Introduction
Mortality from colorectal cancer (CRC) has decreased
over the past 30 years; however, there is a significant
heterogeneity in survival rates that may be mainly explained by
variations in patient and tumor characteristics, host response
factors and applied treatment modalities. At the time of diagnosis,
~25% of the patients have already developed metastases and ~50%
will develop metastases following treatment of the primary CRC
(1).
During cancer development, the cells acquire
multiple point mutations and chromosomal rearrangements.
Chromosomal abnormalities that are found in primary CRC cells are
gains of 8q, 11q, 13q and 20q and losses of 1p, 4, 8p, 17p, 18q and
22q chromosomal regions (2–5).
Van den Broek et al reported a clinical
relevance of chromosomal breaks in metastatic CRC (mCRC) in 2015;
they identified 1605 genomic breakpoint locations and 748
breakpoint genes with recurrence in multiple CRC samples. None of
the individual breakpoint genes was significantly associated with
overall survival (OS), but four CRC subtypes were revealed based on
recurrent breakpoint genes and mutation status of commonly affected
CRC genes [adenomatous polyposis coli, tumor protein p53, Kirsten
rat sarcoma viral oncogene homolog (KRAS),
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α,
B-Raf proto-oncogene, serine/threonine kinase, neuroblastoma RAS
viral oncogene homolog, mothers against decapentaplegic homolog 4
and F-box/WD repeat-containing protein 7] (6).
Chromothripsis, a massive chromosome fragmentation
occurring in one catastrophic event, is observed in 2–3% of cancers
(7). Such genomic rearrangements may
drive the development of cancer through several mechanisms,
including deletion of tumor suppressor genes and increased copy
number of oncogenes. The prevalence of chromothripsis and its
effect on prognosis and metastasis is unclear. The incidence of
chromothripsis ranges from 1.3% in multiple myeloma (8) to 33% in osteosarcoma (7). Furthermore, chromothripsis has been
associated with poor patient survival in the aggressive
triple-negative subtype of breast cancer, multiple myeloma and
pediatric medulloblastoma (8–10),
indicating its potential relevance as a prognostic marker, and
suggesting chromothripsis to be a characteristic of certain
particularly aggressive types of cancer.
The incidence of chromothripsis and its effect on
survival in CRC is unclear, but it may be a prevalent genomic
rearrangement in this type of cancer (11).
Patients and methods
Study population
A total of 19 mCRC patients who received
chemotherapy at the Clinic of Oncology of Pauls Stradins Clinical
University Hospital (Riga, Latvia) between August, 2011 and
October, 2012 were selected. The study was performed with approval
from the Ethics Committee of the Riga Stradins University and
written informed consent was obtained from all the patients who
were investigated. Tissue samples were acquired as part of a series
of routine diagnostic and pathological analyses at the hospital.
The patients were followed up for 3–48 months.
The clinical and biological characteristics of the
patients are summarized in Table I.
Of the 19 patients, 15 had primary metastatic cancer (stage IV),
whereas the 4 remaining patients developed metastases after
treatment of the primary cancer. A total of 10 patients developed
metastases only to the liver; 16 patients underwent primary tumor
surgery, whereas in 3 patients biopsy alone was performed. In 7
patients, the carcinoembryonic antigen (CEA) level was ≤5.5 ng/ml
prior to chemotherapy.
 | Table I.Clinical and biological
characteristics of the patients (n=19). |
Table I.
Clinical and biological
characteristics of the patients (n=19).
| Characteristics | No. (%) |
|---|
| Age, years [mean
(range)] | 63.15 (38–78) |
| Gender |
|
| Male | 11 (57.89) |
|
Female | 8 (42.11) |
| Tumor
localization |
|
| Left side
(sigmoid colon, rectal cancer) | 11 (57.89) |
| Right
side (colon cancer) | 8 (42.11) |
| Grade |
|
|
Unknown | 1 (5.26) |
| G2 | 13 (68.42) |
| G3 | 5 (26.32) |
| Metastases |
|
|
Synchronous | 15 (78.95) |
|
Metachronous (median time
tometastasis, 12.25 months; range, 9–18 months) | 4 (21.05) |
| Stage II
(n=3) |
|
| Stage III
(n=1) |
| Metastases |
|
| Liver
only | 10 (52.63) |
|
Other | 9 (47.37) |
| KRAS status in
primary tumor |
|
|
Wild-type | 10 (52.64) |
| Mutation
in 12 codon | 7 (36.84) |
| Mutation
in 13 codon | 1 (5.26) |
| Not
known | 1 (5.26) |
| Serum CEA prior to
chemotherapy |
|
| CEA ≤5.5
ng/ml | 7 (36.84) |
| CEA
>5.5 ng/ml | 12 (63.16) |
| Level of
CEA, ng/ml [mean (range)] | 232.1 (7.1959.8) |
| Median follow-up
(months) | 25.5 (348) |
| mPFS | 8 months |
|
1-year | 33.3% |
|
2-year | 5.6% |
| mOS | 21 months |
|
1-year | 78.9% |
|
2-year | 42.1% |
|
3-year | 21.1% |
A total of 18 patients received FOLFOX first-line
chemotherapy (1 patient declined chemotherapy). After disease
progression, 13 patients received irinotecan-containing second-line
chemotherapy, 1 patient was rechallenged with FOLFOX, 1 patient
received oral fluoropyrimidine therapy with Ftorafur (tegafur) and
1 patient received best supportive care. Only 5 patients (26.3%)
received third-line therapy [irinotecan, oxaliplatin or
5-fluorouracil (5FU)]. One patient underwent hepatic surgery for
CRC metastases after discontinuation of second-line chemotherapy, 2
patients received salvage transcatheter arterial chemoembolization
of CRC liver metastases by irinotecan-eluting microspheres, and 1
patient received palliative radiotherapy for local rectal cancer.
Data on clinical follow-up were obtained until August, 2016.
Genotyping
DNA was extracted from formalin-fixed
paraffin-embedded (FFPE) samples with QIAamp DNA Mini kit (Qiagen,
Hilden, Germany) according to the manufacturer's instructions.
Quality was evaluated using the Illumina FFPE QC kit (Illumina, San
Diego, CA, USA) by reverse transcription-polymerase chain reaction.
DNA was restored with the Illumina DNA restoration kit (Illumina).
Microarray analysis was performed using the Infinium
HumanOmniExpress-12 v1.0 FFPE BeadChip kit (Illumina). BeadChip was
scaned on HiScan (Illumina). Analysis was performed by GenomeStudio
software (Illumina) and R version 3.1.2. (https://www.r-project.org/). Copy number variation and
breakpoints on the chromosomes were analyzed using the DNA copy
package (http://bioconductor.org/packages/release/bioc/html/DNAcopy.html).
OS and progression-free survival (PFS) rates were
estimated using the Kaplan-Meier method. The log-rank test was used
to calculate any significant difference between the subgroups by
univariate analysis. Significance levels were set at P<0.05. All
statistical analyses were performed using MedCalc software, version
16.4.8 (MedCalc Software, Ostend, Belgium).
Results
Breakpoint count per chromosome
The aim of this study was to evaluate the total
number of chromosomal breaks and association of chromothripsis
(>100 breakpoints detected in one chromosome) with PFS in mCRC.
The total number of breakpoints per genome in cancer tissue
[breakpoint instability index (BPI)] was 368–4,009. The highest
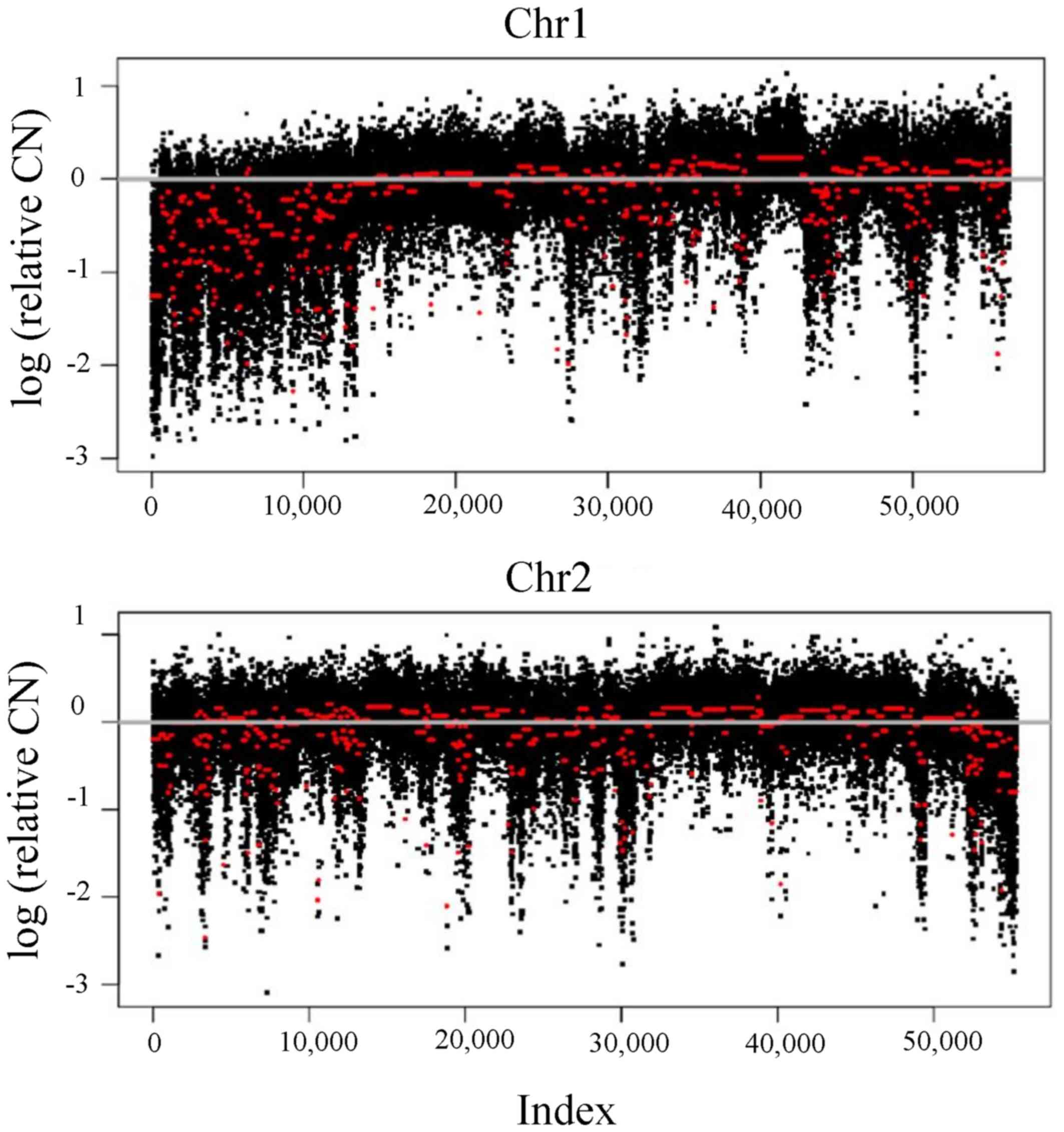
breakpoint count was seen in chromosome 1 (27–365 breakpoints),
followed by chromosome 2 (25–315) (Table II), indicating the crucial role of
these chromosomes in CRC development. The lowest density of breaks
occurred in chromosome 21 (7–99 breakpoints; Fig. 1). Of note, a similar pattern of
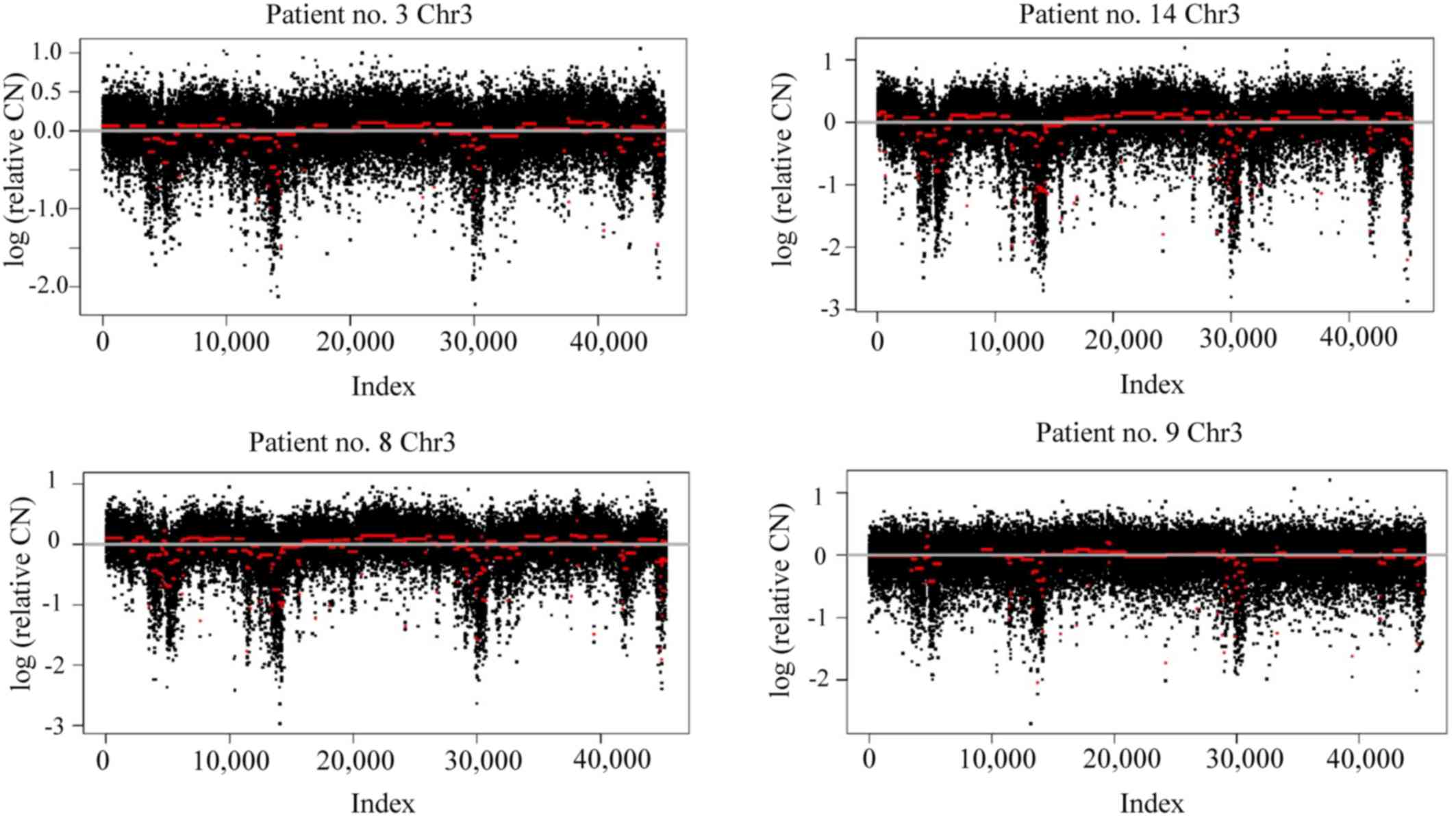
chromosomal rearrangements was observed in several patients for
chromosomes 1, 3 and 8 (Fig. 2).
 | Table II.Chromosomes affected by chromothripsis
and total breakpoint count per chromosome. |
Table II.
Chromosomes affected by chromothripsis
and total breakpoint count per chromosome.
| Chromosome | Breakpoint no., range
(median) | Chromothripsis, no.
of patients (%) |
|---|
| 1 | 27–365 (150.2) | 10 (52.6) |
| 2 | 25–315 (136.8) | 10 (52.6) |
| 3 | 21–234 (94.5) | 7 (36.8) |
| 4 | 10–232 (73.9) | 5 (26.3) |
| 5 | 20–189 (87.1) | 5 (26.3) |
| 6 | 22–298 (118.9) | 10 (52.6) |
| 7 | 16–195 (82.7) | 5 (26.3) |
| 8 | 16–199 (81.6) | 5 (26.3) |
| 9 | 11–205 (79.2) | 5 (26.3) |
| 10 | 21–275 (106.8) | 7 (36.8) |
| 11 | 34–252 (104.1) | 8 (42.1) |
| 12 | 18–206 (84.2) | 6 (31.6) |
| 13 | 14–166 (54.1) | 3 (15.8) |
| 14 | 12–152 (58.6) | 4 (21.1) |
| 15 | 3–166 (58.9) | 4 (21.1) |
| 16 | 9–158 (60.5) | 4 (21.1) |
| 17 | 10–206 (66.8) | 4 (21.1) |
| 18 | 10–149 (50.5) | 2 (10.5) |
| 19 | 7–137 (44.2) | 2 (10.5) |
| 20 | 9–124 (42.3) | 2 (10.5) |
| 21 | 7–99 (24.7) | 0 (0.0) |
| 22 | 4–131 (37) | 1 (5.3) |
In 10 of the tumor samples (52.6%), multiple
chromosomal fragmentations were found, found, referred to as
chromothripsis, a recently reported catastrophic genetic event. The
most commonly affected chromosomes were chromosomes 1, 2 and 6
(52.6% of the patients; Table II).
The maximal count of chromosomes affected by chromothripsis was 20,
which was observed in 1 patient.
Association of breakpoint number with
clinicopathological characteristics
No association of BPI value and chromothripsis with
cancer localization (rectal or colon cancer), CEA level, KRAS
mutational status in primary cancer and stage [synchronous
metastatic disease (stage IV) vs. metachronous metastatic disease]
was observed.
Association of breakpoint number with
survival
PFS and OS were measured for all the patients in
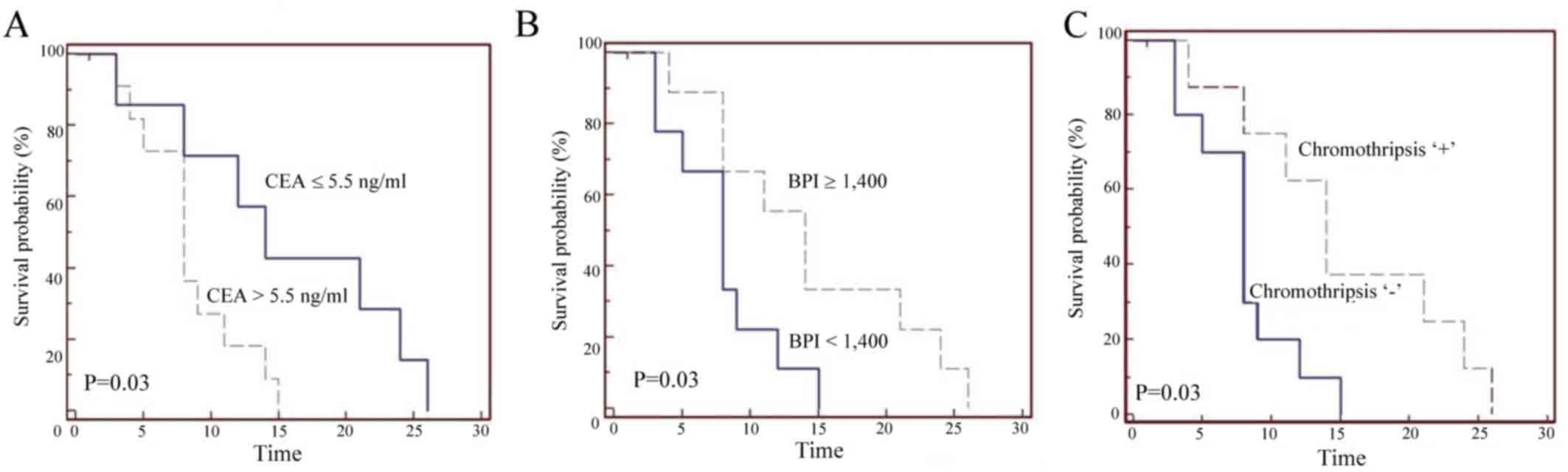
study. Decreased PFS was observed in patients with left-sided
metastatic cancer (sigmoid colon and rectal cancer), elevated CEA
level prior to chemotherapy (Fig.
3A) and KRAS mutations.
A positive correlation between high BPI
(chromothripsis) and increased PFS was observed (Fig. 3B and C). Due to the observed better
PFS in different clinical subgroups by CEA, KRAS status and primary
tumor location, the effect of BPI and chromothripsis on clinical
findings was analyzed; however, the number of patients in each
subgroup was insufficient to establish a statistically significant
correlation. In patients with detected chromothripsis and CEA ≤5.5
ng/ml (n=4), a median PFS (mPFS) of 22.5 months was observed; in
patients without chromothripsis and elevated CEA level (n=7), an
mPFS of 8 months was observed, but these findings did not reach
statistical significance. A statistically significant effect of any
clinical or biological factors on OS was not observed.
Discussion
The effect of chromosomal rearrangements and
mutations on pathogenesis, prognosis and resistance to treatment
are widely described in mCRC studies. The prognostic and predictive
role of breakpoint instability index and chromothripsis remains
unclear.
In the present study, a correlation between massive
DNA fragmentation (chromothripsis) and PFS in mCRC was observed. As
opposed to recent studies suggesting chromothripsis to be
associated with worse prognosis, we found chromothripsis to be a
positive predictive factor for first-line chemotherapy. It may be
hypothesized that cancer cells exhibiting radical DNA
rearrangements, such as chromothripsis, are more sensitive to
nucleic acid-damaging therapy with 5FU and oxaliplatin.
Breakpoint instability index was previously reported
in breast cancer study by Przybytkowski et al (10); they reported a BPI of 25–300 in
breast cancer tissue, with the highest density of breaks in
chromosome 17 and the lowest density in chromosome 4. The BPI
appeared to be different in different breast cancer molecular
subtypes, with the highest breakpoint count in aggressive
triple-negative breast cancer. In comparison, in our study on CRC
tissue, the highest breakpoint density was found in chromosomes 1
and 2 and the lowest in chromosome 21, but the BPI was
significantly higher (368–4,009). In addition, 10 tumor samples
(52.6%) exhibited a chromothripsis pattern on ≥3 chromosomes, which
was higher compared with previous reports (7,8,10). The high BPI value and high prevalence
of chromothripsis in our study may be attributed to the fact that
all the patients had late-stage metastatic disease, which is
consistent with the prevalence of chromothripsis reported in
high-risk aggressive tumors.
In conclusion, the present study demonstrated that
chromothripsis is associated with increased PFS, but not with OS in
mCRC. Further studies with larger sample sizes are required to
determine whether chromothripsis and BPI value may be used as a
prognostic or predictive factor for oxaliplatin-containing
first-line chemotherapy for mCRC.
Acknowledgements
The present study was partly supported by the
National Research Program ‘Biomedicine for Public Health
(BIOMEDICINE)’.
References
|
1
|
Khatri VP, Petrelli NJ and Belghiti J:
Extending the frontiers of surgical therapy for hepatic colorectal
metastases: Is there a limit? J Clin Oncol. 23:8490–8497. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sayagués JM, Fontanillo C, Mdel M Abad,
González-González M, Sarasquete ME, Chillon MdC, Garcia E,
Bengoechea O, Fonseca E, Gonzalez-Diaz M, et al: Mapping of genetic
abnormalities of primary tumours from metastatic CRC by
high-resolution SNP arrays. PLoS One. 5:e137522010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sylvester BE and Vakiani E: Tumor
evolution and intratumor heterogeneity in colorectal carcinoma:
Insights from comparative genomic profiling of primary tumors and
matched metastases. J Gastrointest Oncol. 6:668–675.
2015.PubMed/NCBI
|
|
4
|
González-González M, Muñoz-Bellvis L,
Mackintosh C, Fontanillo C, Gutiérrez ML, Abad MM, Bengoechea O,
Teodosio C, Fonseca E, Fuentes M, et al: Prognostic impact of del
(17p) and del (22q) as assessed by interphase FISH in sporadic
colorectal carcinomas. PLoS One. 7:e426832012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ali Hassan NZ, Mokhtar NM, Kok Sin T, Rose
I Mohamed, Sagap I, Harun R and Jamal R: Integrated analysis of
copy number variation and genome-wide expression profiling in
colorectal cancer tissues. PLoS One. 9:e925532014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
van den Broek E, Dijkstra MJ, Krijgsman O,
Sie D, Haan JC, Traets JJ, van de Wiel MA, Nagtegaal ID, Punt CJ,
Carvalho B, et al: High prevalence and clinical relevance of genes
affected by chromosomal breaks in colorectal cancer. PLoS One.
10:e01381412015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Stephens PJ, Greenman CD, Fu B, Yang F,
Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA,
et al: Massive genomic rearrangement acquired in a single
catastrophic event during cancer development. Cell. 144:27–40.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Magrangeas F, Avet-Loiseau H, Munshi NC
and Minvielle S: Chromothripsis identifies a rare and aggressive
entity among newly diagnosed multiple myeloma patients. Blood.
118:675–678. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rausch T, Jones DT, Zapatka M, Stütz AM,
Zichner T, Weischenfeldt J, Jäger N, Remke M, Shih D, Northcott PA,
et al: Genome sequencing of pediatric medulloblastoma links
catastrophic DNA rearrangements with TP53 mutations. Cell.
148:59–71. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Przybytkowski E, Lenkiewicz E, Barrett MT,
Klein K, Nabavi S, Greenwood CM and Basik M: Chromosome-breakage
genomic instability and chromothripsis in breast cancer. BMC
Genomics. 15:5792014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kloosterman WP, Hoogstraat M, Paling O,
Tavakoli-Yaraki M, Renkens I, Vermaat JS, van Roosmalen MJ, van
Lieshout S, Nijman IJ, Roessingh W, et al: Chromothripsis is a
common mechanism driving genomic rearrangements in primary and
metastatic colorectal cancer. Genome Biol. 12:R1032011. View Article : Google Scholar : PubMed/NCBI
|