Introduction
Gallbladder cancer (GBC) is the one of the highest
incidence and a particularly aggressive biliary tract cancer
(1). Epidemiologically, it tends
to be more commonly found in women, with high rates of recurrence
and mortality, which is reflected by it having the worst overall
survival (OS) rate among all other biliary tract cancer types
(2-6). In 2018, ~219,420 new cases and
165,087 cases of mortality due to GBC were reported globally
(7). Although surgical treatment
is currently the most effective method for treating GBC, the
majority of patients with GBC are typically already at metastatic
or late clinical stages at diagnosis on presentation, limiting the
curative treatment options (8-10).
Therefore, it remains in high demand to develop novel precise
targeted treatment methods to inhibit the progression of GBC whilst
also improving the prognosis of treatment strategies. The diverse
characteristics of GBC pathogenesis leads to poor prognoses
according to a previous gene set enrichment analysis, specifically
due to epithelial-mesenchymal transition (EMT), angiogenic immune
suppression and hypoxia (11).
According to a previous Kyoto Encyclopedia of Genes and Genomes
analysis, overactivation of the TGF-β pathway in GBC can lead to
significantly poorer survival outcomes (11,12).
However, the detailed mechanism underlying EMT and the TGF-β
pathway in GBC require further study.
Members of the B7/CD28 family have been previously
found to regulate the immune response and cancer invasion, in both
positive and negative manners (13). Human endogenous retrovirus-H long
terminal repeat-associating protein 2 (HHLA2; also known as B7H7 or
B7H5) is the only member of the B7 protein family found in humans
(14). HHLA2 cannot be found in
mice and is mainly expressed in human breast, lung, thyroid,
melanoma, pancreas, ovary, liver, bladder, colon, prostate, kidney
and esophagus cancer cells, activated myeloid cells,
monocyte-derived macrophages and dendritic cells (14). Previous studies have found that
HHLA2 can activate antigen-presenting cells in response to
inflammation (14,15). In addition, HHLA2 interacts with
the co-receptor CD28H to stimulate T cell proliferation,
differentiation and the production of cytokines, including IFN-γ,
IL-2 and IL-10 (14,15). However, HHLA2 can also bind to
another inhibitory receptor, three immunoglobulin domains and long
cytoplasmic tail 3 (KIR3DL3), which inhibits T cell action
(16). In terms of cancer, higher
expression levels of HHLA2 were previously reported to be
associated with poorer prognoses or higher degrees of tumour
invasion in lung carcinoma, hepatocellular carcinoma and prostate
cancer (17-19). By contrast, in epithelial ovarian
cancer, higher HHLA2 expression levels were demonstrated to be
associated with superior survival outcomes and reduced tumor growth
(20). Therefore, HHLA2 can serve
opposite physiological roles in cancer depending on the malignancy
of interest. However, the expression profile of HHLA2 and its
relationship with GBC progression remain unclear.
Long noncoding RNAs (lncRNAs) are non-coding
transcripts that are >200 bp in length and server important
roles in regulating cellular protein expression and function
(21). LncRNAs have also been
previously found regulate the progression of various cancers.
LncRNA H19 (H19) is one such lncRNA, which was one of the earliest
discovered lncRNAs (22,23). H19 has been observed to serve a
significant role in EMT progression in colorectal cancer, breast
cancer and gallbladder cancer (22,23).
Furthermore, previous studies have reported that H19 can function
as a molecular sponge to restrain the functions of miR-342-3p and
promote EMT in GBC (24,25). In the present study, the role of
H19-induced upregulation on HHLA2 expression in the molecular
regulatory network underlying EMT in GBC was investigated.
Additionally, in the present study, the aim was to
investigate the role of HHLA2 in GBC progression and to further
evaluate the potential of HHLA2 as a potentially novel therapeutic
target.
Materials and methods
Patients and specimens
GBC specimens were collected from patients at
Shengjing Hospital of China Medical University (Shenyang, China)
from October 20, 2011 to December 10, 2020. Patients diagnosed with
GBC and confirmed by pathologists according to the American Joint
Committee on Cancer (AJCC) cancer staging system (8th edition)
(26) and the 2019 (5th edition)
WHO classification of tumours of the digestive system (27), who underwent surgery as the primary
treatment and with comprehensive clinical and follow-up data, were
included. Patients with preoperative chemotherapy, other concurrent
malignant tumors, unsuccessful follow-up or those whose survival
data cannot be obtained were excluded from this study. A total of
89 patients were selected from our patient database. Among these
patients, 37 were males (42%) and 52 were females (58%). The mean
age was 62.34±10.19 years, ranging from 30 to 89 years. This
research arm of the present study was approved by the Ethics
Committee of Shengjing Hospital of China Medical University
(approval no. 2019PS036K). Written consent was obtained from all
the patients involved in the present study. Each patient's age,
sex, differentiation, Nevin stage (28), tumor invasion, regional lymph node
metastasis, distant metastasis and histological stage were
obtained.
Follow-up arrangement
The final date of the last follow-up was November 1,
2021. OS was defined as the interval either between the initial
diagnosis and mortality or between the initial diagnosis and the
date of the last observation for the surviving patients. Data were
censored at the date of the last follow-up for any living
patients.
Immunohistochemical staining and
evaluation
HHLA2 expression in the GBC specimens was measured
by immunohistochemistry. The GBC specimens were fixed in a 10%
formalin solution for 24 h at room temperature, before they were
embedded in paraffin, sliced into 4 µm-thick sections and
placed onto glass slides. They were then dehydrated in an ascending
ethanol gradient and cleared with xylene. The slices were treated
with 95°C Sodium Citrate Antigen Retrieval Solution (cat. no.
C1031; Beijing Solarbio Science & Technology Co., Ltd.) for 10
min. The slices were then treated with 3%
H2O2 at 37°C for 10 min and blocked in 3% BSA
(Beyotime Institute of Biotechnology) solution for 30 min at room
temperature. Immunohistochemical staining were subsequently
performed. Rabbit anti-HHLA2 monoclonal antibodies (1:100, cat. no.
ab214327; Abcam) were used as the primary antibody and incubated at
4°C for 10 h, whereas HRP-conjugated goat anti-rabbit IgG was used
as the secondary antibody (1:100, cat. no. ab288151; Abcam) and
incubated at room temperature for 30 min. Diaminobenzidine (cat.
no. DA1010; Beijing Solarbio Science & Technology Co., Ltd.)
incubating for 5 min at room temperature was used to develop color
reaction. Paraffin sections were counterstained with Haematoxylin
(Beijing Solarbio Science & Technology Co., Ltd.) lasting for 2
min at room temperature.
Immunohistochemical staining results were assessed
by two different pathologists using light microscopy
(magnifications, ×200 and ×400; Olympus Corporation). In cases
where there was disagreement, the results were evaluated again. If
a consensus could not be reached, another pathologist participated
in the evaluation. All pathologists were blinded to the patient's
clinical data. In summary, ≥ two pathologists must agree with each
result. Immunostaining in the tumor tissues was semi-quantified
using the ImageJ software (version 1.52; National Institutes of
Health), for the staining intensity. The staining intensity was
divided into four levels as follows: i) Negative, 0; ii) low
positive, 1; iii) positive, 2; and iv) high positive, 3. In total,
five high-power fields (magnification, ×400) of each slice were
randomly selected to judge the corresponding percentage of positive
cells. The H-score of each case was measured by multiplying the
staining intensity by the corresponding percentage of positive
cells (range from 0 to 168). To associate the relationship between
HHLA2 and patient outcome, the existence of tumor classifications
should be revealed. Since H-score was determined to be continuous
variables, the optimal cut-off value was determined using X-tile
software (version 3.6.1; https://medicine.yale.edu/lab/rimm/research/software/)
to divide the specimens into HHLA2-high group and the HHLA2-low
group. The X-tile program performed a single, global assessment of
every possible method of dividing the population. In the
assessment, each H-score value of all the specimens was used as a
cut-off value to divide the specimens into two groups.
Subsequently, every χ2 value of each grouping grouped by
different H-score values were calculated using the Kaplan-Meier
method respectively to find the highest χ2 value, which
was 7.16 (ranging from 0.40 to 7.16). The grouping with a
χ2 value of 7.16 was regarded as the optimal grouping,
where the cut-off value of the optimal grouping was determined as
the optimal cut-off value, which was 80 (29). Therefore, specimens with H-score
>80 were included in the HHLA2-high group and specimens with
H-score ≤80 specimens were included in the HHLA2-low group.
Cell culture
The GBC cell line GBC-SD (cat. no. CSTR:19375
.09.3101HUMTCHu16) was purchased from The Cell Bank of Type Culture
Collection of the Chinese Academy of Sciences. The cells were
cultured in RPMI1640 medium (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.)
and were treated with or without TGF-β1 (PeproTech, Inc). The
TGF-β1 cytokine was dissolved in PBS containing 5% Trehalose as
storage solution at a concentration of 20 µg/ml. The storage
solution of TGF-β1 was added into the culture medium to reach a
concentration of 10 ng/ml for treatment for 48 h at 37°C, whilst
the control group received the same amount of vehicle treatment
(PBS containing 5% Trehalose). For culturing, 5% CO2 and
37°C was applied for the cell lines.
Western blotting
The cells were lysed with RIPA buffer (Beyotime
Institute of Biotechnology) for 2 h on ice with a protein inhibitor
cocktail (Beyotime Institute of Biotechnology). This mixture was
then centrifuged at 12,000 × g for 30 min at 4°C, before the
supernatant was collected and the protein concentration was
measured using a BCA kit (Beyotime Institute of Biotechnology). In
total, 5X loading buffer was added to the protein solution at a 1:4
ratio and heated at 95°C for 5 min. After SDS-PAGE in 5% stacking
gel and in 10% separating gel (15 µg protein per lane) and
transfer onto polyvinyl difluoride membranes (Thermo Fisher
Scientific, Inc.), before blocked with 5% skimmed milk powder
solution (diluted in Tris-buffered saline containing 0.1% Tween-20)
for 2 h under room temperature. After blocking, the membranes were
incubated with the corresponding primary antibodies against GAPDH
(1:1,000; cat. no. ab181602; Abcam), vimentin (1:1,000; Abcam; cat.
no. ab92547), α-smooth muscle actin (SMA; 1:1,000; Abcam; cat. no.
ab124964), HHLA2 (1:1,000; Abcam; cat. no. ab214327), E-cadherin
(1:1,000; Abcam; cat. no. ab40772), N-cadherin (1:1,000; Abcam;
cat. no. ab18203) and Collagen I (Col-I; 1:1,000; Abcam; cat. no.
ab34710) for 10 h at 4 °C. After incubating with the primary
antibodies, the membranes were incubated with HRP-conjugated goat
anti-rabbit IgG secondary antibodies (1:5,000; cat. no. ab6721;
Abcam) for 1 h at room temperature. Protein expression levels were
detected by Chemistar™ High-sig ECL Western Blotting substrate
(cat. no. 180-5001; Tanon Science and Technology Co., Ltd.). The
western blotting results were quantified using the ImageJ software
(version 1.52; National Institutes of Health).
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated using RNAiso Plus (Takara
Bio, Inc.) according to the manufacturer's protocol and reverse
transcribed into cDNA using the PrimeScript™ RT Master Mix kit
(Takara Bio, Inc.). The temperature protocol was 37°C for 15 min,
85°C for 5 sec and 4°C for 1 h. qPCR was performed using the TB
Green™ Premix Ex Taq™ II (Takara Bio, Inc.) and a Roche 96
instrument (Roche Diagnostics) to measure mRNA expression. The
thermocycling conditions were as follows: Initial denaturation at
95°C for 30 sec; followed by 40 cycles at 95°C for 5 sec and 60°C
for 30 sec; Relative mRNA expression levels were determined using
the 2−ΔΔCq method (30), where the experiments were repeated
three times. GAPDH was used as the internal control. The GAPDH
primers were purchased from Guangzhou RiboBio Co., Ltd. The
sequences for GAPDH primers were: Forward, 5′-GAA CGG GAA GCT CAC
TGG-3′, and reverse, 5′-GCC TGC TTC ACC ACC TTC T-3′, The sequences
of the H19 primers were H19-forward, 5′-CGT GAC AAG CAG GAC ATG
ACA-3′ and reverse, 5′-CCA TAG TGT GCC GAC TCC G-3′.
Immunofluorescence staining
Cultured cells were first fixed in 4%
paraformaldehyde at room temperature for 30 min, before they were
blocked with 5% BSA (Beyotime Institute of Biotechnology) at room
temperature for 1 h and permeabilized with 0.1% Triton X-100 at
room temperature for 15 min. Next, the cells were incubated with
1:200 diluted primary antibodies against vimentin (cat. no.
ab92547; Abcam), α-smooth muscle actin (cat. no. ab124964; Abcam),
HHLA2 (cat. no. ab214327; Abcam), E-cadherin (cat. no. ab40772;
Abcam), N-cadherin (cat. no. ab18203; Abcam) and Col-I (cat. no.
ab34710; Abcam) for 10 h at 4°C. After washing three times with PBS
containing 0.1% Tween-20 (PBST), the cells were incubated in 1:200
diluted Goat anti-Rabbit IgG (H+L) Highly Crossed-Adsorbed
Secondary Antibody, Alexa Fluor™ 488 (cat. no. A-11034; Thermo
Fisher Scientific, Inc.) for 1 h at room temperature, protected
from light. DAPI (Thermo Fisher Scientific, Inc.) at the
concentration of 300 nM was used to stain nuclei for 5 min at room
temperature. Protein expression was observed and recorded by
fluorescence microscopy (magnification, ×200; Olympus
Corporation).
Construction of stably transfected cell
lines
Lentiviruses encoding the short hairpin (sh)RNA
silencing HHLA2 and non-target shRNA, the HHLA2 (GenBank accession
no. NM_007072) or H19 (GenBank accession no. NR_ (NR_002196)
overexpression vector and empty control lentiviral vector were
designed and synthesized by Shanghai GeneChem Co., Ltd. The
overexpression vector was sent for sequencing and designated GV348
[Ubi-MCS-SV40-Puromycin]. The knockdown vector was sent for
sequencing and designated GV112 [hU6-MCS-CMV-Puromycin]. In brief,
the 20 µg GV348 vectors were co-transfected with 15
µg pHelper 1.0 (envelope plasmid) and 10 µg pHelper
2.0 (packaging plasmid) in the 2nd generation transfection system
into 293T cells (The Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences) cultured in DMEM (Hyclone; Cytiva)
with 10% FBS (Hyclone; Cytiva) at 37°C with 5% CO2.
After 48 h culturing at 37°C, the supernatant of the 293T cells
were collected and the lentivirus was concentrated through
centrifugation of 25,000 g at 4°C for 2 h. Subsequently, the GBC-SD
cell lines were plated into six-well plates until reaching 80%
confluence, before the lentivirus was added and co-cultured with
the cells for 24 h at 37°C (multiplicity of infection, 10). The
medium was then replaced prior to another 48 h of incubation in the
5% CO2 and 37°C incubator. Subsequently, 25 µg/ml
puromycin treatment for 2 weeks at 37°C was used to select for the
stably transfected cell lines before the knockdown efficiency was
confirmed by western blotting or RT-qPCR after the cell line
reached a confluence of 80%. In the rescue experiments, HHLA2-shRNA
or NC-shRNA was transfected into H19-overexpressing GBC-SD cells.
In total, 24 h after transfection, the cells were harvested for
subsequent experiments. The target sequence of shRNA for HHLA2 was
5′-ATC CCG ATT CTC ATG GAA CAA-3′, whereas the target sequence of
NC shRNA was 5′-TTC TCC GAA CGT GTC ACG T-3′.
Wound healing assay
The GBC-SD cells were seeded into six-well plates,
and cultured until 100% confluence. A 20-µl sterile yellow
plastic pipette tip was then used to scratch the cell monolayer.
After 48 h of incubation with or without 10 ng/ml TGF-β1 at 37°C
with serum-free RPMI1640 medium, the scratches were imaged by light
microscopy (magnification, ×200) and analysed by ImageJ software
(version 1.52; National Institutes of Health). Migration area was
calculated by multiplying the migration distance (scratch width
observed at 0 h-scratch width observed at 48 h) with the width of
field of view (500 µm). Relative migration area was
calculated by dividing the migration area by the migration area of
each control group.
Transwell assay
Transwell (Corning, Inc.) chambers were fist
pre-coated at 37°C for 30 min with Matrigel (cat. no. 356234; BD
Biosciences) that was diluted at 1:8 with serum-free RPMI1640
medium. In total of 2×104 GBC-SD cells suspended with
serum-free RPMI-1640 medium with or without 10 ng/ml TGF-β1 were
seeded into the upper surfaces of each Transwell chambers, whilst
the lower chambers were added with RPMI-1640 medium with 10% FBS.
while the lower chambers were filled with culture medium with 10%
FBS. After 48 h invasion at 37°C, the cells that had not passed
through the chamber membrane were gently removed with a wet cotton
ball. The membranes were then fixed with 4% paraformaldehyde at
room temperature for 30 min and stained with 0.1% crystal violet at
room temperature for 15 min and photographed under a light
microscope (magnification, ×200). In total, five high-power fields
(magnification, ×200) of each membrane were randomly selected for
calculating the cell numbers by ImageJ software (version 1.52;
National Institutes of Health).
EdU assay
An EdU assay was performed to evaluate the
proliferation levels of the cells using Cell-Light Edu Apollo 488
In Vitro Kit (cat. no. C10310-3; Guangzhou RiboBio Co.,
Ltd.). Briefly, GBC-SD cells were labelled with 50 µM EdU
(100 µl/well) for 2 h in a 5% CO2 and 37°C
incubator. After fixation with 4% paraformaldehyde at room
temperature for 30 min, the cells were permeabilized with 0.5%
Triton X-100 at room temperature for 10 min. Next, the cells were
incubated with 1X Apollo staining solution of the EdU kit at room
temperature for 30 min before the nuclei were stained with DAPI
(Thermo Fisher Scientific, Inc.) at the concentration of 300 nM for
5 min at room temperature. The cells were observed and imaged by
fluorescence microscopy (magnification, ×200).
Xenograft mouse model
The mouse xenograft experimental protocol was
approved by the Ethics Committee of Shengjing Hospital of China
Medical University (approval no. 2019PS325K) and performed in
accordance with the Laboratory Animal Guideline for Ethical Review
of Animal Welfare (31). All mice
were housed in an animal room at a constant temperature of 25°C
with 30-40% humidity, under a 12-h light/dark cycle and had free
access to food and water.
A total of 24 6-week-old female BALB/c nude mice
(weight, 19-22 g) were used to establish the GBC xenograft models.
The mice were randomly divided into the HHLA2 overexpression group,
NC overexpression group, HHLA2-shRNA group and NC-shRNA group, with
6 mice in each group. After the cells in each group reached a
confluence of 80-90%, they were digested with 0.25% trypsin. Next,
the cells were resuspended in PBS to form a cellular suspension at
a density of 1×106 cells in 100 µl. Under
isoflurane inhalation anaesthesia (1-2%), each mouse was gradually
and slowly inoculated subcutaneously with 100 µl this cell
suspension in the right armpit. The health and behaviour of the
mice were monitored once a day to determine if there were
difficulties eating or drinking, symptoms of pain or long-term
problems without recovery. If the tumor volume was found to be
>2,000 mm3, the animal would be euthanized as a
humane endpoint. In total, 18 days after inoculation, all mice were
sacrificed by cervical dislocation after isoflurane inhalation (5%
for 5 min). Cardiac arrest was then used to verify death by pulse
palpation. Tumor tissues were collected for measurement and
analysis of the volume and weight. The maximum tumor volume
observed was 1,680 mm3 and the maximum tumour diameter
was 15 mm.
Statistical analysis
All count data were analysed using the χ2
test, continuity correction χ2 test or Fisher's exact
test. Survival differences were compared using the Kaplan-Meier
method and log-rank test. Multivariate and univariate analyses were
performed using a Cox stepwise proportional hazard model. Data for
experiments performed in triplicate are expressed as the mean ±
standard deviation. Comparisons between two groups were made using
the unpaired Student's t-test. Differences among groups were tested
using one-way analysis of variance (ANOVA) and Tukey's post hoc
test. SPSS software version 26.0 (IBM Corp.) or GraphPad Prism 8.0
(GraphPad Software, Inc.) was used to perform the statistical
analysis. P<0.05 was considered to indicate a statistically
significant difference.
Results
HHLA2 is upregulated in progressed
gallbladder cancer, and high expression of HHLA2 is associated with
a poor prognosis
The basic clinical characteristics of 89 patients
with GBC are summarized in Table
I. In total, 37 (42%) of the patients with GBC had a lower
Nevin staging of I-III, whereas 58% (n=52) had a high grade (IV-V).
According to the AJCC 8th edition cancer staging system, the tumour
invasions were classified into two groups, namely the low group
(stage I-II; 36 cases; 40%) and the high group (stage III-IV; 53
cases; 60%). Tumour invasions were also classified into two groups,
where the low group included T1 and T2 (44 cases; 49%) and the high
group included T3 and T4 (45 cases; 51%). In terms of regional
lymph node metastasis, 31 (34%) patients had regional lymph node
metastasis, whilst 46 (52%) patients did not. For distant
metastasis, 28 (31%) patients had distant metastasis (to the liver,
intestine and peritoneum), whereas 61 (69%) patients did not. At
the end of the follow-up period, 50 patients had succumbed to the
disease, where the median follow-up duration was 70 months (range,
1 to 114).
 | Table IRelationship between HHLA2 expression
and clinical characteristics in patients with gall bladder cancer
(N=89). |
Table I
Relationship between HHLA2 expression
and clinical characteristics in patients with gall bladder cancer
(N=89).
| Parameter | Total | HHLA2 expression
| P-value |
|---|
| Low | High |
|---|
| Patients (n) | 89 | 44 | 45 | |
| Sex | | | | 0.244 |
| Male | 37 | 21 | 16 | |
| Female | 52 | 23 | 29 | |
| Age (years) | | | | 0.899 |
| <62 | 49 | 24 | 25 | |
| ≥62 | 40 | 20 | 20 | |
|
Differentiation | | | | 0.219 |
| Poorly or
moderately | 21 | 13 | 8 | |
| Well | 48 | 22 | 26 | |
| Nevin stage | | | | 0.014 |
| I-III | 37 | 24 | 13 | |
| IV-V | 52 | 20 | 32 | |
| American Joint
Committee on Cancer stage | | | | 0.007 |
| I-II | 36 | 24 | 12 | |
| III-IV | 53 | 20 | 33 | |
| Tumor invasion | | | | |
| T1-T2 | 44 | 27 | 17 | 0.026 |
| T3-T4 | 45 | 17 | 28 | |
| Regional node
metastasis | | | | |
| Absent | 46 | 29 | 17 | 0.018 |
| Present | 31 | 11 | 20 | |
| Distant
metastasis | | | | |
| Absent | 61 | 34 | 27 | 0.079 |
| Present | 28 | 10 | 18 | |
To detect the HHLA2 expression spectrum in the GBC
tissues, immunohistochemistry was performed. As shown in Fig. 1A, which shows a cross section of
the GBC tissue, HHLA2 was expressed in the membrane and/or
cytoplasm in GBC tissues and corresponding adjacent noncancerous
tissues. The intense staining was predominately seen in apical
surfaces of the papillary epithelium and luminal surfaces of the
glands (black arrows). Compared with tissues with low HHLA2
expression, HHLA2 high expression tissue showed higher staining
intensity and percentage of positively staining cells. Positive
HHLA2 staining was observed in all 89 cases (100%). To evaluate the
association between HHLA2 expression and various
clinicopathological parameters of GBC progression, the cases were
subdivided into two groups: HHLA2-high group (45 cases, 51%) and
the HHLA2-low group (44 cases, 49%; Fig. 1A and B). HHLA2 expression was found
to be positively associated with tumour progression (Fig. 1; Table
I). Specifically, 32 cases in the HHLA2-high group reached
Nevin stage IV-V, accounting for 71% (Fig. 1C). In addition, 33 cases in the
HHLA2-high group reached AJCC stage III-IV, accounting for 73%
(Fig. 1D), whilst 28 cases in the
HHLA2-high group exhibited tumor invasion T3-T4, accounting for 62%
(Fig. 1E). In terms of regional
lymph node metastasis, 20 cases in the HHLA2-high group were
demonstrated to be positive, accounting for 54% (Fig. 1F). According to Table I, higher HHLA2 expression was
significantly associated with Nevin stage, AJCC stage, tumour
invasion and regional lymph node metastasis.
Subsequently, univariate analysis of OS was
performed to analyse the clinical prognosis of the patients with
GBC. The OS of the HHLA2-high group was found to be shorter
compared with that in the HHLA2-low group (hazard ratio=2.15; 95%
CI=1.20-3.83; P=0.01), where patients with GBC and higher HHLA2
expression tended to have significantly shorter survival times
(Fig. 1G).
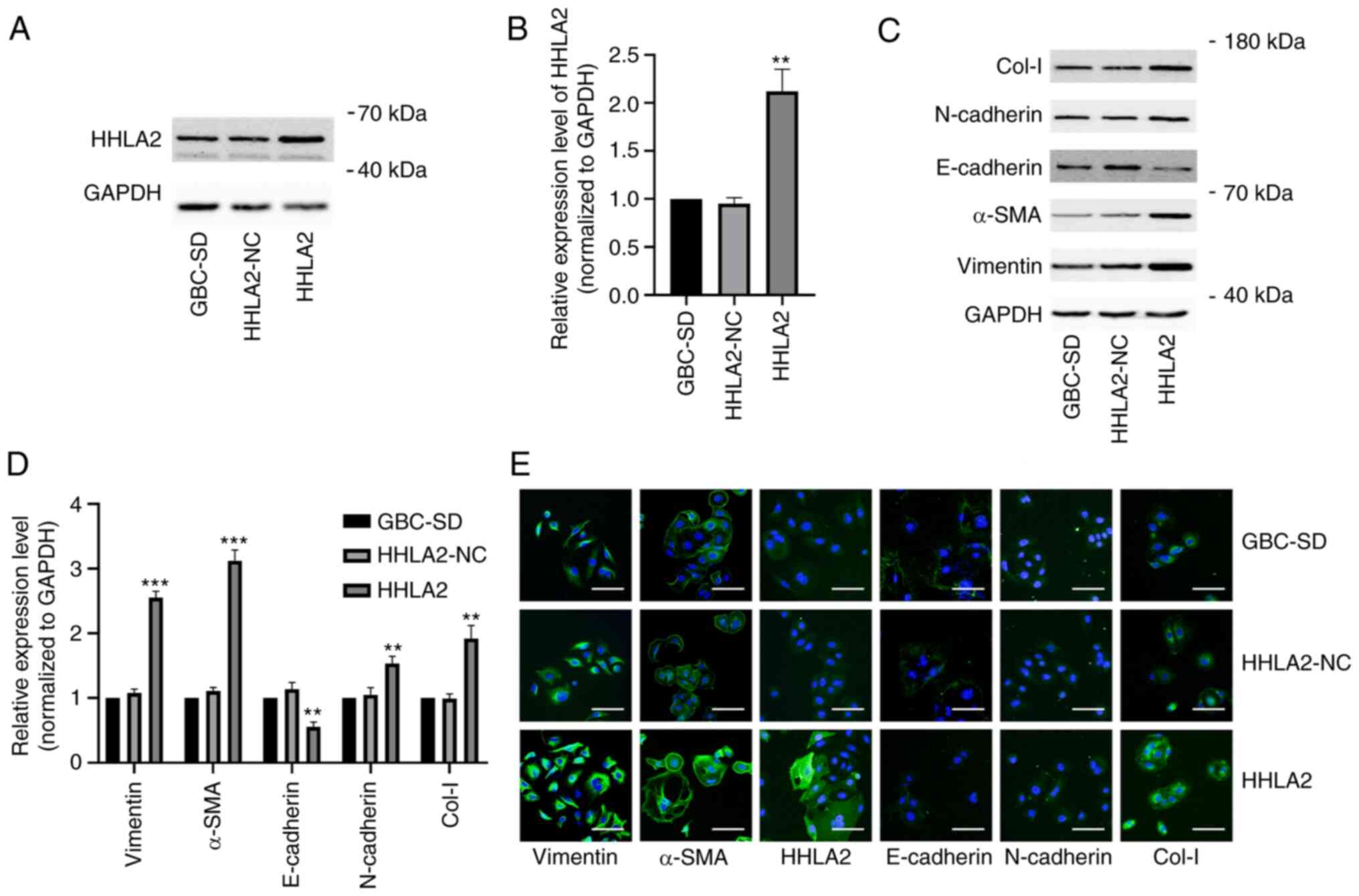
HHLA2 overexpression promotes EMT in GBC
in vitro
To further explore the function of HHLA2 in the
GBCs, the GBC-SD cell line were transfected with the HHLA2 plasmid
for overexpression. The efficiency of this HHLA2 overexpression in
GBC-SD cells was verified by western blot analysis (Fig. 2A and B). Subsequently, the
expression of EMT markers α-SMA, vimentin, N-cadherin and Col-I
were all significantly increased, whereas the expression of
E-cadherin was significantly decreased, in HHLA2-overexpressing
GBC-SD cells compared with those in the HHLA2-NC cells (Fig. 2C and D). The relative expression
levels of these markers were then observed through
immunofluorescence staining, which yielded similar observations
compared with those from western blotting. These results suggest
that HHLA2 can promote EMT in GBC (Fig. 2E).
HHLA2 exerts oncogenic functions in
GBC
Changes in the migratory ability of GBC-SD cells
after HHLA2 overexpression was next measured using wound-healing
assays. Overexpression of HHLA2 led to significantly more potent
migratory capabilities compared with those in the HHLA2-NC group
(Fig. 3A and B). The invasive
abilities of these HHLA2-overexpressing cells were next explored
through Transwell assays. The results demonstrated a significantly
higher invasive ability in the HHLA2-overexpressing cell line
compared with that in the HHLA2-NC group (Fig. 3C and D). An EdU assay was used to
investigate the proliferation of GBC-SD cells, which revealed that
there was a significant increase in the proliferation of the
HHLA2-overexpressing cell line compared with that in the HHLA2-NC
group (Fig. 3E and F). These
results suggest that HHLA2 can mediate oncogenic functions in GBC
in vitro.
 | Figure 3Overexpression of HHLA2 promotes the
proliferation, migration and invasion of GBC in vitro whilst
promoting tumor formation in vivo. (A) Wound healing assay
and (B) subsequent quantification showed that HHLA2 overexpression
enhanced cell migration. Scale bars, 100 µm. (C) HHLA2
overexpression was found to increase invasive capability, (D) which
was quantified. Scale bars, 100 µm. (E) The proliferative
abilities of the cells overexpressing HHLA2 were higher compared
with those of the HLLA2-NC group, (F) according to the
quantification. Scale bars, 100 µm. Green, EdU; Blue, DAPI.
(G) Representative images of mice in each group harboring the
tumors. (H) Overexpression of HHLA2 increases the diameters and the
weights of GBC tumors according to the mouse xenograft assay, (I)
which was qualified (n=6). **P<0.01 and
***P<0.001 (HHLA2 group vs. HHLA2-NC group). HHLA2,
human endogenous retrovirus-H long terminal repeat-associating
protein 2; GBC, gall bladder cancer; NC, negative control. |
To determine whether HHLA2 can regulate the
tumorigenic ability of GBC in vivo, a xenograft mouse tumor
model was established. HHLA2 is not expressed in the mouse genome
(14), negating the requirement
for measuring the protein or mRNA expression levels in these
samples prior to the establishment of this xenograft model. The GBC
cell line stably overexpressing HHLA2 and the negative control
group were injected into the left flanks of six nude mice in each
group. After 18 days, compared with those in the negative control
group, tumors in the HHLA2 overexpression group exhibited a
significantly larger volume and were heavier in weight (Fig. 3G-I). Taken together, these results
suggest that HHLA2 can promote the tumorigenic ability of GBC in
vivo.
HHLA2 knockdown impedes EMT in GBC
To determine if HHLA2 has the potential to become a
therapeutic target in GBC, GBC-SD cells were transfected with
HHLA2-shRNA to knock down HHLA2 expression in GBC-SD cells. The
knockdown efficiency was validated by western blotting (Fig. 4A and B). According to western
blotting and immunofluorescence staining data, HHLA2 knockdown led
to significant decreases in the expression of Col-I, N-cadherin,
α-SMA and vimentin but significant increases in the expression of
E-cadherin in GBC-SD cells (Fig.
4C-E). TGF-β1-induced EMT in vitro model was next
established (Fig. S1A and B).
There was a significant increase in the expression of HHLA2 in the
GBC cells (Fig. S1A and B), which
was reversed by transfection with HHLA2-shRNA. In addition,
transfection with HHLA2-shRNA significantly reduced the expression
of the EMT biomarkers compared with that in the TGF + sh-NC group
(Fig. S1C-E). These results
suggest that downregulation of HHLA2 can inhibit EMT in GBC both
alone and/or in the presence of TGF-β1.
 | Figure 4HHLA2 knockdown inhibits the EMT
process in GBC cells in vitro. (A) HHLA2 expression was
significantly decreased after transfection with the HHLA2 shRNA
(n=3), (B) according to the quantification. (C) Expression of EMT
markers Col-I, N-cadherin, α-SMA and vimentin were decreased whilst
that of E-cadherin was increased in GBC cells transfected with the
HHLA2 shRNA, (D) according to subsequent quantification. (E)
Immunofluorescence staining revealed similar results to western
blotting. Scale bars, 100 µm. Green, target protein; Blue,
DAPI.**P<0.01 and ***P<0.001 (shHHLA2
group vs. sh-NC group). HHLA2, human endogenous retrovirus-H long
terminal repeat-associating protein 2; EMT, epithelial-mesenchymal
transition; Col-I, collagen I; α-SMA, α-smooth muscle actin; shRNA
or sh, short hairpin RNA; GBC, gall bladder cancer; NC, negative
control. |
HHLA2 knockdown negatively regulates the
tumorigenic ability of GBC cells
The effects of HHLA2 knockdown on GBC-SD migration,
invasion and proliferation was subsequently assessed. Results of
the wound healing assay revealed that HHLA2-shRNA transfection
significantly decreased the migratory ability of GBC-SD cells both
alone (Fig. 5A and B) and in the
presence of TGF-β1 (Fig. 6A and
B). Transwell assay results demonstrated that HHLA2 knockdown
in GBC-SD cells significantly inhibited cell invasion (Fig. 5C and D) both alone and in the
presence of TGF-β1 (Fig. 6C and
D). In addition, results of the EdU assay showed significantly
lower proliferation levels in the HHLA2-shRNA group compared with
those in the sh-NC group (Fig. 5E and
F). In the TGF-β1-treated in vitro model, knocking down
HHLA2 expression was found to significantly reduce the
proliferation of GBC-SD cells (Fig. 6E
and F).
 | Figure 5Knocking down HHLA2 expression
inhibits the proliferation, migration and invasion of GBC cells
in vitro and tumour formation in vivo. (A) According
to the wound-healing assay, which was (B) quantified, HHLA2
knockdown suppressed cell migration. Scale bars, 100 µm. (C)
HHLA2 knockdown decreased cell invasion, (D) according to
subsequent quantification. Scale bars, 100 µm. (E) Cell
proliferation ability in HHLA2 knockdown group was evaluated using
EdU assay, which was (F) quantified. Scale bars, 100 µm.
Green: EdU; Blue: DAPI. (G) Representative images of mice harboring
GBC tumors. HHLA2 knockdown reduced the tumorigenic ability of GBC
cells according to the mouse xenograft assay both (H) in terms of
size and (I) weight (n=6). **P<0.01 and
***P<0.001 (shHHLA2 group vs. sh-NC group). HHLA2,
human endogenous retrovirus-H long terminal repeat-associating
protein 2; GBC, gall bladder cancer; shRNA or sh, short hairpin
RNA; NC, negative control. |
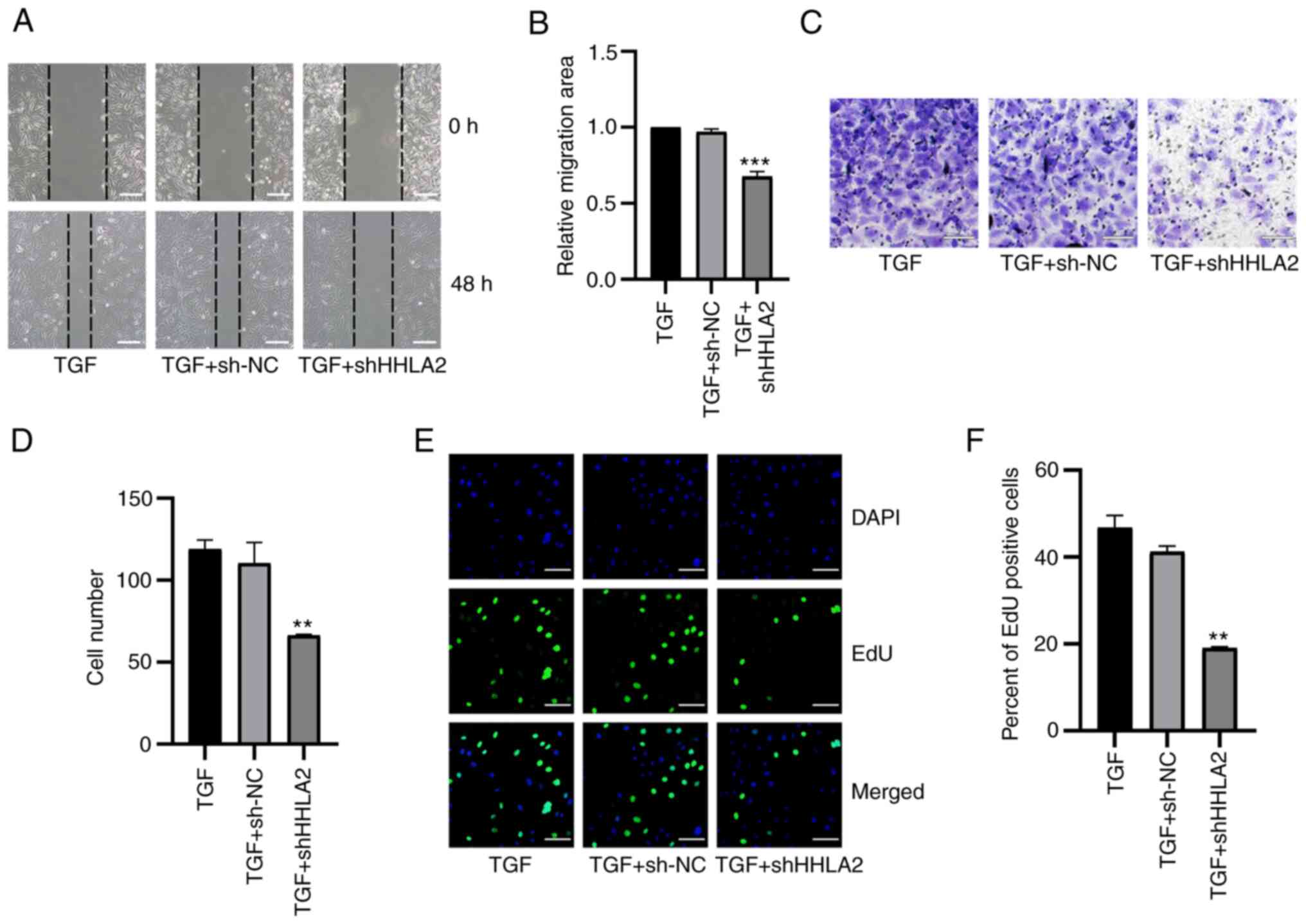 | Figure 6HHLA2 knockdown inhibits
TGF-β1-induced proliferation, migration and invasion in the gall
bladder cancer cell lines. (A) According to the wound-healing
assay, (B) which was quantified, after treatment with TGF-β1 for 48
h, cell migration in the HHLA2 knockdown group was decreased
compared with that of the NC group. Scale bar, 100 µm. (C)
HHLA2 knockdown downregulated decreased cell invasion, (D)
according to subsequent quantification. Scale bars, 100 µm.
(E) The proliferative ability of cells after HHLA2 knockdown was
decreased compared with that in the TGF + sh-NC group, as
determined by EdU assay and (F) was quantified. Scale bars, 100
µm. Green, EdU; Blue, DAPI. **P<0.01 and
***P<0.001 (TGF + shHHLA2 group vs. TGF + sh-NC
group). HHLA2, human endogenous retrovirus-H long terminal
repeat-associating protein 2; shRNA or sh, short hairpin RNA; NC,
negative control. |
GBC-SD stably transfected with HHLA2-shRNA was then
used to establish a xenograft model to further investigate its
potential role in vivo. After 18 days, the tumors in the
HHLA2-shRNA group exhibited a significantly light in weight and
smaller volume (Fig. 5G-I). These
observations suggest that HHLA2 can serve as a therapeutic target
for inhibiting the tumorigenicity of GBC.
HHLA2 expression is positively regulated
by H19 in GBC
A previous study reported the oncogenic role of H19
in GBC, showing that H19 can promote EMT, migration and invasion in
GBC cells (25). After stably
transfected with H19 pc-DNA, GBC-SD cells were found with
significantly higher expression levels of Col-I, N-cadherin, α-SMA
and vimentin but lower expression levels of E-cadherin compared
with those in the the H19-NC group (Fig. S2A and B). The H19 overexpression
group also showed higher migration and invasion capabilities
compared with those in the H19-NC group (Fig. S2C-F). However, the mechanism
underlying this require further exploration. Therefore, a GBC cell
line stably overexpressing H19 was constructed. The efficiency of
H19 overexpression was verified using reverse
transcription-quantitative PCR (Fig.
7A). According to the western blotting results, it was found
that H19 overexpression significantly increased HHLA2 expression in
the GBC-SD cells compared with that in the H19-NC group (Fig. 7B and C).
 | Figure 7Knockdown of HHLA2 inhibits H19
overexpression-induced EMT in GBC in vitro. (A) Reverse
transcription-quantitative PCR was used to examine the
overexpression efficiency of H19. (B) Western blot analysis showed
that H19 overexpression increased HHLA2 expression, (C) according
to the quantification. (D) After the knockdown of HHLA2 in GBC
cells stably overexpressing H19, the knockdown efficiency of HHLA2
and EMT marker expression were examined using western blotting, (E)
which were quantified. Expression of EMT markers Col-I, N-cadherin,
α-SMA and vimentin were decreased whilst that of E-cadherin was
increased in the HHLA2 knockdown group. (n=3). (F) Similar results
were observed according to the immunofluorescence staining images.
Scale bars, 100 µm. Green, target protein; Blue, DAPI.
**P<0.01 and ***P<0.001 (H19 + shHHLA2
group vs. H19 + sh-NC group). HHLA2, human endogenous retrovirus-H
long terminal repeat-associating protein 2; EMT,
epithelial-mesenchymal transition; GBC, gall bladder cancer; Col-I,
collagen I; α-SMA, α-smooth muscle actin; shRNA or sh, short
hairpin RNA; NC, negative control. |
To explore the possible association between H19 and
HHLA2 in EMT, GBC-SD cells stably overexpressing H19 were
transiently transfected with HHLA2-shRNA. Both western blotting and
immunofluorescence revealed marked downregulation of HHLA2
expression and the expression of EMT markers in the HHLA2 knockdown
group, namely that of α-SMA, vimentin, N-cadherin and Col-I
(Fig. 7D-F). By contrast, HHLA2
knockdown significantly increased the expression of E-cadherin
(Fig. 7D-F). These results suggest
that HHLA2 is a key factor in H19-induced GBC-EMT.
H19 promotes GBC migration, invasion and
proliferation through HHLA2
Following the observation that HHLA2 is a key factor
in H19-induced EMT in GBC-SD, additional assays were performed to
investigate the role of HHLA2 in H19 overexpression-induced
migration, invasion and proliferation in vitro. Results from
wound healing assay showed that cells with HHLA2 expression knocked
down exhibited significantly lower migratory ability compared with
that in the H19 + sh-NC group (Fig. 8A
and B). Subsequently, results from the Transwell assay revealed
that the invasive ability of cells transfected with HHLA2-shRNA was
significantly decreased (Fig. 8C and
D). In addition, results of the EdU assay suggested that
knockdown of HHLA2 led to a significant reduction of GBC-SD cell
proliferation (Fig. 8E and F).
 | Figure 8HHLA2 knockdown inhibits H19
overexpression-induced proliferation, migration and invasion in GBC
cell lines. (A) From the images of wound healing assay, (B) which
were quantified, HHLA2 knockdown reversed H19
overexpression-induced promotion of GBC migration. Scale bars, 100
µm. (C) After transient transfection with HHLA2 shRNA, the
H19-overexpressing GBC cell line showed decreased invasive
capabilities (D) following quantification. Scale bars, 100
µm. (E) Knocking down HHLA2 expression inhibited the H19
overexpression-induced cell proliferation according to EdU assay,
(F) which was quantified. Scale bars, 100 µm. Green, EdU;
Blue, DAPI. ***P<0.001 (H19 + shHHLA2 group vs. H19 +
sh-NC group). HHLA2, human endogenous retrovirus-H long terminal
repeat-associating protein 2; GBC, gall bladder cancer; shRNA or
sh, short hairpin RNA; NC, negative control. |
Discussion
The present study aimed to explore the clinical
significance of HHLA2 in GBC, one of the most aggressive biliary
tract malignancies. In addition, the present study aimed to
validate the biological role of HHLA2 in GBC progression, whilst
exploring its potential as a novel therapeutic target for GBC
treatment. It was first found that in GBC tissues from patients
with poorer prognoses and more advanced stage GBC, higher
expression of HHLA2 was observed. In vitro, overexpression
of HHLA2 was found to enhance the aggressive phenotype of GBC. By
contrast, knockdown of HHLA2 expression suppressed the progression
of GBC both in vitro and in vivo. These findings
suggest that HHLA2 is a potential therapeutic target for GBC.
In the present study, the relationship between the
HHLA2 expression and its biological role in GBC was investigated.
It was found that higher expression levels of HHLA2 in patients
with GBC were associated with higher Nevin stages, more advanced
AJCC stages, more severe tumor invasion and node metastasis, in
addition to worse patient OS. The clinical data strongly suggest
that HHLA2 can contribute to the progression of GBC. This
observation is consistent with that in previous studies, which
revealed that HHLA2 can mediate immunosuppressive roles in lung
cancer (32), prostate cancer
(19) and renal carcinoma
(16). However, previous studies
also reported ambiguous findings on the role of HHLA2 in tumor
progression. In pancreatic, ampullary and ovarian cancers, patients
with higher expression of HHLA2 achieved superior prognoses
(20,33). These results serve as a reminder
that HHLA2 can more than likely mediate different functions
depending on the cellular context. To verify the reliability of the
clinical results, the function of HHLA2 on GBC was next explored on
a cellular level.
To date, 14 protein subtypes in the B7/CD28 family
have been discovered (34), the
majority which function as immune checkpoint regulators during
tumour progression (35-38). However, previous studies have shown
that the expression of programmed death-ligand 1 (PD-L1), one of
the ligands in the B7 family, is associated with the EMT process
and immune evasion in breast cancer, squamous cell carcinoma,
non-small-cell lung cancer and cancer stem-like cells (39-42).
EMT serves a critical role during early GBC progression (43). Following EMT activation, epithelial
cells acquire more mesenchymal properties, resulting in reduced
intercellular adhesion and higher migratory and invasive phenotypes
(44). Recently, Cao et al
(45) reported that PD-L1 can
promote EMT and aggressiveness in GBC. Therefore, since it appears
to be part of the similar signaling pathway, HHLA2 may also promote
GBC progression by directly regulating EMT in GBC cells. Results
from the in vivo and in vitro experiments appeared to
support this hypothesis and observations from the clinical data,
which showed that overexpression of HHLA2 promoted GBC
progression.
Subsequently, by TGF-β1 treatment, an apparently
more aggressive GBC-SD cell line (46), was generated, which increased HHLA2
expression. To validate the role of HHLA2 in GBC progression, the
GBC cell lines were transfected with the HHLA2 plasmid to
overexpress it. Cell proliferation, migration and invasion in GBC
cells overexpressing HHLA2 group were all significantly increased,
which concurred with the results of the clinical data. Since EMT
serves such a key role in the early stages of GBC progression
(43), EMT marker expression was
next measured. The expression of EMT markers N-cadherin, α-SMA,
fibronectin, vimentin and Col-I was all found to be increased,
whereas that of E-cadherin was significantly decreased, in the
HHLA2-overexpressing GBC-SD cells. In vivo, overexpression
of HHLA2 resulted in larger tumors being formed. Consistently,
investigations into other protein members in the B7 family revealed
similar functions in colorectal cancer, pancreatic cancer and
hepatocellular cancer (47-49).
These EMT mediators confirmed by previous reports will also likely
mediate HHLA2-induced EMT in GBC. Kang et al (49) previously reported that B7-H3
promoted EMT in hepatocellular carcinoma through the slug pathway,
which may also mediate an EMT-promoting role in GBC. Indeed, Lee
et al (50) previously
found that slug was a mediator of EMT in GBC.
To test the therapeutic potential of HHLA2 in GBC, a
GBC-SD cell line with HHLA2 expression knocked down was then
constructed. Parameters of progression, including proliferation,
invasion and migration, were all found to be significantly reduced
in the GBC-SD cells after HHLA2 expression was knocked down. This
also occurred even in the presence of TGF-β1. In the in vivo
experiments, HHLA2 knockdown was also observed to reduce GBC tumor
size and weight.
In addition to TGF-β1, the present study also
investigated another regulatory molecule in HHLA2 expression. Wang
et al (24,25) previously reported on two occasions
that the lncRNA H19 can promote the progression of GBC and its EMT
process. Based on these previous observations, the potential
regulatory relationship between lncRNA H19 and HHLA2 in GBC was
next assessed. In the GBC-SD cell line, overexpression of lncRNA
H19 significantly increased HHLA2 expression. In addition, in the
rescue experiments, knockdown of HHLA2 inhibited GBC progression
induced by lncRNA H19 overexpression, revealing a regulatory
network between H19 and HHLA2 and suggesting the therapeutic
potential of HHLA2 in GBC progression. However, it is likely that a
complex, as yet unexplored signalling mechanism exist between
lncRNA H19 and HHLA2 in the EMT process. Wu et al (51) and Yan et al (52) previously found that H19 can promote
cancer EMT through the Wnt/β-catenin canonical pathway, which was
summarized further in a review (51-53).
This raises the hypothesis that HHLA2 may be involved in the
H19/Wnt/β-catenin axis. Jang et al (54) revealed that by recruiting Smad2/3
to the promoter of PD-L1, another member of the B7-family,
polo-like kinase 1, enhanced the expression of PD-L1 and promoted
EMT in lung adenocarcinoma. This finding also provides further
support for the future analysis of the signaling pathways involved
in the H19/HHLA2 axis upstream of EMT regulation. In addition to
HHLA2, lncRNA H19 may regulate other members of the B7 family
upstream of EMT in GBC and other tumors, which require further
exploration in the future.
In conclusion, the present study revealed the
clinical and biological significance of HHLA2 in GBC. Higher
expression levels of HHLA2 were found to associate with worse
clinicopathological parameters in patients with GBC and more
advanced types of GBC. By contrast, HHLA2 knockdown suppressed GBC
progression induced by TGF-β1 and lncRNA H19 overexpression,
suggesting that HHLA2 is a potential therapeutic target and
prognostic marker for GBC.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YT and YZ designed the present study. HL and YZ
performed the experiments. CL, YL and FX collected tissue samples
and the clinical data. YY, ZL, YS and QL performed the in
vivo experiments. BW, JZ and CZ analysed and interpreted the
data. YL, YS and JZ completed the figures and tables. FX and YT
reviewed the manuscript. YT and YZ can authenticate all raw data.
All the authors read and approved the final manuscript.
Ethics approval and consent to
participate
Ethical approval of animal experiments was approved
by the Ethics Committee of Shengjing Hospital of China Medical
University (approval no. 2019PS325K; Shenyang, China). All
participants consented to an institutional review broad-approved
protocol that allows for the comprehensive analysis of tissue
samples (Ethics Committee Shengjing Hospital of China Medical
University; approval no. 2019PS036K). Informed written consent was
obtained from all the participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors would like to thank Dr Xingqi Guo
(Department of General Surgery, Cancer Hospital of Dalian
University of Technology, Shenyang, China) and Dr Huixin Hu
(Department of General Surgery, Shengjing Hospital of China Medical
University, Shenyang, China) for their assistance in IHC
evaluation.
Funding
The present study was supported by the Natural Science
Foundation of China (grant no. 81974377 to YT), The Scientific
Research Project of Education Department of Liaoning Province
(grant no. JC2019017 to YT) and 345 Talent Project of Shengjing
Hospital (grant no. 2019-2021 to YT).
References
|
1
|
Castro FA, Koshiol J, Hsing AW and Devesa
SD: Biliary tract cancer incidence in the United States-Demographic
and temporal variations by anatomic site. Int J Cancer.
133:1664–1671. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Acharya M, Patkar S, Parray A and Goel M:
Management of gallbladder cancer in India. Chin Clin Oncol.
8:352019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Song X, Hu Y, Li Y, Shao R, Liu F and Liu
Y: Overview of current targeted therapy in gallbladder cancer.
Signal Transduct Targeted Ther. 5:2302020. View Article : Google Scholar
|
|
4
|
Jin L, Cai Q, Wang S, Wang S, Wang J and
Quan Z: Long noncoding RNA PVT1 promoted gallbladder cancer
proliferation by epigenetically suppressing miR-18b-5p via DNA
methylation. Cell Death Dis. 11:8712020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
McNamara M, Lopes A, Wasan H, Malka D,
Goldstein D, Shannon J, Okusaka T, Knox JJ, Wagner AD, André T, et
al: Landmark survival analysis and impact of anatomic site of
origin in prospective clinical trials of biliary tract cancer. J
Hepatol. 73:1109–1117. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jackson S, Adami H, Andreotti G,
Beane-Freeman LE, de González AB, Buring JE, Fraser GE, Freedman
ND, Gapstur SM, Gierach G, et al: Associations between reproductive
factors and biliary tract cancers in women from the biliary tract
cancers pooling project. J Hepatol. 73:863–872. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Regmi P, Hu HJ, Chang-Hao Y, Liu F, Ma WJ,
Ran CD, Wang JK, Paudyal A, Cheng NS and Li FY: Laparoscopic
surgery for oncologic extended resection of T1b and T2 incidental
gallbladder carcinoma at a high-volume center: A single-center
experience in China. Surg Endos. 35:6505–6512. 2020. View Article : Google Scholar
|
|
9
|
Rizzo A, Tavolari S, Ricci AD, Frega G,
Palloni A, Relli V, Salati M, Fenocchio E, Massa A, Aglietta M and
Brandi G: Molecular features and targeted therapies in extrahepatic
cholangiocarcinoma: Promises and failures. Cancers (Basel). 12. pp.
32562020, View Article : Google Scholar
|
|
10
|
Valle JW, Kelley RK, Nervi B, Oh DY and
Zhu AX: Biliary tract cancer. Lancet. 397:428–444. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ebata N, Fujita M, Sasagawa S, Maejima K,
Okawa Y, Hatanaka Y, Mitsuhashi T, Oosawa-Tatsuguchi A, Tanaka H,
Miyano S, et al: Molecular classification and tumor
microenvironment characterization of gallbladder cancer by
comprehensive genomic and transcriptomic analysis. Cancers.
13:7332021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nepal C, Zhu B, O'Rourke C, Bhatt DK, Lee
D, Song L, Wang D, Van Dyke AL, Choo-Wosoba H, Liu Z, et al:
Integrative molecular characterisation of gallbladder cancer
reveals micro-environment-associated subtypes. J Hepatol.
74:1132–1144. 2021. View Article : Google Scholar
|
|
13
|
Zou W and Chen L: Inhibitory B7-family
molecules in the tumour microenvironment. Nat Rev Immunol.
8:467–477. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhu Y, Yao S, Iliopoulou BP, Han X,
Augustine MM, Xu H, Phennicie RT, Flies SJ, Broadwater M, Ruff W,
et al: B7-H5 costimulates human T cells via CD28H. Nat Commun.
4:20432013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Crespo J, Vatan L, Maj T, Liu R, Kryczek I
and Zou W: Phenotype and tissue distribution of CD28H immune cell
subsets. Oncoimmunology. 6:e13625292017. View Article : Google Scholar
|
|
16
|
Bhatt R, Berjis A, Konge JC, Mahoney KM,
Klee AN, Freeman SS, Chen CH, Jegede OA, Catalano PJ, Pignon JC, et
al: KIR3DL3 is an inhibitory receptor for HHLA2 that mediates an
alternative immunoinhibitory pathway to PD1. Cancer Immunol Res.
9:156–169. 2021. View Article : Google Scholar :
|
|
17
|
Dong Z, Zhang L, Xu W and Zhang G: EGFR
may participate in immune evasion through regulation of B7-H5
expression in non-small cell lung carcinoma. Mol Med Rep.
18:3769–3779. 2018.PubMed/NCBI
|
|
18
|
Luo M, Lin Y, Liang R, Li Y and Ge L:
Clinical significance of the HHLA2 protein in hepatocellular
carcinoma and the tumor microenvironment. J Inflamm Res.
14:4217–4228. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou Q, Li K, Lai Y, Yao K, Wang Q, Zhan
X, Peng S, Cai W, Yao W, Zang X, et al: B7 score and T cell
infiltration stratify immune status in prostate cancer. J
Immunother Cancer. 9:e0024552021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu G, Shi Y, Ling X, Wang D, Liu Y, Lu H,
Peng Y and Zhang B: HHLA2 predicts better survival and exhibits
inhibited proliferation in epithelial ovarian cancer. Cancer Cell
Int. 21:2522021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Winkle M, El-Daly SM, Fabbri M and Calin
GA: Noncoding RNA therapeutics-challenges and potential solutions.
Nat Rev Drug Discov. 20:629–651. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rainier S, Johnson LA, Dobry CJ, Ping AJ,
Grundy PE and Feinberg AP: Relaxation of imprinted genes in human
cancer. Nature. 362:747–749. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu SJ, Dang HX, Lim DA, Feng FY and Maher
CA: Long noncoding RNAs in cancer metastasis. Nat Rev Cancer.
21:446–460. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang SH, Ma F, Tang ZH, Wu XC, Cai Q,
Zhang MD, Weng MZ, Zhou D, Wang JD and Quan ZW: Long non-coding RNA
H19 regulates FOXM1 expression by competitively binding endogenous
miR-342-3p in gallbladder cancer. J Exp Clin Cancer Res.
35:1602016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang SH, Wu XC, Zhang MD, Weng MZ, Zhou D
and Quan ZW: Upregulation of H19 indicates a poor prognosis in
gallbladder carcinoma and promotes epithelial-mesenchymal
transition. Am J Cancer Res. 6:15–26. 2016.PubMed/NCBI
|
|
26
|
Giannis D, Cerullo M, Moris D, Shah KN,
Herbert G, Zani S, Blazer DG III, Allen PJ and Lidsky ME:
Validation of the 8th edition American joint commission on cancer
(AJCC) gallbladder cancer staging system: Prognostic discrimination
and identification of key predictive factors. Cancers. 13:5472021.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nagtegaal ID, Odze RD, Klimstra D, Paradis
V, Rugge M, Schirmacher P, Washington KM and Carneiro F: The 2019
WHO classification of tumours of the digestive system.
Histopathology. 76:182–188. 2020. View Article : Google Scholar :
|
|
28
|
Xu X, He M, Wang H, Zhan M and Yang L:
Development and validation of a prognostic nomogram for gallbladder
cancer patients after surgery. BMC Gastroenterol. 22:2002022.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Camp RL, Dolled-Filhart M and Rimm DL:
X-tile: A new bio-informatics tool for biomarker assessment and
outcome-based cut-point optimization. Clin Cancer Res.
10:7252–7259. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
31
|
Clark JM and Sun D: Guidelines for the
ethical review of laboratory animal welfare People's Republic of
China National Standard GB/T 35892-2018[Issued 6 February 2018
Effective from 1 September 2018]. Animal Models Exp Med. 3:103–113.
2020. View Article : Google Scholar
|
|
32
|
Cheng H, Janakiram M, Borczuk A, Lin J,
Qiu W, Liu H, Chinai JM, Halmos B, Perez-Soler R and Zang X: HHLA2,
a new immune checkpoint member of the B7 family, is widely
expressed in human lung cancer and associated with EGFR mutational
status. Clin Cancer Res. 23:825–832. 2017. View Article : Google Scholar :
|
|
33
|
Boor P, Sideras K, Biermann K, Aziz MH,
Levink IJM, Mancham S, Erler NS, Tang X, van Eijck CH, Bruno MJ, et
al: HHLA2 is expressed in pancreatic and ampullary cancers and
increased expression is associated with better post-surgical
prognosis. Br J Cancer. 122:1211–1218. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhong C, Lang Q, Yu J, Wu S, Xu F and Tian
Y: Phenotypical and potential functional characteristics of
different immune cells expressing CD28H/B7-H5 and their
relationship with cancer prognosis. Clin Exp Immunol. 200:12–21.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Seaman S, Zhu Z, Saha S, Zhang XM, Yang
MY, Hilton MB, Morris K, Szot C, Morris H, Swing DA, et al:
Eradication of tumors through simultaneous ablation of
CD276/B7-H3-positive tumor cells and tumor vasculature. Cancer
Cell. 31:501–515.e8. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Schildberg FA, Klein SR, Freeman GJ and
Sharpe AH: Coinhibitory pathways in the B7-CD28 ligand-receptor
family. Immunity. 44:955–972. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Marangoni F, Zhakyp A, Corsini M, Geels
SN, Carrizosa E, Thelen M, Mani V, Prüßmann JN, Warner RD, Ozga AJ,
et al: Expansion of tumor-associated Treg cells upon disruption of
a CTLA-4-dependent feedback loop. Cell. 184:3998–4015.e19. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Nishimura CD, Pulanco MC, Cui W, Lu L and
Zang X: PD-L1 and B7-1 cis-interaction: New mechanisms in immune
checkpoints and immunotherapies. Trends Mol Med. 27:207–219. 2021.
View Article : Google Scholar
|
|
39
|
Chen L, Gibbons DL, Goswami S, Cortez MA,
Ahn YH, Byers LA, Zhang X, Yi X, Dwyer D, Lin W, et al: Metastasis
is regulated via microRNA-200/ZEB1 axis control of tumour cell
PD-L1 expression and intratumoral immunosuppression. Nat Commun.
5:52412014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lee Y, Shin J, Longmire M, Wang H, Kohrt
HE, Chang HY and Sunwoo JB: CD44+ cells in head and neck squamous
cell carcinoma suppress T-cell-mediated immunity by selective
constitutive and inducible expression of PD-L1. Clin Cancer Res.
22:3571–3581. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Alsuliman A, Colak D, Al-Harazi O, Fitwi
H, Tulbah A, Al-Tweigeri T, Al-Alwan M and Ghebeh H: Bidirectional
cross-talk between PD-L1 expression and epithelial to mesenchymal
transition: significance in claudin-low breast cancer cells. Mol
Cancer. 14:1492015. View Article : Google Scholar
|
|
42
|
Hsu JM, Xia W, Hsu YH, Chan L, Yu WH, Cha
JH, Chen CT, Liao HW, Kuo CW, Khoo KH, et al: STT3-dependent PD-L1
accumulation on cancer stem cells promotes immune evasion. Nat
Commun. 9:19082018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xu S, Zhan M and Wang J:
Epithelial-to-mesenchymal transition in gallbladder cancer: From
clinical evidence to cellular regulatory networks. Cell Death
Discov. 3:170692017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Krebs AM, Mitschke J, Losada MA,
Schmalhofer O, Boerries M, Busch H, Boettcher M, Mougiakakos D,
Reichardt W, Bronsert P, et al: The EMT-activator Zeb1 is a key
factor for cell plasticity and promotes metastasis in pancreatic
cancer. Nat Cell Biol. 19:518–529. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Cao L, Bridle KR, Shrestha R, Prithviraj
P, Crawford DHG and Jayachandran A: CD73 and PD-L1 as potential
therapeutic targets in gallbladder cancer. Int J Mol Sci.
23:15652022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shi Y, Sun L, Zhang R, Hu Y, Wu Y, Dong X,
Dong D, Chen C, Geng Z, Li E and Fan Y: Thrombospondin 4/integrin
α2/HSF1 axis promotes proliferation and cancer stem-like traits of
gallbladder cancer by enhancing reciprocal crosstalk between
cancer-associated fibroblasts and tumor cells. J Exp Clin Cancer
Res. 40:142021. View Article : Google Scholar
|
|
47
|
Yin Y, Shi L, Yang J, Wang H, Yang H and
Wang Q: B7 family member H4 induces epithelial-mesenchymal
transition and promotes the proliferation, migration and invasion
of colorectal cancer cells. Bioengineered. 13:107–118. 2022.
View Article : Google Scholar :
|
|
48
|
Kang JH, Jung MY and Leof EB: B7-1 drives
TGF-β stimulated pancreatic carcinoma cell migration and expression
of EMT target genes. PLoS One. 14:e02220832019. View Article : Google Scholar
|
|
49
|
Kang F-B, Wang L, Jia H-C, Li D, Li H-J,
Zhang Y-G and Sun D-X: B7-H3 promotes aggression and invasion of
hepatocellular carcinoma by targeting epithelial-to-mesenchymal
transition via JAK2/STAT3/Slug signaling pathway. Cancer Cell Int.
15:452015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Lee DW, Lee SH, Kim JS, Park J, Cho YL,
Kim KS, Jo DY, Song IC, Kim N, Yun HJ, et al: Loss of NDRG2
promotes epithelial-mesenchymal transition of gallbladder carcinoma
cells through MMP-19-mediated Slug expression. J Hepatol.
63:1429–1439. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wu K, Liang W, Feng L, Pang JX, Waye MMY,
Zhang JF and Fu WM: H19 mediates methotrexate resistance in
colorectal cancer through activating Wnt/β-catenin pathway. Exp
Cell Res. 350:312–317. 2017. View Article : Google Scholar
|
|
52
|
Yan L, Yang S, Yue CX, Wei XY, Peng W,
Dong ZY, Xu HN, Chen SL, Wang WR, Chen CJ and Yang QL: Long
noncoding RNA H19 acts as a miR-340-3p sponge to promote
epithelial-mesenchymal transition by regulating YWHAZ expression in
paclitaxel-resistant breast cancer cells. Environ Toxicol.
35:1015–1028. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wu B, Zhang Y, Yu Y, Zhong C, Lang Q,
Liang Z, Lv C, Xu F and Tian Y: ViaLong noncoding RNA H19: A novel
therapeutic target emerging in oncology regulating oncogenic
signaling pathways. Front Cell Dev Biol. 9:7967402021. View Article : Google Scholar
|
|
54
|
Jang H-R, Shin S-B, Kim C-H, Won J-Y, Xu
R, Kim D-E and Yim H: PLK1/vimentin signaling facilitates immune
escape by recruiting Smad2/3 to PD-L1 promoter in metastatic lung
adenocarcinoma. Cell Death Diff. 28:2745–2764. 2021. View Article : Google Scholar
|