Introduction
Oxygen levels are crucial for cell survival. Hypoxia
(insufficient oxygen) is always present in tumor areas due to the
excessive proliferation of tumor cells and the increasing oxygen
consumption. For solid tumors, hypoxia plays an important role in
promoting tumor development and disease progression via favoring
the formation of a neoplastic microenvironment.
Osteosarcoma is a common, high-risk primary bone
malignancy that mostly presents in the first or second decades of
life (1). Surgery and chemotherapy
are the main treatment strategies for osteosarcoma. However, for
high-grade osteosarcoma, the combination of surgery and
chemotherapy does not always yield satisfactory outcomes due to the
high rate of cancer recurrence and metastasis to the lungs
(2,3). There has been no marked improvement
in the clinical outcomes of osteosarcoma patients to date. This
stagnation in therapeutic advances may be attributed to the
genetic, epigenetic and biological complexities of this tumor
(4). Therefore, identifying new
biomarkers and novel therapeutic targets is crucial for improving
the prognosis of patients with osteosarcoma.
MicroRNAs (miRNAs/miRs) are a family of endogenous
single-stranded, small non-protein-coding RNAs that affect
fundamental cellular behaviors in diverse organisms. Mature miRNAs
are ~22 nt long and regulate gene expression at the
post-transcriptional level by inhibiting the translation and/or
decreasing of the stability of target mRNAs (5,6).
Several hundred miRNAs have been identified and studied thus far,
and they have been found to play key roles in various biological
and pathological processes, such as cell proliferation, apoptosis,
differentiation, metabolism, autophagy, migration and invasion
(7–11).
miR-15a, located at 13q14.3, was identified as a
tumor suppressor gene in chronic lymphocytic leukemia in 2002
(12). Later investigations
indicated that miR-15a exerts its anti-tumor effects through
targeting several oncogenes that are associated with cancer, such
as CCND1 (encoding cyclin D1), WNT3A and B-cell lymphoma 2 (Bcl-2)
(13,14). To date, deletion and/or
downregulation of miR-15a have been reported in several types of
cancer, including breast, colorectal and lung cancer, as well as
osteosarcoma (11,15–17).
Tian et al examined 45 pairs of human osteosarcoma samples
and demonstrated that miR-15a expression was downregulated compared
with that in corresponding adjacent normal tissues (11). However, the role of miR-15a in
osteosarcoma tumor invasion and migration, particularly under
hypoxic conditions, remains largely unknown.
The aim of the present study was to investigate the
role of miR-15a in regulating hypoxia-induced cell invasion and
migration in human osteosarcoma cells, as well as the involvement
of Bcl-2 in this process and the underlying mechanism, in order to
determine whether miR-15a may be of value as a therapeutic target
for the treatment of osteosarcoma, particularly in patients with
high-grade cancer or heavy tumor burden.
Materials and methods
Cell culture
The human osteosarcoma cell lines MG63 and U-2 OS,
and the human osteoblast cell line hFOB1.19, were obtained from
American Type Culture Collection (Manassas, VA, USA). MG63 and U-2
OS cells were maintained in Dulbecco's modified Eagle's medium
(DMEM, Biological Industries, BI, Shanghai, China) supplemented
with 10% fetal bovine serum (FBS; BI, Kibbutz Beit-Haemek, Israel)
and streptomycin (100 mg/ml)/penicillin (100 U/ml; HyClone,
Beijing, China). U-2 OS cells were maintained in DMEM/Ham's F12
medium supple mented with 10% FBS and streptomycin/penicillin.
Cells were incu bated at 37°C with 5% CO2 and 20%
O2 in a humidified incubator (Thermo Fisher Scientific,
Waltham, MA, USA).
Hypoxic culture
For the hypoxic culture, tissue culture plates were
placed in a 37°C humidified CO2 (5%)/O2
(1%)/N2 (94%) incubator (Fisher Scientific Forma; Thermo Fisher
Scientific).
α-amanitin treatment of MG63 cells
MG63 cells were cultured to 70–80% confluence and
treated with 100 µg/ml α-amanitin (Sigma-Aldrich; Merck
KGaA, St. Louis, MO, USA) for 3 h to block nascent pri-miRNA
synthesis. Subsequently, the cells were cultured under hypoxic or
normoxic conditions for another 1, 2, 4 or 8 h. Cells were
collected and total RNA was extracted for the quantitative
polymerase chain reaction (qPCR) assay.
miRNA, siRNA and cell transfection
miR-15a mimics (sequence: 5′-uagcagcaca
uaaugguuugug-3′) and the miRNA scrambles (sequence: 5′-UUGUACUAC
ACAAAAGUACUG-3′, control, CT) were purchased from GenePharma Co.
Ltd. (Shanghai, China). For miRNA transfection, MG63 cells were
transfected with Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific). According to the manufacturer's protocol, 100 pmol
miRNA mimics or miRNA scrambles were added to each well of a 6-well
plate. For DNA transfection, 2.5 µg plasmid DNA was
transfected with Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific) in each well of a 6-well plate.
To knock down the level of Bcl-2, MG63 cells were
transfected with 25 pmol Bcl-2 siRNA or non-targeting (NT) negative
control siRNA (siRNA ID: 214532, Thermo Fisher Scientific) using
Lipofectamine RNAiMAX transfection reagent (Thermo Fisher
Scientific) following the manufacturer's instructions. Knockdown
efficiency was measured by western blotting. All the cell
experiments were performed at 24 post-transfection.
Cell invasion assay
For the cell invasion assay, 1×105 MG63
cells or the transfected cells were seeded in serum-free medium in
a Transwell migration chamber (8-µm pore size; Corning,
Shanghai, China). The upper chamber was coated with Matrigel (BD
Biosciences, San Jose, CA, USA). DMEM containing 10% FBS was loaded
to the lower well. After a 24-h culture in the hypoxic incubator,
the cells that did not invade through the pores were carefully
wiped off with a cotton swab. The cells located on the lower
surface of the chamber were fixed and stained with 0.1% crystal
violet (Sigma-Aldrich; Merck KGaA) at room temperature and counted
under a light microscope (Olympus, Tokyo, Japan) from five random
fields. These experiments were repeat ed three times.
Migratory behavior assay
For the cell migration assay, MG63 cells or the
transfected cells were seeded on a fibronectin- precoated CELLview
cell culture dish (Greiner Bio-One, Stuttgart,
Badenia-Wirtembergia, Germany) and cultured for 12 h in the hypoxic
incubator. Cell images were captured at 10-min intervals with a CCD
camera for 6 h to acquire image stacks. Each image stack was
quantitatively analyzed by NIH ImageJ software.
RNA extraction, reverse transcription
(RT) and qPCR analysis
Total RNA was extracted by Qiagen RNeasy Mini Kit
(Qiagen, Shanghai, China) and reverse-transcribed to cDNA using
Moloney murine leukemia virus reverse transcriptase (M-MLV,
Invitrogen; Thermo Fisher Scientific). For Bcl-2 mRNA detection,
800 ng total RNA was reverse-transcribed using oligo(dT) primers
and random hexamer primers. For deleted in lymphocytic leukemia 2
(DLEU2) RNA detection, 800 ng total RNA was reverse-transcribed
using gene-specific reverse primer (GSRP). For microRNA detection,
1,500 ng total RNA was reverse-transcribed using microRNA-specific
reverse primers (MSRP). The levels of β-actin and U6 were used to
normalize gene and microRNA expression. The GSRP sequence was as
follows: 5′-TCTCATACAGGTTACAGTTC-3′. The MSRP sequences were as
follows: U6
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAATA-3′; miR-15a
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCACAAA-3′. qPCR
analysis for the Bcl-2 and DLEU2 genes and miR-15a was performed
using an aliquot of first-strand cDNA as the template in a
20-µl reaction system containing 10 µl 2X SYBR
premixed buffer (Roche, Shanghai, China), 2 µl forward and
reverse primers. The primers were as follows: Bcl-2 forward,
5′-CTGCACCTGACGCCCTTCACC-3′ and reverse,
5′-CACATGACCCCACCGAACTCAAAGA-3′; DLEU2 forward,
5′-TCTGGAGAACAGCCTCACTTC-3 and reverse, 5′-TGCT
GAGCTAAGTAGAGGTCTC-3′ (18);
β-actin forward, 5′-CTGGCTCCTAGCACCATGAAGAT-3′ and reverse,
5′-GGTGGACAGTGAGGCCAGGAT-3′ (19);
U6 forward, 5′-CTCGCTTCGGCAGCACATA-3′ and reverse,
5′-CAGTGCAGGGTCCGAGGTA-3′; miR-15a forward,
5′-CGCCTAGCAGCACATAATGG-3′ and reverse, 5′-AGTGCAGGGTCCGAGGTAT-3′
(20). The PCR amplification
process was as follows: 5 min denaturation at 95°C followed by 40
cycles at 95°C for 20 sec, 58°C for 15 sec, and 72°C for 15 sec
(ABI StepOnePlus, Beijing, China). Data analysis was performed
using the 2−ΔΔCq method.
Western blot analysis
Protein was extracted from MG63 cells with RIPA
buffer (KeyGen Biotech. Co., Ltd., Nanjing, China) supplemented
with cocktail protease inhibitor (Roche). Equal amounts of protein
from each sample were separated by 12% sodium dodecyl sulfate
polyacrylamide gel electrophoresis and transferred to
polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA,
USA). The blots were then blocked with 5% BSA (Sigma-Aldrich; Merck
KGaA) in TBST at 37°C for 1 h and incubated with primary antibodies
[Bcl-2 rabbit polyclonal antibody, 1:1,000, ab59348;
hypoxia-inducible factor (HIF)-1α mouse monoclonal antibody,
1:1,000, ab1; Abcam, Shanghai, China. Matrix metalloproteinase
(MMP)-2 rabbit monoclonal antibody, 1:1,000, 40994; MMP-9 rabbit
monoclonal antibody, 1:1,000, 13667; Cell Signaling Technology,
Shanghai, China] overnight at 4°C. After washing 3 times in TBST,
the membranes were incubated with horseradish peroxidase-coupled
goat anti-rabbit or goat anti-mouse secondary antibody (sc-2004 and
sc-2005, 1:3,000, Santa Cruz Biotechnology, Shanghai, China) at
room temperature for 1 h. The chemiluminescence signals were
detected with a chemiluminescence system (ECL; EMD Millipore).
Densitometric analysis was conducted using Quantity One software,
version 4.6 (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Construction of luciferase reporter
vectors
For luciferase reporter vector construction, a 3′UTR
segment of 546 bp of the 3′UTR of the Bcl-2 gene (UTR-WT) was
amplified by PCR from human genomic DNA and inserted into the
pGL3-control vector (Promega Corporation, Beijing, China), using
the XbaI site downstream from the stop codon of luciferase.
The following primers were used to generate pGL3-UTR-WT: UTR-WT
forward, 5′-CTAGTCTAGAGCCTCAGGGAACAGAATGATCAG-3′ and reverse,
5′-CTAGTCTAGAAAGCGTCCACGTTCTTCATTG-3′ (21). We also generated a lucif-erase
reporter plasmid with mutant Bcl-2 3′UTR (UTR-MU) by using the Fast
Site-Directed Mutagenesis kit (Tiangen, Beijing, China). The
following primers were used to generate pGL3-UTR-MU: UTR-MU
forward, 5′-ATCCAATCCTGAGGTCCGATCCTGCCAAA-3′ and reverse,
5′-TTTGGCAGGATCGGACCTCAGGATTGGAT-3′. Both UTR-WT and UTR-MU were
confirmed by sequencing.
Luciferase activity assay
Briefly, 1×105 MG63 cells were
co-transfected with 0.5 µg pGL3-UTR-WT or pGL3- UTR-MU
luciferase vectors and 50 pmol miRNAs using Lipofectamine 2000 in
24-well plates. Renilla luciferase reporter plasmid pRL-SV40
(0.05 µg, Promega Corporation) was co-transfected for
monitoring transfection efficiency. Approximately 24 h later, cells
were rinsed with phosphate-buffered saline and lysed with passive
lysis buffer. Luminescence was measured by Dual-Luciferase Reporter
assay system (Glomax 20/20, Promega Corporation). The luciferase
activity was calculated as the ratio between the luminescence of
firefly luciferase and the Renilla luciferase. Each
experiment was repeated in triplicate.
Statistical analysis
Statistical analysis was conducted with SPSS 17.0
software (SPSS Inc., Chicago, IL, USA). All data were expressed as
arithmetic mean ± standard deviation. Statistical analysis was
performed with one-way analysis of variance or Student's t-test.
Results were considered statistically significant for P-values
<0.05.
Results
Hypoxia represses miR-15a expression and
stimulates MG63 cell invasion
In order to understand the expression of miR-15a in
osteosarcoma cells, its levels in two osteosarcoma cell lines, MG63
and U-2 OS, were measured by qPCR. As a comparison, we also
measured the level of miR-15a in the normal osteoblast cell line
hFOB1.19. As shown in Fig. 1A, the
level of miR-15a in the two osteosarcoma cell lines was
significantly lower compared with that in the normal osteoblast
cell line (P<0.05).
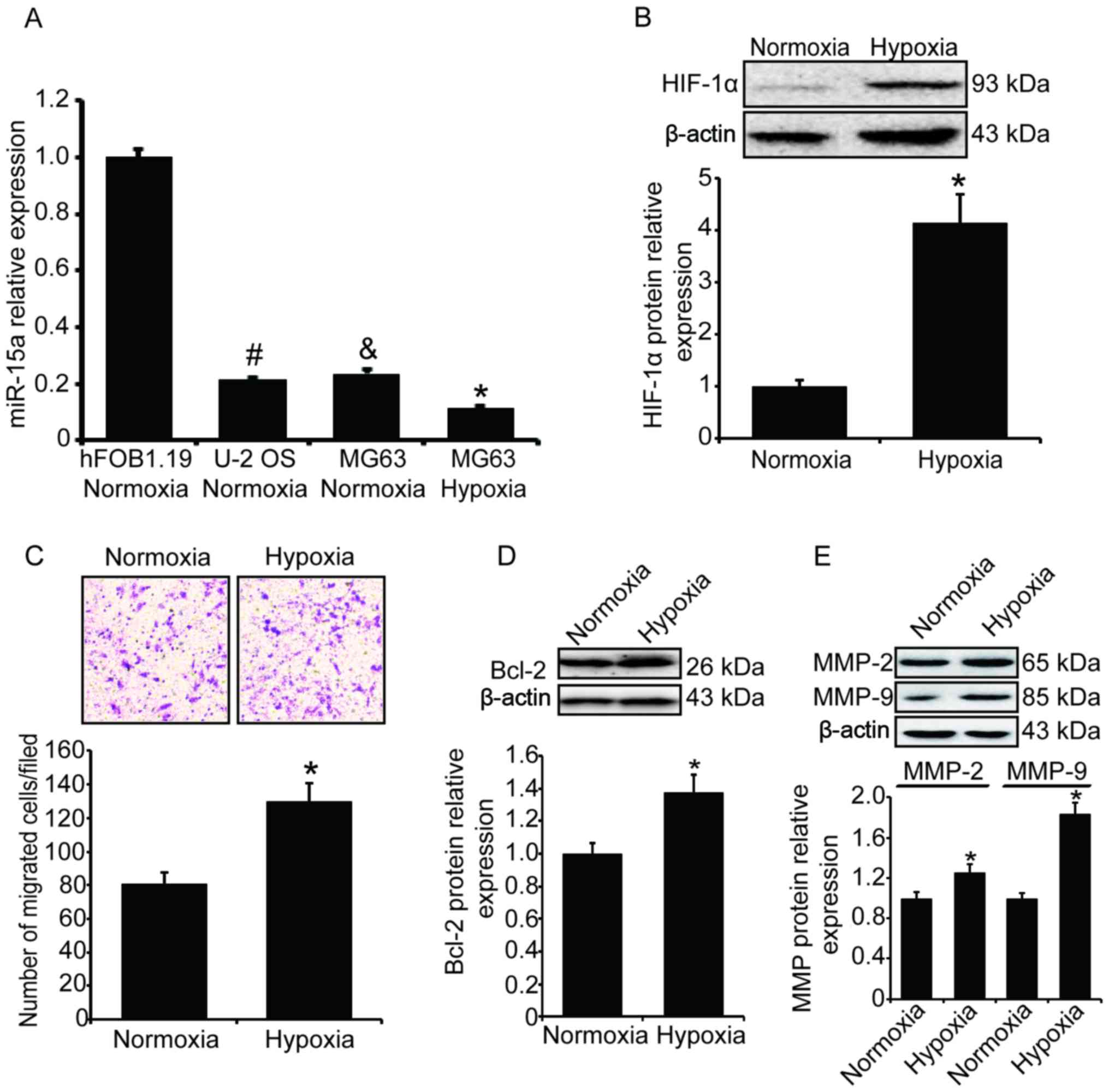 | Figure 1Hypoxia represses miR-15a expression
and stimulates cell invasion in osteosarcoma. The levels of miR-15a
in two human osteosarcoma cell lines (MG63 and U-2 OS) and one
human osteoblast cell line (hFOB1.19) were measured and compared.
MG63 cells were exposed to hypoxia (1% O2) for 24 h. The
expression of miR-15a was measured by qPCR, the expression of
HIF-1α, Bcl-2 and MMPs was measured by western blotting, and the
cell invasion ability was measured by the Transwell assay. (A)
Compared with the normal osteoblast cells, the levels of miR-15a in
osteosarcoma cells were significantly lower. Additionally, the
level of miR-15a decreased significantly after MG63 cells were
exposed to hypoxia. All the results were normalized to U6 and
expressed as fold-change. Statistical results were based on one-way
analysis of variance; #&P<0.05 vs. normal
osteoblast cells; *P<0.05 vs. MG63 cells cultured
under normoxic conditions. (B) Hypoxia-induced HIF-1α upregulation
in MG63 cells. Upper panel, western blotting; lower panel, relative
band density of western blotting. (C) The number of MG63 cells
migrating through the Transwell chamber inserts was significantly
increased after cells were exposed to hypoxia. Upper panel,
invading MG63 cells were stained with crystal violet; lower panel,
mean number of invading MG63 cells. (D) Hypoxia-induced Bcl-2
upregulation in MG63 cells. Upper panel, western blotting; lower
panel, relative band density of the western blotting. (E)
Hypoxia-induced upregulation of MMP-2 and MMP-9 in MG63 cells.
Upper panel, western blotting; lower panel, relative band density
of the western blotting. qPCR, quantitative polymerase chain
reaction analysis; HIF-1α, hypoxia-inducible factor-1α; Bcl-2,
B-cell lymphoma 2; MMP, matrix metalloproteinase. |
We further selected the MG63 cell line to
investigate the effect of hypoxia on miR-15a expression and cell
invasion in human osteosarcoma. MG63 cells were placed in a hypoxic
incubator with 1% O2 and cultured for 24 h. To determine
whether the hypoxia was effective, the expression of HIF-1α was
measured, and the western blotting results demonstrated that the
level of HIF-1α in MG63 cells was significantly increased (Fig. 1B). The miR-15a level under hypoxic
stress was also measured by qPCR. Compared with the cells cultured
under normoxic conditions, the level of miR-15a in MG63 cells
cultured in hypoxia decreased significantly (Fig. 1A, P<0.05).
The invasion ability of MG63 cells was measured by
the Transwell experiment and cells migrating through the Transwell
chamber inserts were counted. Following exposure to hypoxia, the
number of migrated cells was significantly increased compared with
the normoxia group (P<0.05, Fig.
1C). We also detected the expression of Bcl-2 and matrix
metalloproteinases by western blotting, and found that the levels
of Bcl-2, MMP-2 and MMP-9 were significantly increased (Fig. 1D and E) under hypoxic
conditions.
Overexpression of miR-15a represses the
invasion and migration of MG63 cells under hypoxia
To investigate whether the hypoxia-induced cell
invasion is correlated with the down-regulation of miR-15a, we
introduced exogenous miR-15a into MG63 cells by transfection of
miR-15a mimics. At 48 h post-transfection, the total RNA was
extracted and the level of miR-15a was measured by qPCR. As shown
in Fig. 2A, the total miRA-15a was
markedly increased after the transfection (P<0.05).
Subsequently, the invasion ability of the transfected MG63 cells
under hypoxic condition was measured. After the transfected MG63
cells were exposed to hypoxia for 24 h, the number of cells
migrating through the Transwell chamber inserts was counted and
illustrated. As shown in Fig. 2B,
the number of migrated cells under hypoxia was markedly decreased
after miR-15a was overexpressed (P<0.05). We also analyzed the
migratory behavior of the transfected MG63 cells by tracking the
migration paths and analyzing the accumulated distance of the
individual cells. As shown in Fig.
2C, the average accumulated distance of the miR-15a-transfected
cells was 26.1 µm, whereas the average accumulated distance
of the miR-scramble-transfected cells was 51.2 µm
(P<0.05). The cancer cell invasion and migration required MMP-2
and MMP-9, which were also detected by western blotting, and the
results demonstrated that introduction of miR-15a significantly
suppressed the expression of MMP-2 and MMP-9 under hypoxic
conditions (Fig. 2D,
P<0.05).
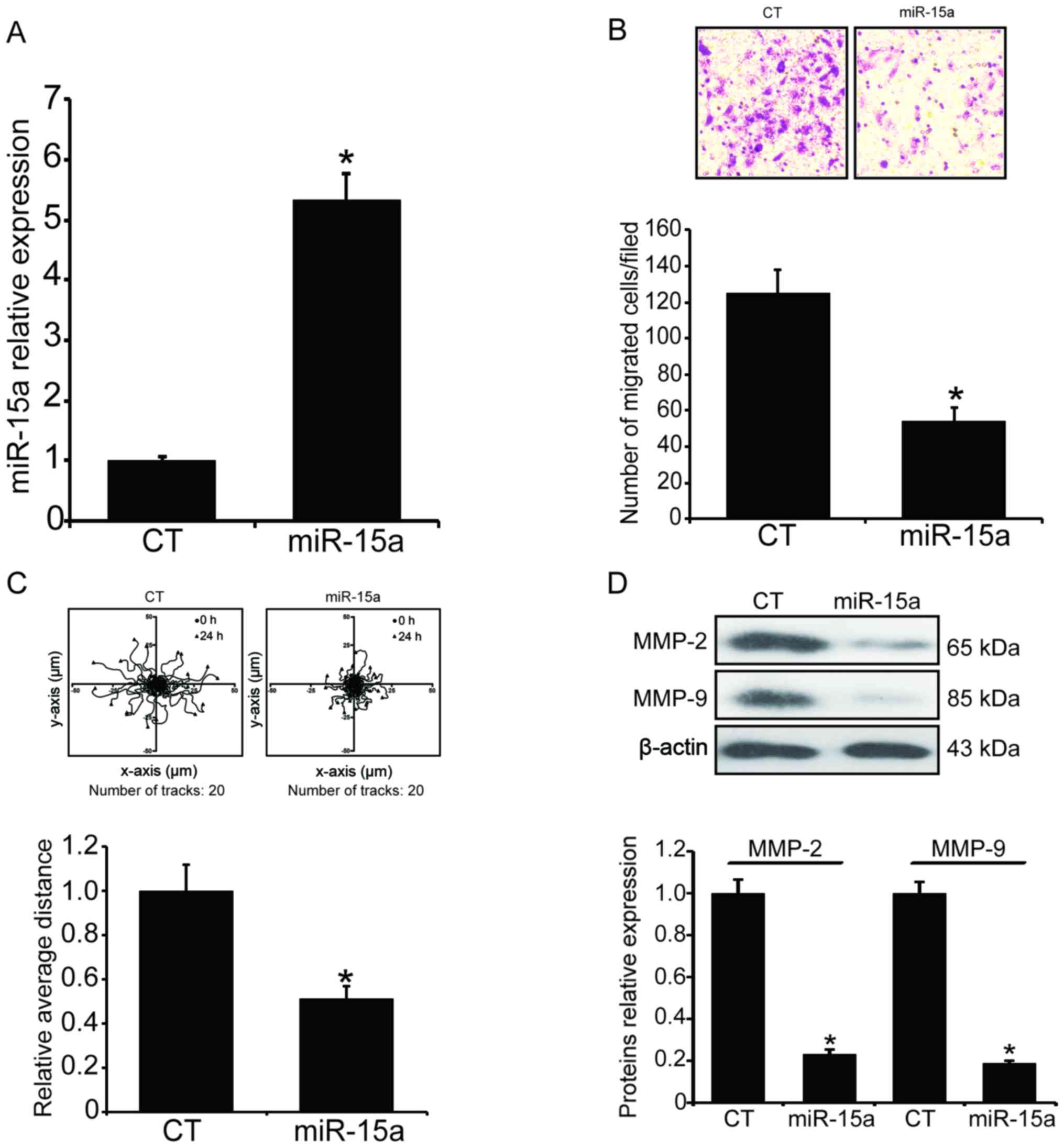 | Figure 2Overexpression of miR-15a inhibits
the invasion and migration of MG63 cells under hypoxic conditions.
Exogenous miR-15a was introduced into MG63 cells by transfection of
miR-15a mimics using Lipofectamine 2000. After the transfection,
the invasion and migration ability of the transfected MG63 cells
under hypoxia was assessed by the Transwell assay and the tracking
of the migration paths. (A) The level of total miR-15a was measured
by qPCR. The results were normalized to U6 and expressed as
fold-change relative to the control. (B) The number of cells
migrating through the Transwell chamber inserts was markedly
decreased. Upper panel, invading MG63 cells were stained by crystal
violet; lower panel, mean number of invading MG63 cells. (C) Mean
accumulated distance of the miR-15a-transfected MG63 cells. Upper
panel, the cell migration paths were tracked by a CCD camera; lower
panel, the accumulated distance of the individual cells was
analyzed by ImageJ software. (D) MMP-2 and MMP-9 levels were
determined by western blotting. Upper panel, western blotting;
lower panel, relative band density of the western blotting.
*P<0.05 vs. the miR-scramble-transfection group.
qPCR, quantitative polymerase chain reaction analysis; MMP, matrix
metalloproteinase; CT, control. |
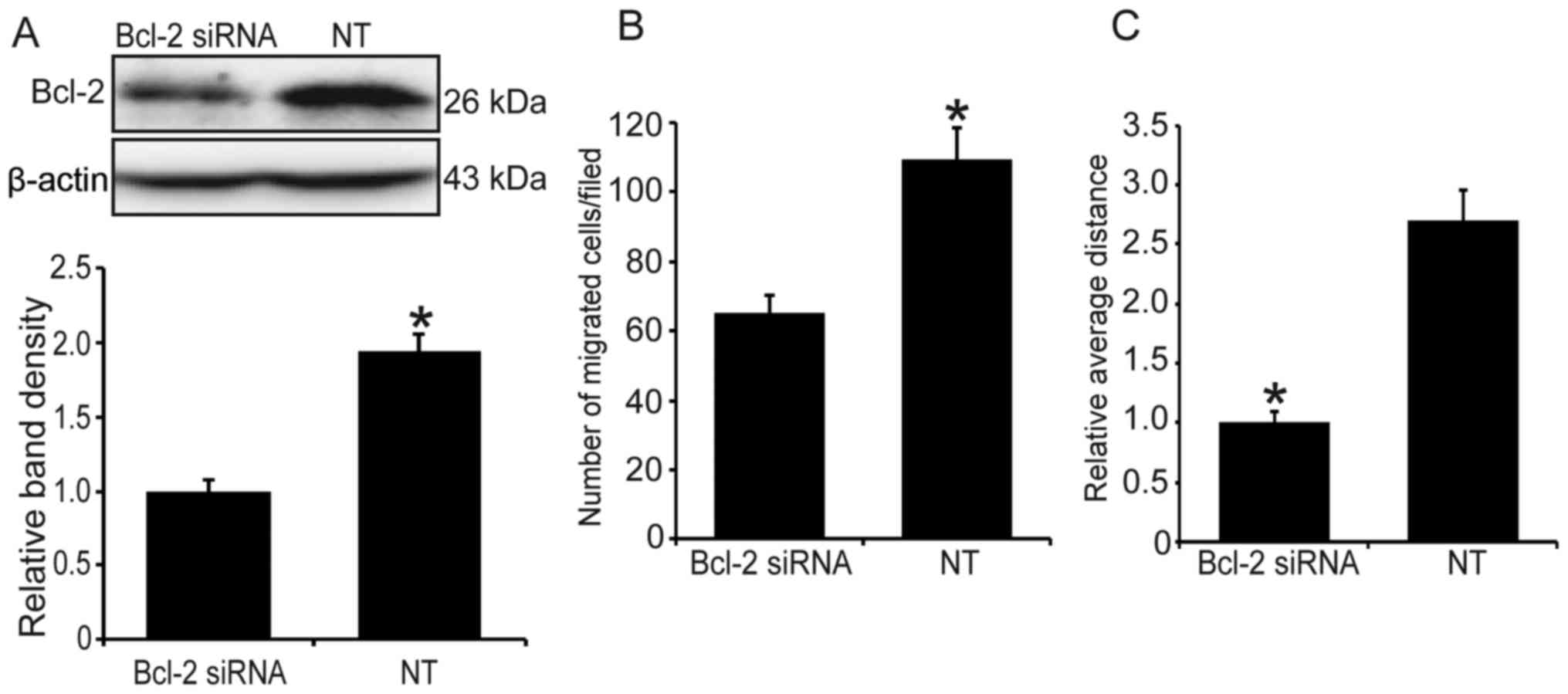
Knockdown of Bcl-2 represses cell
invasion and migration in MG63 cells under hypoxia
Since hypoxia can stimulate the expression of
endogenous Bcl-2, we used Bcl-2 siRNA to knock down the level of
Bcl-2 and determine whether it will reduce the hypoxia-induced cell
invasion and migration. As shown in Fig. 3A, following transfection of Bcl-2
siRNA, the level of Bcl-2 was significantly decreased (P<0.05).
Additionally, compared with the control group, Bcl-2 knockdown
significantly suppressed hypoxia-induced cell invasion and
migration (Fig. 3B and C).
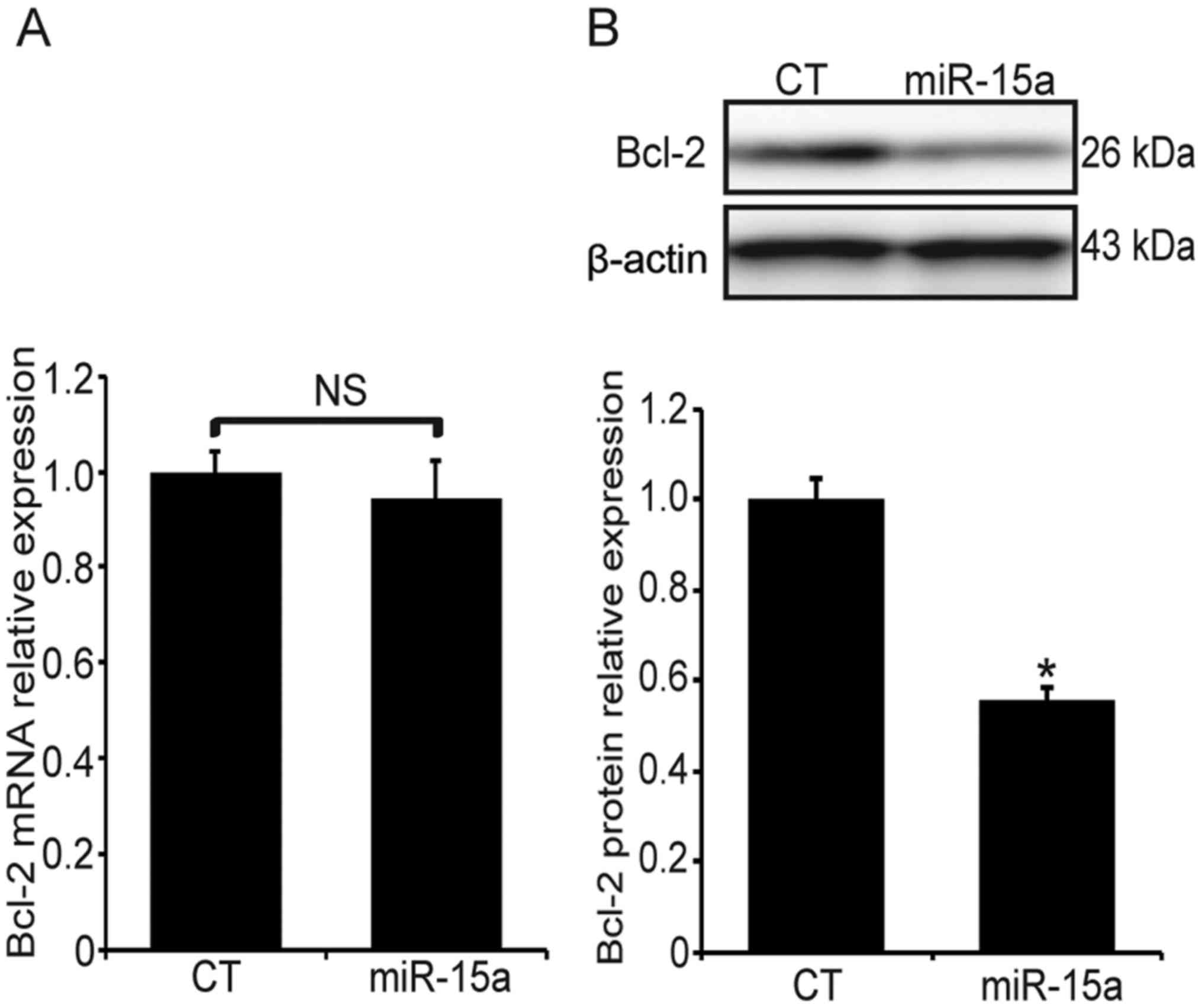
miR-15a represses the expression of Bcl-2
at the post-transcriptional level
The miR-15a-transfected MG63 cells were exposed to
hypoxia for 24 h and the expression of Bcl-2 was examined at the
transcriptional and post-transcriptional levels. The Bcl-2 mRNA
expression was measured by qPCR, and there was no significant
difference between the miR-15a and the miR-scramble transfection
groups (Fig. 4A, P>0.05).
However, the Bcl-2 protein level in the miR-15a transfection group
was significantly decreased (Fig.
4B, P<0.05). These findings suggest that miR-15a may
suppress the expression of Bcl-2 at the post-transcriptional
level.
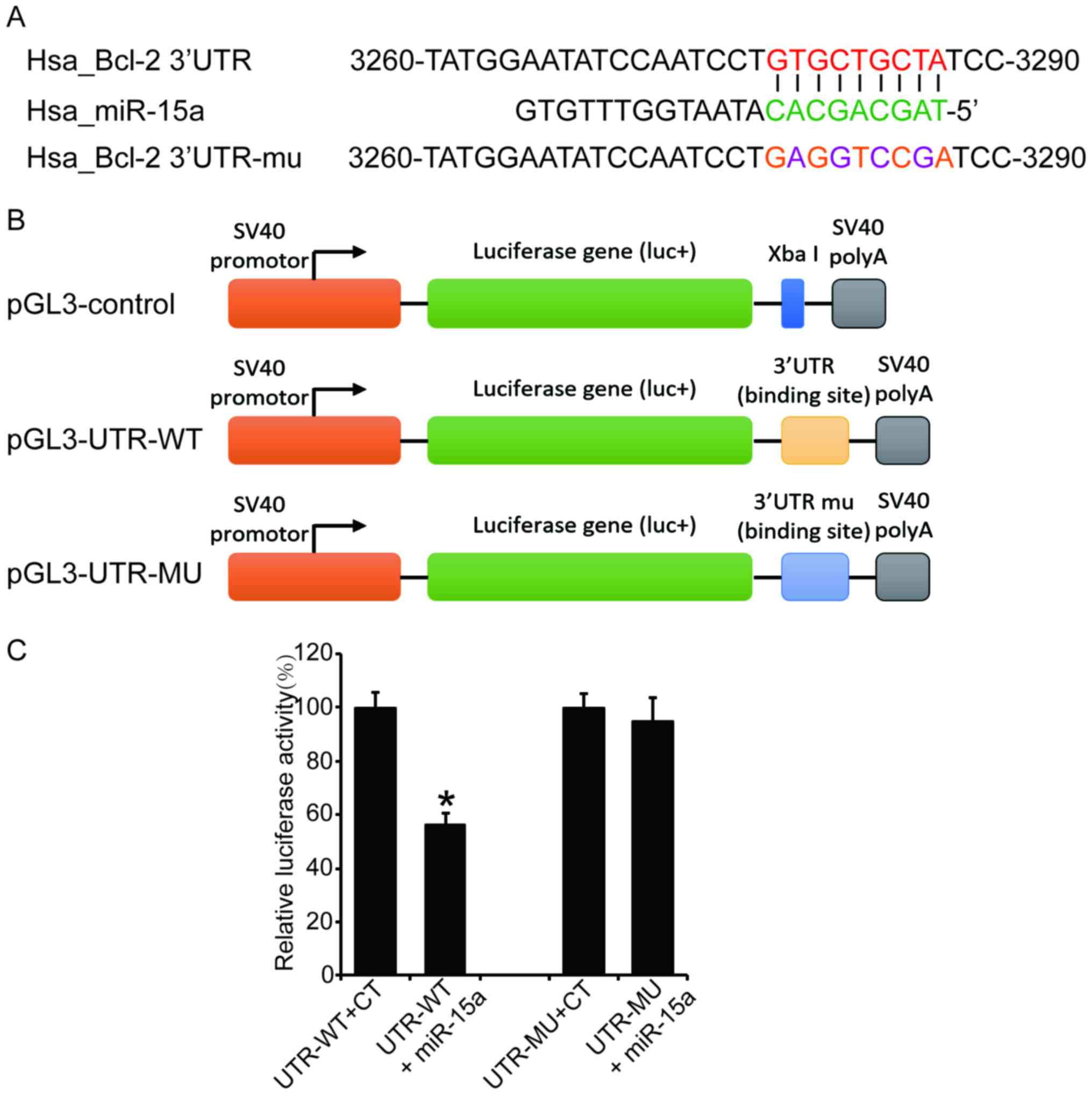
Bcl-2 mRNA is a direct target of
miR-15a
It was reported that the Bcl-2 mRNA is a target of
the miR-15 family (21), and our
current experiment results also revealed that miR-15a suppresses
the expression of Bcl-2 at the post-transcriptional level. In order
to confirm the direct binding of miR-15a to the Bcl-2 mRNA in MG63
cells, the 3′UTR fragment containing wild-type or mutant seed
region of the miR-15a was cloned into a luciferase reporter vector
(Fig. 5A and B). The luciferase
activity assay revealed that the wild-type reporter vector
exhibited a significant reduction of luciferase activity when
co-transfected with miR-15a compared with the control group; by
contrast, the mutation vector abrogated this inhibitory effect of
miR-15a (Fig. 5C, P<0.05). This
result suggests that miR-15a suppresses Bcl-2 expression through
direct binding to its 3′UTR.
Hypoxia decreases DLEU2
transcription
After proving that hypoxia represses miR-15a
expression in MG63 cells, we further investigated whether miR-15a
was downregulated at the transcriptional or post-transcriptional
level. MG63 cells were first treated with α-amanitin to block the
nascent pri-miRNA synthesis, and then the levels of miR-15a at 1,
2, 4 or 8 h under hypoxia were measured. As shown in Fig. 6A, following treatment of the MG63
cells with α-amanitin, the level of miR-15a decreased by ~25% after
the cells were cultured for 8 h, regardless of whether the cells
were under hypoxic or normoxic conditions (P>0.05).
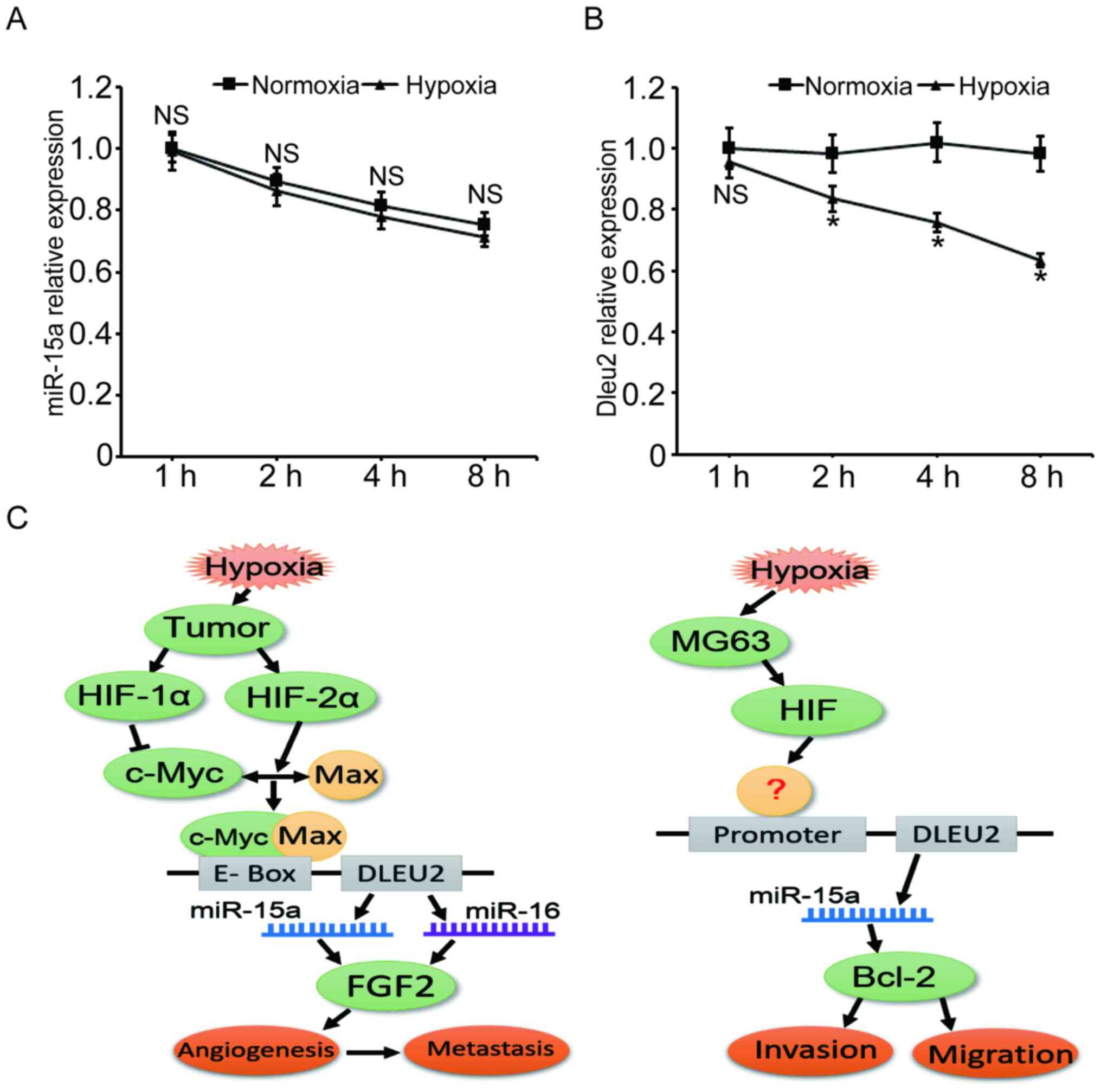 | Figure 6Hypoxia decreases the level of
miR-15a through inhibiting the transcription of the DLEU2 gene.
miR-15a expression under hypoxic stress was investigated at the
transcriptional and post-transcriptional levels. (A) The
degradation velocity of miR-15a was almost the same under hypoxic
and normoxic conditions. MG63 cells were treated with 100
µg/ml α-amanitin for 3 h to block nascent pri-miRNA
synthesis and cells were cultured under hypoxia or normoxia for
another 1, 2, 4 or 8 h. The miR-15a level was measured by qPCR. (B)
The DLEU2 gene transcription level was significantly decreased
under hypoxic stress. MG63 cells were cultured in hypoxia or
normoxia, and the DLEU2 mRNA level was measured by qPCR at 1, 2, 4
or 8 h time points. (C) Sketch maps for the mechanisms of
miR-15a-mediated cell activities under hypoxia. Left panel,
c-Myc-HIF-mediated repression of miR-15-16 in hypoxia promotes
tumor angiogenesis and metastasis (34); right panel, putative mechanism of
miR-15a mediated cell invasion and migration in MG63 cells. DLEU2,
deleted in lymphocytic leukemia 2; qPCR, quantitative polymerase
chain reaction analysis; HIF, hypoxia-induciblefactor; FGF,
fibroblast growth factor. |
Since DLEU2 is the host gene of miR-15a, the mRNA
levels of this gene under hypoxia were measured to estimate the
transcriptional level of miR-15a. The qPCR results demonstrated
that, after the cells were cultured in hypoxia for 2, 4 and 8 h,
the mRNA level of DLEU2 gene decreased by 15.0, 25.6 and 35.6%,
respectively, compared with the normoxia control group. These
findings suggest that hypoxia represses the level of miR-15a mainly
at the transcriptional level.
Discussion
Inadequate oxygen supply is common in tissues
harboring a heavy tumor burden. Tumor cells alter DNA transcription
in response to hypoxic stress and, therefore, cell behaviors
change. HIF family members are considered to be oxygen sensors, and
HIF-involved hypoxia signaling pathways are among the most popular
research targets. In addition to HIF, other genes, such as vascular
endothelial growth factor, angiogenin, p53, nuclear factor-κB, p38
and Bcl-2, also play key roles in hypoxia-induced cellular
activities (22-27).
Hypoxia promotes cell invasion, survival,
angiogenesis and other cellular activities by modifying or
regulating cellular molecules. miRNAs are recognized as a class of
regulators affecting gene expression and are involved in several
normal and pathological cellular processes. Numerous studies have
demonstrated that miRNAs are correlated with various types of human
cancer. In vivo studies demonstrated that restoration of miR-15a in
tumors may ameliorate disease manifestations (28,29).
However, how miRNAs interact with certain conditions in the tumor
microenvironment, such as hypoxia, has not yet been fully
elucidated (30). In recent years,
it was demonstrated that miRNAs play important roles in several
pivotal signaling pathways that respond to hypoxia and are
associated with hypoxic adaptation. For example, Zuo et al
reported that miR-210 inhibits cell proliferation via inducing cell
cycle arrest and apoptosis in human laryngeal carcinoma (31), whereas Li et al observed
that miR-137 inhibits mitophagy via the regulation of the mitophagy
receptors FUNDC1 and NIX (32).
Growing evidence indicates that the miR-15 family
members, which include miR-15a, miR-15b, miR-16, miR-195, miR-424
and miR-497, are implicated in oncogenesis, cardiovascular and
neurodegenerative diseases. Recent studies reported that miR-15a
was also involved in hypoxia-induced gene expression regulation and
cellular hypoxic adaptation. Yang et al found that hypoxia
increases the level of miR-15a in rat cardiomyoblasts, and the
inhibition of miR-15a significantly reduces cardiomyocyte apoptosis
and the release of lactate dehydrogenase and malondialdehyde
(33); Xue et al found that
miR-15-16 transcription is repressed by hypoxia, which promotes
tumor angiogenesis and hematogenous metastasis in tumor cells
(34). We herein demonstrated that
hypoxia enhances cell invasion and migration in the human
osteosarcoma cell line MG63 through the downregulation of miR-15a
and the upregulation of Bcl-2 (Fig.
1). However, as tumor cells are always heterogeneous and
different types of tumors have distinct genetical backgrounds, more
osteosarcoma cell lines and clinical osteosarcoma tissues should be
investigated in future studies to draw more definitive
conclusions.
Bcl-2 is one of the key participants in the
protection of cells from apoptotic death. Abnormally high levels of
Bcl-2 occur in over half of all human cancers (35). In addition to conferring a survival
advantage on cells, Bcl-2 is key to a wide variety of biological
processes, including tumor invasion and metastasis (36,37).
For example, Ricca et al proved that Bcl-2 overexpression
induces MMP-9 transcription and enhances cell metastatic potential
in human MCF7 breast cancer cells (38). miRNAs have been estimated to
regulate up to 30% of all human genes (39). However, how miRNAs regulate Bcl-2
during the process of tumor metastasis, particularly under hypoxic
conditions, remains unclear. Wang et al reported that
miR-429 inhibits cell invasion and promotes apoptosis by targeting
Bcl-2 and SP1 in esophageal carcinoma cells (40); Wu et al reported that miR-16
inhibits cell proliferation, invasion and metastasis of HepG2 cells
through upregulation of Bax, downregulation of Bcl-2, and decrease
of MMP-2 and MMP-9 (41). In the
present study, we demonstrated that miR-15a suppresses the
expression of Bcl-2 at the post-transcriptional level and partially
abolishes the hypoxia-induced cell invasion and migration (Figs. 2 and 4). Since the miR-15a and Bcl-2 mRNA
sequences share a complementary homology, we demonstrated that
Bcl-2 is a post-transcriptional target of miR-15a tumor suppressor,
and miR-15a directly binds to the 3′UTR of Bcl-2 mRNA under hypoxia
(Fig. 5). These results suggest
that the interaction between miR-15a and Bcl-2 plays a pivotal role
in hypoxia-induced MG63 cell invasion and migration. Our findings
are consistent with those of Wang et al and Wu et
al.
miR-15a has been recognized as one of the most
important miRNAs in multiple human tissues, and has been identified
as downregulated or deleted in several types of cancer; however,
little is known on its transcriptional regulation. Since DLEU2 is
the host gene of miR-15a, the level of mature miR-15a is associated
with the transcription of DLEU2. Lerner et al reported that
DLEU2 overexpression blocks cellular proliferation and inhibits the
colony-forming ability of tumor cells in a
miR-15a/miR-16-1-dependent manner (42); Xue et al reported that
hypoxia promotes the heterodimerization of c-Myc/Max and
downregulates the transcription of DLEU2/miR-15-16, which enhances
angiogenesis and tumor metastasis (Fig. 6, left panel) (34). In the present study, we observed a
similar phenomenon, in that hypoxia decreases the transcription of
DLEU2/miR-15a. However, in our study, the target gene of miR-15a,
which regulates cell invasion and migration, was Bcl-2 rather than
FGF2 (Fig. 6, right panel).
Moreover, the mechanism through which hypoxia stimulates the
transcription of DLEU2 and whether HIF is involved in this process
must be fully elucidated through further investigation.
In conclusion, our study demonstrated that low
concentration of oxygen represses the transcription of the DLEU2
gene, decreases the level of miR-15a, and promotes MG63 cell
invasion and migration. Overexpression of miR-15a represses cell
migration and invasion via the regulation of Bcl-2 and MMPs. Our
findings indicate that miR-15a may be a valuable target for the
treatment of osteosarcoma, particularly in patients with high-grade
cancer or heavy tumor burden. Of note, the present study is a
preliminary study, and these results are based on an in vitro cell
model. In order to obtain more solid experimental data, an in vivo
investigation using an osteosarcoma-bearing nude mice model is
required.
Acknowledgments
Not applicable.
Notes
[1]
Funding
This study was supported in part by grants from the
National Science Foundation of China (no. 81670221 to Yuguang
Zhao).
[2] Availability
of data and materials
The analysed data sets generated during the study
are available from the corresponding authors on reasonable
request.
[3] Authors'
contributions
ZW contributed to the design of the study and wrote
the manuscript. JL performed the experiments. QS analyzed the data.
YZ helped perform the analysis with constructive discussions. All
authors have read and approved this manuscript.
[4] Ethics
approval and consent to participate
Not applicable.
[5] Consent for
publication
Not applicable.
[6] Competing
interests
The authors declare that they have no competing
interests.
References
|
1
|
Berner K, Johannesen TB and Bruland OS:
Clinical epidemiology of low-grade and dedifferentiated
osteosarcoma in Norway during 1975 and 2009. Sarcoma.
2015:9176792015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Longhi A, Errani C, De Paolis M, Mercuri M
and Bacci G: Primary bone osteosarcoma in the pediatric age: State
of the art. Cancer Treat Rev. 32:423–436. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Anderson ME: Update on survival in
osteosarcoma. Orthop Clin North Am. 47:283–292. 2016. View Article : Google Scholar
|
|
4
|
Morrow JJ and Khanna C: Osteosarcoma
genetics and epigenetics: Emerging biology and candidate therapies.
Crit Rev Oncog. 20:173–197. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Van Wynsberghe PM, Chan SP, Slack FJ and
Pasquinelli AE: Analysis of microRNA expression and function.
Methods Cell Biol. 106:219–252. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Farh KK, Grimson A, Jan C, Lewis BP,
Johnston WK, Lim LP, Burge CB and Bartel DP: The widespread impact
of mammalian MicroRNAs on mRNA repression and evolution. Science.
310:1817–1821. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mendell JT: MicroRNAs: Critical regulators
of development, cellular physiology and malignancy. Cell Cycle.
4:1179–1184. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pourrajab F, Babaei Zarch M, BaghiYazdi M,
Hekmatimoghaddam S and Zare-Khormizi MR: MicroRNA-based system in
stem cell reprogramming; differentiation/dedifferentiation. Int J
Biochem Cell Biol. 55:318–328. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dumortier O, Hinault C and Van Obberghen
E: MicroRNAs and metabolism crosstalk in energy homeostasis. Cell
Metab. 18:312–324. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Frankel LB and Lund AH: MicroRNA
regulation of autophagy. Carcinogenesis. 33:2018–2025. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tian X, Zhang J, Yan L, Dong JM and Guo Q:
miRNA-15a inhibits proliferation, migration and invasion by
targeting TNFAIP1 in human osteosarcoma cells. Int J Clin Exp
Pathol. 8:6442–6449. 2015.PubMed/NCBI
|
|
12
|
Calin GA, Dumitru CD, Shimizu M, Bichi R,
Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al:
Frequent deletions and down-regulation of micro- RNA genes miR15
and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad
Sci USA. 99:15524–15529. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bonci D, Coppola V, Musumeci M, Addario A,
Giuffrida R, Memeo L, D'Urso L, Pagliuca A, Biffoni M, Labbaye C,
et al: The miR-15a-miR-16-1 cluster controls prostate cancer by
targeting multiple oncogenic activities. Nat Med. 14:1271–1277.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Aqeilan RI, Calin GA and Croce CM: miR-15a
and miR-16-1 in cancer: Discovery, function and future
perspectives. Cell Death Differ. 17:215–220. 2010. View Article : Google Scholar
|
|
15
|
Luo Q, Li X, Li J, Kong X, Zhang J, Chen
L, Huang Y and Fang L: miR-15a is underexpressed and inhibits the
cell cycle by targeting CCNE1 in breast cancer. Int J Oncol.
43:1212–1218. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shi L, Jackstadt R, Siemens H, Li H,
Kirchner T and Hermeking H: p53-induced miR-15a/16-1 and AP4 form a
double-negative feedback loop to regulate epithelial-mesenchymal
transition and metastasis in colorectal cancer. Cancer Res.
74:532–542. 2014. View Article : Google Scholar
|
|
17
|
Bandi N and Vassella E: miR-34a and
miR-15a/16 are co-regulated in non-small cell lung cancer and
control cell cycle progression in a synergistic and Rb-dependent
manner. Mol Cancer. 10:552011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang H, Li L, Liu F, Zhang WY, Wei YZ, Wu
XJ, Xi YY, Zhou YR, Chen HX and Lin YL: Proliferation, migration
and invasion of cervical cancer cells. Xiandai Shengwu Yixue
Jinzhan. 16:2801–2805. 2016.In Chinese.
|
|
19
|
Liu G, Li Y and Gao XG: microRNA-181a is
upregulated in human atherosclerosis plaques and involves in the
oxidative stress-induced endothelial cell dysfunction through
direct targeting Bcl-2. Eur Rev Med Pharmacol Sci. 20:3092–3100.
2016.PubMed/NCBI
|
|
20
|
Zhu DX, Miao KR, Zhu YD, Zhu HY, Fan L,
Liu P, Xu W and Li JY: Detection of miRNA levels in leukemia
patients by real-time quantitative PCR. Zhongguo Shi Yan Xue Ye Xue
Za Zhi. 18:757–761. 2010.In Chinese. PubMed/NCBI
|
|
21
|
Cimmino A, Calin GA, Fabbri M, Iorio MV,
Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et
al: miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl
Acad Sci USA. 102:13944–13949. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu MZ, Chen SF, Nieh S, Benner C, Ger LP,
Jan CI, Ma L, Chen CH, Hishida T, Chang HT, et al: Hypoxia drives
breast tumor malignancy through a TET-TNFα-p38-MAPK signaling axis.
Cancer Res. 75:3912–3924. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Deacon K, Onion D, Kumari R, Watson SA and
Knox AJ: Elevated SP-1 transcription factor expression and activity
drives basal and hypoxia-induced vascular endothelial growth factor
(VEGF) expression in non-small cell lung cancer. J Biol Chem.
287:39967–39981. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hartmann A, Kunz M, Köstlin S, Gillitzer
R, Toksoy A, Bröcker EB and Klein CE: Hypoxia-induced up-regulation
of angiogenin in human malignant melanoma. Cancer Res.
59:1578–1583. 1999.PubMed/NCBI
|
|
25
|
Timani KA, Liu Y, Fan Y, Mohammad KS and
He JJ: Tip110 regulates the cross talk between p53 and
hypoxia-inducible factor 1α under hypoxia and promotes survival of
cancer cells. Mol Cell Biol. 35:2254–2264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu L, Salnikov AV, Bauer N,
Aleksandrowicz E, Labsch S, Nwaeburu C, Mattern J, Gladkich J,
Schemmer P, Werner J, et al: Triptolide reverses hypoxia-induced
epithelial-mesenchymal transition and stem-like features in
pancreatic cancer by NF-κB downregulation. Int J Cancer.
134:2489–2503. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shimizu S, Eguchi Y, Kosaka H, Kamiike W,
Matsuda H and Tsujimoto Y: Prevention of hypoxia-induced cell death
by Bcl-2 and Bcl-xL. Nature. 374:811–813. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kasar S, Salerno E, Yuan Y, Underbayev C,
Vollenweider D, Laurindo MF, Fernandes H, Bonci D, Addario A,
Mazzella F, et al: Systemic in vivo lentiviral delivery of
miR-15a/16 reduces malignancy in the NZB de novo mouse model of
chronic lymphocytic leukemia. Genes Immun. 13:109–119. 2012.
View Article : Google Scholar :
|
|
29
|
Dai L, Wang W, Zhang S, Jiang Q, Wang R,
Dai L, Cheng L, Yang Y, Wei YQ and Deng HX: Vector-based
miR-15a/16-1 plasmid inhibits colon cancer growth in vivo. Cell
Biol Int. 36:765–770. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shen G, Li X, Jia YF, Piazza GA and Xi Y:
Hypoxia-regulated microRNAs in human cancer. Acta Pharmacol Sin.
34:336–341. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zuo J, Wen M, Lei M, Peng X, Yang X and
Liu Z: miR-210 links hypoxia with cell proliferation regulation in
human Laryngocarcinoma cancer. J Cell Biochem. 116:1039–1049. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li W, Zhang X, Zhuang H, Chen HG, Chen Y,
Tian W, Wu W, Li Y, Wang S, Zhang L, et al: MicroRNA-137 is a novel
hypoxia-responsive microRNA that inhibits mitophagy via regulation
of two mitophagy receptors FUNDC1 and NIX. J Biol Chem.
289:10691–10701. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang Y, Ding S, Xu G, Chen F and Ding F:
MicroRNA-15a inhibition protects against
hypoxia/reoxygenation-induced apoptosis of cardiomyocytes by
targeting mothers against decapentaplegic homolog 7. Mol Med Rep.
15:3699–3705. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xue G, Yan HL, Zhang Y, Hao LQ, Zhu XT,
Mei Q and Sun SH: c-Myc-mediated repression of miR-15-16 in hypoxia
is induced by increased HIF-2α and promotes tumor angiogenesis and
metastasis by upregulating FGF2. Oncogene. 34:1393–1406. 2015.
View Article : Google Scholar
|
|
35
|
Reed JC: Regulation of apoptosis by bcl-2
family proteins and its role in cancer and chemoresistance. Curr
Opin Oncol. 7:541–546. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pinkas J, Martin SS and Leder P:
Bcl-2-mediated cell survival promotes metastasis of EpH4 betaMEKDD
mammary epithelial cells. Mol Cancer Res. 2:551–556.
2004.PubMed/NCBI
|
|
37
|
Planas-Silva MD, Bruggeman RD, Grenko RT
and Smith JS: Overexpression of c-Myc and Bcl-2 during progression
and distant metastasis of hormone-treated breast cancer. Exp Mol
Pathol. 82:85–90. 2007. View Article : Google Scholar
|
|
38
|
Ricca A, Biroccio A, Del Bufalo D, Mackay
AR, Santoni A and Cippitelli M: bcl-2 over-expression enhances
NF-kappaB activity and induces mmp-9 transcription in human
MCF7(ADR) breast-cancer cells. Int J Cancer. 86:188–196. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang Y, Li M, Zang W, Ma Y, Wang N, Li P,
Wang T and Zhao G: miR-429 up-regulation induces apoptosis and
suppresses invasion by targeting Bcl-2 and SP-1 in esophageal
carcinoma. Cell Oncol (Dordr). 36:385–394. 2013. View Article : Google Scholar
|
|
41
|
Wu WL, Wang WY, Yao WQ and Li GD:
Suppressive effects of microRNA-16 on the proliferation, invasion
and metastasis of hepatocellular carcinoma cells. Int J Mol Med.
36:1713–1719. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lerner M, Harada M, Lovén J, Castro J,
Davis Z, Oscier D, Henriksson M, Sangfelt O, Grandér D and Corcoran
MM: DLEU2, frequently deleted in malignancy, functions as a
critical host gene of the cell cycle inhibitory microRNAs miR-15a
and miR-16-1. Exp Cell Res. 315:2941–2952. 2009. View Article : Google Scholar : PubMed/NCBI
|