potentially regulates the expression of CXCR4 in
inducing EMT processes.
Expression of Vimentin was positively
associated with CD133/CXCR4 co-expression
To investigate the role of CD133, CXCR4 and
EMT-related molecules (E-cadherin and Vimentin) in NSCLC
metastasis, we detected the expression levels of CD133 and CXCR4,
and investigated the correlative expression of EMT-related
molecules (E-cadherin and Vimentin) with CD133/CXCR4 co-expression
in 64 NSCLC patients by immunohistochemistry. Our results revealed
that there was a significant increase in the Vimentin levels, while
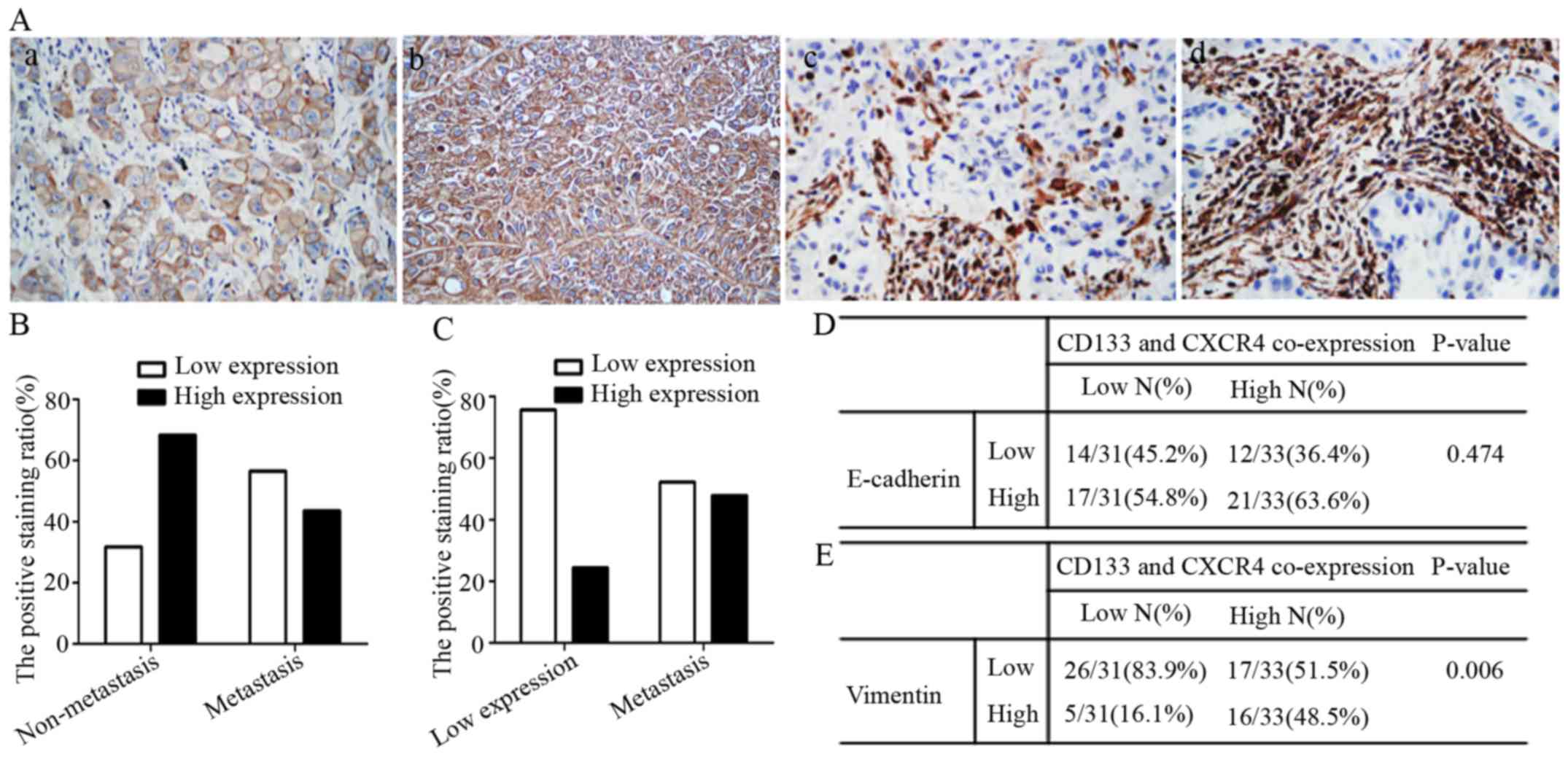
the expression levels of E-cadherin decreased as shown in Fig. 6A. NSCLC metastatic patients were
categorized as either low (56.5%) or high (43.5%) E-cadherin
expression groups and non-metastatic patients were categorized as
either low (31.6%) or high (68.4%) E-cadherin expression groups. In
terms of Vimentin expression, NSCLC metastatic patients were
categorized as either low (52.2%) or high (47.8%) expression
groups, while non-metastatic patients were further categorized as
either low (75.6%) or high (24.4%) expression groups (Fig. 6B and C).
We found E-cadherin expression did not correlate
with CD133/CXCR4 co-expression (Fig.
6D), while Vimentin had a positive correlative expression with
CD133/CXCR4 co-expression in NSCLC patients (p=0.006, Fig. 6E). Taken together these results
suggest that Vimentin is involved in CD133/CXCR4 induced NSCLC
metastasis.
Discussion
Lung cancer is one of the leading causes of cancer
mortality worldwide. Being the most dominating type of lung cancer,
NSCLC cases are being diagnosed at more advanced stages and hence
has been associated with a poor prognosis in patients (4). So far, little success has been made
in significantly improving patient survival rates, as the 5-year
survival rates have remained around 18% (34,35).
The main challenges for the NSCLC patient prognosis
are tumor metastasis and chemoresistance. Metastasis is a
multi-step process, which begins when primary tumor cells break
away from their neighboring cells invading the basement membrane,
subsequently entering the circulation, either directly or via the
lymphatics and eventually homing in distant organs. Tumor cells
that successfully adapt to the new micro-environments proliferate
from micro-metastases into clinical detectable metastatic tumor
(36). The CSCs in tumor
microenvironment are vital for the acquisition of the metastatic
potential as they initiate, drive carcinogenesis and
differentiation. In essence contributing to tumor cellular
heterogeneity through the deregulation of the self-renewal
processes, thus CSC populations may be a risk factor for
carcinogenesis (37).
Previous studies have identified a number of
cellular markers which include CD133 (38–40)
and CXCR4 (41,42), that may contribute to the
properties exhibited by lung CSCs. Chemokines are a family of small
secreted proteins of ~70–80 amino acids. CXCR4 is an α-chemokine
receptor specific for the CXCL12. Unlike other chemokine receptors
that have several ligands, CXCL12 is the only known specific legend
for the receptor CXCR4 (43). We
previously reported that CXCR4 was upregulated in NSCLC patients,
and that specific downregulation of CXCR4 inhibited cell growth,
invasiveness and migration. Additionally, we also reported that
MMP2 and MMP9 could be upregulated after treatment with CXCL12 in
A549 cells (20,21), and that CXCR4 was highly expressed
in chemo-resistant NSCLC patients and induced anti-apoptosis to
cisplatin in a CYP1B1 manner (data not shown).
In this study, we found that CD133 and CXCR4 are
highly expressed in NSCLC metastatic patients as compared to
non-metastatic patients. We further used Cox regression analysis to
verify whether CD133/CXCR4 could be a potential prognostic marker
for NSCLC patients, of which univariate Cox regression results
indicated that the N stage, M stage and CD133/CXCR4 co-expression
significantly associated with the patient prognosis. Moreover,
multivariate Cox regression results indicated that only the N stage
and CD133/CXCR4 co-expression significantly correlated with the
patient prognosis, with a CD133/CXCR4 co-expression hazards ratio
of 2.48, thus suggesting that CD133/CXCR4 may be an independent
prognostic marker in NSCLC patients. In the same line, other
researchers have also shown that CD133/CXCR4 co-expression is
highly associated with poor prognosis in patients (18). We also investigated the role of
CD133 in promoting NSCLC tumorigenesis as a potential CSCs marker,
as in some types of tumors, the stem cells exert increased
proliferation (44,45), furthermore, CD133-positive cells
showed proliferative characters in various tumors (46–48),
which are consistent with our results, so these results indicated
that CXCR4 might be modulated in part via CD133, as shown by the
CXCR4 mRNA and protein expression downregulation upon CD133
silencing, but, CXCR4 silencing did not influence the CD133
expression after transfected with siCXCR4 or siNC for 48 h in A549
cells.
In recent years, EMT has emerged as a potential
mechanism underlying cancer progression and metastasis. EMT
processes are accompanied by profound changes in cell
characteristics, which enable the epithelial cell to detach from
tight junctions, change their shape and polarity, delaminate and
migrate. As a result, mesenchymal cells exhibit a front-rear
polarity, become flat and spindle-shaped, and become loosely
associated with neighboring mesenchymal cells (49). We therefore also explored the role
of CD133/CXCR4 as a CSCs marker in mediating EMT processes and
observed that CD133+CXCR4+ cells had a
stronger migration ability accompanied by a lower E-cadherin
expression and a higher Vimentin expression as compared to
CD133−CXCR4+ and
CD133+CXCR4− A549 cells. Knapp et al
also used immunohistochemical analysis of CXCR4 and E-cadherin on a
melanoma tissue microarray which included 110 primaries, 73
local/regional metastatic and 44 distant metastatic patients, and
observed that there was no significant E-cadherin expression
(50).
Furthermore, we detected the expression of
EMT-related molecules (E-cadherin, Vimentin) and transcription
factors (Snail, Slug and Twist) after CXCR4 silencing or inhibition
by a CXCR4 antagonist. Our results showed that E-cadherin was
upregulated, while Vimentin, Snail, Slug and Twist were
downregulated as a result of CXCR4 knockdown or inhibition, thus
suggesting a potential role for CXCR4 in regulating EMT
processes.
In addition, Vimentin expression was observed to
have a positive correlation with CD133/CXCR4 co-expression in NSCLC
patients and survival analysis results suggested that Vimentin high
expression might be significantly associated with poor patients'
survival rates, thus suggesting that Vimentin may be involved in
CD133/CXCR4 induced NSCLC metastasis. Vimentin expressing
metastatic lung cancer cells have been reported to be more motile
and invasive while acting via a VAV2-Rac1 pathway to control focal
adhesion kinase activity (51). In
conclusion, this study shows that CXCR4 is involved in
CD133-induced EMT processes in NSCLC. Thus, the CD133/CXCR4/EMT
axis could be used as a potential therapeutic target in NSCLC
metastasis.
Acknowledgments
This study was supported by National Natural Science
Foundation of China (nos. 81270607 and 81541027) and Natural
Science Foundation of Hubei (nos. 2014CFA070 and 2015CFB653).
Abbreviations:
|
NSCLC
|
non-small cell lung cancer
|
|
EMT
|
epithelial-mesenchymal transition
|
|
CSC
|
cancer stem cell
|
References
|
1
|
Miller KD, Siegel RL, Lin CC, Mariotto AB,
Kramer JL, Rowland JH, Stein KD, Alteri R and Jemal A: Cancer
treatment and survivorship statistics, 2016. CA Cancer J Clin.
66:271–289. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chaffer CL and Weinberg RA: A perspective
on cancer cell metastasis. Science. 331:1559–1564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cai C, Yu JW, Wu JG, Lu RQ, Ni XC, Wang SL
and Jiang BJ: CD133 promotes the invasion and metastasis of gastric
cancer via epithelial-mesenchymal transition. Zhonghua Wei Chang
Wai Ke Za Zhi. 16:662–667. 2013.In Chinese. PubMed/NCBI
|
|
8
|
Bock C, Kuhn C, Ditsch N, Krebold R,
Heublein S, Mayr D, Doisneau-Sixou S and Jeschke U: Strong
correlation between N-cadherin and CD133 in breast cancer: Role of
both markers in metastatic events. J Cancer Res Clin Oncol.
140:1873–1881. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ding Q, Miyazaki Y, Tsukasa K, Matsubara
S, Yoshimitsu M and Takao S: CD133 facilitates
epithelial-mesenchymal transition through interaction with the ERK
pathway in pancreatic cancer metastasis. Mol Cancer. 13:152014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nomura A, Banerjee S, Chugh R, Dudeja V,
Yamamoto M, Vickers SM and Saluja AK: CD133 initiates tumors,
induces epithelial-mesenchymal transition and increases metastasis
in pancreatic cancer. Oncotarget. 6:8313–8322. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Long H, Xiang T, Qi W, Huang J, Chen J, He
L, Liang Z, Guo B, Li Y, Xie R, et al: CD133+ ovarian
cancer stem-like cells promote non-stem cancer cell metastasis via
CCL5 induced epithelial-mesenchymal transition. Oncotarget.
6:5846–5859. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Koren A, Rijavec M, Kern I, Sodja E,
Korosec P and Cufer T: BMI1, ALDH1A1, and CD133 transcripts connect
epithelial-mesenchymal transition to cancer stem cells in lung
carcinoma. Stem Cells Int. 2016:97143152016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Latorre E, Carelli S, Raimondi I,
D'Agostino V, Castiglioni I, Zucal C, Moro G, Luciani A, Ghilardi
G, Monti E, et al: The ribonucleic complex HuR-MALAT1 represses
CD133 expression and suppresses epithelial-mesenchymal transition
in breast cancer. Cancer Res. 76:2626–2636. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee SO, Yang X, Duan S, Tsai Y, Strojny
LR, Keng P and Chen Y: IL-6 promotes growth and
epithelial-mesenchymal transition of CD133+ cells of
non-small cell lung cancer. Oncotarget. 7:6626–6638. 2016.
|
|
15
|
Moon Y, Kim D, Sohn H and Lim W: Effect of
CD133 overexpression on the epithelial-to-mesenchymal transition in
oral cancer cell lines. Clin Exp Metastasis. 33:487–496. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fargeas CA, Florek M, Huttner WB and
Corbeil D: Characterization of prominin-2, a new member of the
prominin family of pentaspan membrane glycoproteins. J Biol Chem.
278:8586–8596. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sowa T, Menju T, Sonobe M, Nakanishi T,
Shikuma K, Imamura N, Motoyama H, Hijiya K, Aoyama A, Chen F, et
al: Association between epithelial-mesenchymal transition and
cancer stemness and their effect on the prognosis of lung
adenocarcinoma. Cancer Med. 4:1853–1862. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bertolini G, D'Amico L, Moro M, Landoni E,
Perego P, Miceli R, Gatti L, Andriani F, Wong D, Caserini R, et al:
Microenvironment-modulated metastatic
CD133+/CXCR4+/EpCAM− lung
cancer-initiating cells sustain tumor dissemination and correlate
with poor prognosis. Cancer Res. 75:3636–3649. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Teicher BA and Fricker SP: CXCL12
(SDF-1)/CXCR4 pathway in cancer. Clin Cancer Res. 16:2927–2931.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dai X, Mao Z, Huang J, Xie S and Zhang H:
The CXCL12/CXCR4 autocrine loop increases the metastatic potential
of non-small cell lung cancer in vitro. Oncol Lett. 5:277–282.
2013.
|
|
21
|
Xie S, Zeng W, Fan G, Huang J, Kang G,
Geng Q, Cheng B, Wang W and Dong P: Effect of CXCL12/CXCR4 on
increasing the metastatic potential of non-small cell lung cancer
in vitro is inhibited through the downregulation of CXCR4 chemokine
receptor expression. Oncol Lett. 7:941–947. 2014.PubMed/NCBI
|
|
22
|
Nikzaban M, Hakhamaneshi MS, Fakhari S,
Sheikhesmaili F, Roshani D, Ahsan B, Kamali F and Jalili A: The
chemokine receptor CXCR4 is associated with the staging of gastric
cancer. Adv Biomed Res. 3:162014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Okuyama Kishima M, de Oliveira CE,
Banin-Hirata BK, Losi-Guembarovski R, Brajão de Oliveira K,
Amarante MK and Watanabe MA: Immunohistochemical expression of
CXCR4 on breast cancer and its clinical significance. Anal Cell
Pathol (Amst). 2015:8910202015.
|
|
24
|
Teng F, Tian WY, Wang YM, Zhang YF, Guo F,
Zhao J, Gao C and Xue FX: Cancer-associated fibroblasts promote the
progression of endometrial cancer via the SDF-1/CXCR4 axis. J
Hematol Oncol. 9:82016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hu TH, Yao Y, Yu S, Han LL, Wang WJ, Guo
H, Tian T, Ruan ZP, Kang XM, Wang J, et al: SDF-1/CXCR4 promotes
epithelial-mesenchymal transition and progression of colorectal
cancer by activation of the Wnt/β-catenin signaling pathway. Cancer
Lett. 354:417–426. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li X, Li P, Chang Y, Xu Q, Wu Z, Ma Q and
Wang Z: The SDF-1/CXCR4 axis induces epithelial-mesenchymal
transition in hepatocellular carcinoma. Mol Cell Biochem.
392:77–84. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lv B, Yang X, Lv S, Wang L, Fan K, Shi R,
Wang F, Song H, Ma X, Tan X, et al: CXCR4 signaling induced
epithelial-mesenchymal transition by PI3K/AKT and ERK pathways in
glioblastoma. Mol Neurobiol. 52:1263–1268. 2015. View Article : Google Scholar
|
|
28
|
Yang P, Wang G, Huo H, Li Q, Zhao Y and
Liu Y: SDF-1/CXCR4 signaling up-regulates survivin to regulate
human sacral chondrosarcoma cell cycle and epithelial-mesenchymal
transition via ERK and PI3K/AKT pathway. Med Oncol. 32:3772015.
View Article : Google Scholar
|
|
29
|
Liao A, Shi R, Jiang Y, Tian S, Li P, Song
F, Qu Y, Li J, Yun H and Yang X: SDF-1/CXCR4 axis regulates cell
cycle progression and epithelial-mesenchymal transition via
up-regulation of survivin in glioblastoma. Mol Neurobiol.
53:210–215. 2016. View Article : Google Scholar
|
|
30
|
Zhang SS, Han ZP, Jing YY, Tao SF, Li TJ,
Wang H, Wang Y, Li R, Yang Y, Zhao X, et al: CD133(+)CXCR4(+) colon
cancer cells exhibit metastatic potential and predict poor
prognosis of patients. BMC Med. 10:852012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Silinsky J, Grimes C, Driscoll T, Green H,
Cordova J, Davis NK, Li L and Margolin DA: CD 133+ and
CXCR4+ colon cancer cells as a marker for lymph node
metastasis. J Surg Res. 185:113–118. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li XF, Guo XG, Yang YY and Liu AY: Effect
of CXCR4 and CD133 co-expression on the prognosis of patients with
stage II–III colon cancer. Asian Pac J Cancer Prev. 16:1073–1076.
2015. View Article : Google Scholar
|
|
33
|
Lu C, Xu F, Gu J, Yuan Y, Zhao G, Yu X and
Ge D: Clinical and biological significance of stem-like
CD133(+)CXCR4(+) cells in esophageal squamous cell carcinoma. J
Thorac Cardiovasc Surg. 150:386–395. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
DeSantis CE, Lin CC, Mariotto AB, Siegel
RL, Stein KD, Kramer JL, Alteri R, Robbins AS and Jemal A: Cancer
treatment and survivorship statistics, 2014. CA Cancer J Clin.
64:252–271. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sun Y and Ma L: The emerging molecular
machinery and therapeutic targets of metastasis. Trends Pharmacol
Sci. 36:349–359. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kakarala M and Wicha MS: Implications of
the cancer stem-cell hypothesis for breast cancer prevention and
therapy. J Clin Oncol. 26:2813–2820. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen Y, Zhang F, Tsai Y, Yang X, Yang L,
Duan S, Wang X, Keng P and Lee SO: IL-6 signaling promotes DNA
repair and prevents apoptosis in CD133+ stem-like cells
of lung cancer after radiation. Radiat Oncol. 10:2272015.
View Article : Google Scholar
|
|
39
|
Roudi R, Korourian A, Shariftabrizi A and
Madjd Z: Differential expression of cancer stem cell markers ALDH1
and CD133 in various lung cancer subtypes. Cancer Invest.
33:294–302. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu QF, Zhang ZF, Hou GJ, Yang GY and He
Y: Polymorphisms of the stem cell marker gene CD133 and the risk of
lung cancer in Chinese population. Lung. 194:393–400. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Nian WQ, Chen FL, Ao XJ and Chen ZT: CXCR4
positive cells from Lewis lung carcinoma cell line have cancer
metastatic stem cell characteristics. Mol Cell Biochem.
355:241–248. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jung MJ, Rho JK, Kim YM, Jung JE, Jin YB,
Ko YG, Lee JS, Lee SJ, Lee JC and Park MJ: Upregulation of CXCR4 is
functionally crucial for maintenance of stemness in drug-resistant
non-small cell lung cancer cells. Oncogene. 32:209–221. 2013.
View Article : Google Scholar
|
|
43
|
Wang Z, Sun J, Feng Y, Tian X, Wang B and
Zhou Y: Oncogenic roles and drug target of CXCR4/CXCL12 axis in
lung cancer and cancer stem cell. Tumour Biol. 37:8515–8528. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Jiang YX, Yang SW, Li PA, Luo X, Li ZY,
Hao YX and Yu PW: The promotion of the transformation of quiescent
gastric cancer stem cells by IL-17 and the underlying mechanisms.
Oncogene. Aug 15–2016.Epub ahead of print. View Article : Google Scholar
|
|
45
|
Fish KM: Mesenchymal stem cells drive
cardiac stem cell chemotaxis, proliferation, and phenotype via
CXCR4 and ckit signaling. Circ Res. 119:891–892. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang C, Zhou C, Wu XJ, Yang M, Yang ZH,
Xiong HZ, Zhou CP, Lu YX, Li Y and Li XN: Human CD133-positive
hematopoietic progenitor cells initiate growth and metastasis of
colorectal cancer cells. Carcinogenesis. 35:2771–2777. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Won C, Kim BH, Yi EH, Choi KJ, Kim EK,
Jeong JM, Lee JH, Jang JJ, Yoon JH, Jeong WI, et al: Signal
transducer and activator of transcription 3-mediated CD133
up-regulation contributes to promotion of hepatocellular carcinoma.
Hepatology. 62:1160–1173. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhao W, Luo Y, Li B and Zhang T:
Tumorigenic lung tumorospheres exhibit stem-like features with
significantly increased expression of CD133 and ABCG2. Mol Med Rep.
14:2598–2606. 2016.PubMed/NCBI
|
|
49
|
Yuan X, Wu H, Han N, Xu H, Chu Q, Yu S,
Chen Y and Wu K: Notch signaling and EMT in non-small cell lung
cancer: Biological significance and therapeutic application. J
Hematol Oncol. 7:872014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Knapp CF, Sayegh Z, Schell MJ, Rawal B,
Ochoa T, Sondak VK and Messina JL: Expression of CXCR4, E-cadherin,
Bcl-2, and survivin in Merkel cell carcinoma: An
immunohistochemical study using a tissue microarray. Am J
Dermatopathol. 34:592–596. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Havel LS, Kline ER, Salgueiro AM and
Marcus AI: Vimentin regulates lung cancer cell adhesion through a
VAV2-Rac1 pathway to control focal adhesion kinase activity.
Oncogene. 34:1979–1990. 2015. View Article : Google Scholar
|