Introduction
Multiple myeloma (MM) is a type of malignant plasma
cell and has a high incidence in malignant tumors of the blood
system, accounting for ~2% of the total mortality of cancer
(1). In China, the incidence of MM
accounted for 10% of total hematopoietic system cancers with an
upward trend year by year. Although treatment strategies changed
from traditional chemotherapy and autologous hematopoietic stem
cell transplantation to novel targeted drug therapy, the outcome
was not improved (2). Therefore,
the identification of mechanisms underlying the regulation of the
malignant behavior of MM and key genes in disease progression has
the greatest significance for the establishment of novel
therapeutic strategies and improvement of the prognosis in
patients. In-depth study of small non-coding RNA molecules
confirmed that microRNA (miR) has an important regulatory role in
cell proliferation, differentiation, metabolism, apoptosis and
development processes (3). Murphy
et al (4) found that miR-21
is closely associated with the tumor and is able to adjust SPRY2
expression. SPRY2 is a member of the signaling pathway-specific
inhibition protein sprouty (SPRY) family. According to their
differential sequences, SPRY proteins were divided into four
subtypes (SPRY1, -2, -3 and -4). The SPRY2 protein contains 315
human amino acid residues (35 kDa), with the C-terminal residues
178–282 being rich in cysteine. Due to its significant biological
effects (5–8), SPRY2 has become a research hotspot.
The present study intended to establish MM cell lines with stably
silenced SPRY2 using RNA interference technology. Under in
vitro conditions, changes in the proliferation and invasion
ability were detected in myeloma cells. To investigate the
occurrence, development and transfer process of MM, a novel
molecular targeted therapy was established to provide a reliable
basis for research.
Materials and methods
Instruments and reagents
ABI7500 real-time polymerase chain reaction (PCR)
instrument (Applied Biosystems Inc., Life Technologies, Thermo
Fisher Scientific, Waltham, MA, USA). A NanoPhotometer nucleic acid
and protein ultraviolet detector (NanoPhotometer® Pearl;
Implen GmbH, Munich, Germany) and a 3K18 type low temperature high
speed centrifuge (Sigma, Osterode am Harz, Germany) were used. The
UVP GelDoc-It 310 gel imaging analysis system was purchased from
Shanghai Kunke Co., Ltd. (Shanghai, China). TRIzol reagent, LA Taq
DNA polymerase and lipid Lipofectamine 2000 (Invitrogen Life
Technologies, Carlsbad, CA, USA) were used. The miRNeasy Mini kit
serum total RNA extraction kit was from QIAGEN Inc. (Hilden,
Germany). For cell culture, 10% FBS RPMI 1640 medium and DMEM
culture medium (Hyclone, GE Healthcare, Little Chalfont, UK) were
used. Agarose gel extraction kit and mir-21qPCR primer kit were
purchased from Takara Bio Inc. (Otsu, Japan). Lentiviral vector
LV-anti-miR-21 and control vector were from Shanghai SBO Medical
Biotechnology Co. (Shanghai, China). SPRY2 eukaryotic expression
vector was purchased from Origene (Rockville, MD, USA) and
microRNA-21 mimics and inhibitors were from Biomics Biotechnologies
(Nantong) Co., Ltd. (Nantong, China).
Construction of plasmids
Prior to construction of the miR-21 lentiviral
expression vector LV-anti-miR-21, the miR-21 precursor pre-miR-21
sequence was obtained using the miRBase (http://www.mirbase.org) database. Primer synthesis was
performed by Shanghai Jierui Bio-Engineering Co., Ltd. (Shanghai,
China). The upstream primer was miR-21 forward,
5′-CCGGTTCAACATCAGTCTGATAAGCTATTTTTTG-3′, and the downstream primer
was mir-21 reverse, 5′-AATTCAAAAAATAGCTTATCAG-3′. DNA containing
the pre-amplified sequence was used as the template for PCR
amplification and the target-resulting fragment was used for
Xho I and BamH I (Promega Corp., Madison, WI, USA)
double digestion. The fragment was connected with the lentiviral
expression vector LV-anti plasmid, and the connection reaction
conditions were as follows: 1 µl digested product, 1
µl LV-anti plasmid vector, 6 µl nuclease-free water,
1 µl 10x ligase buffer and 1 µl T4 DNA ligase
(Promega Corp.) for 22°C in a water bath overnight. The ligation
products were transfected into competent DH5 cells, the monoclonal
colonies were selected and inoculated, and they were placed in a
37°C-thermostat overnight. Plasmids were extracted using a plasmid
extraction kit, and Xho I and BamH I were prepared
for restriction enzyme digestion. The reaction conditions were as
follows: 17 µl DNA, 2 µl 10xPCR buffer, 0.5 µl
Hind III and 0.5 µl BamH I. The reaction was
performed at 37°C for 3 h. The bacterial liquid was sent to
Invitrogen Life Technologies for sequencing.
Establishment of stably SPRY2-silenced MM
cells
LV-anti-miR-21 expression vector and unloaded
cytomegalo-virus (CMV)-green fluorescent protein (GPF)-LV were
added to a 24-well culture plate containing U266 cells (500
µl/well; 7.0×104 cells/ml) for 8 h of infection.
The viral supernatant was replaced with appropriate medium, and
24–48 h after infection, green fluorescence was observed under a
fluorescence microscope (BX51; Olympus Corporation, Tokyo, Japan).
The culture was expanded following infection and cells in the
logarithmic growth phase were collected and subjected to Aldefluor
screening (Biowish Co., Ltd., Jiangsu, China) for 24 h. Cells
containing the target gene and the cells transfected with an empty
vector were isolated and re-cultured. When the cells were in the
logarithmic growth phase, they were divided into three groups,
namely: The U266/un group (untreated), the U266/GFP group
containing the unloaded virus CMV-GFP-LV, and the LV-anti-miR-21
group, which was infected with the target gene. Following 24 h,
G418 (1 mg/ml; Cian Wolsen Biotechnology Co., Ltd., Shanxi, China)
screening was performed for 24 h and the clones were further
cultured for subsequent experiments.
Cell culture, transfection and G418
screening
The human myeloma cell lines U266, KM3 and RPMI8226
were provided by the Department of Cell Biology of China Medical
University (Shenyang, China) The cells were between passages 1:2
and 1:4. RPMI 1640 medium containing 10% FBS was used for the
culture of the myeloma cell lines U266, KM3 and RPMI8226, and they
were placed in a humidified incubator at 37°C and 5% CO2
for subculture. When cells reached 80% confluency, cells at the
logarithmic growth phase were collected. Lipofectamine 2000 was
used to mediate G418 transfection of U266 cells. The cells were
added and cultured in complete medium. Cells in the logarithmic
growth phase were seeded in 12-well plates at a density of
1×105 cells, and were divided into four groups: miR-21
mimics group (transfected with miR-21 mimics), miR-21 inhibitor
group (transfected with anti-miR-21), untreated group
(untransfected cells) and small interfering (si)RNA negative
control (NC) group. Three wells were set for each group. The
transfection concentration was 100 nmol/l, and 6 h after
transfection, the medium was replaced with normal medium. 24 h
later, DMEM containing 10% FBS and G418 was added for selection,
and clones were obtained after two weeks of screening, followed by
culturing of the clones for the subsequent experiments.
miR-21 expression detected by real-time
quantitative (RT-q)PCR
The TRIzol method was used to extract the total
length RNA from the cells. The methods were according to the miR-21
RT-qPCR method by Chen et al (9). Briefly, the process was as follows:
According to instructions of QIAGEN’s miRNeasy Mini kit, 200
µl serum was used to extract total RNA from the cells, and
RNA was stored at −80°C. The reverse transcription kit (Promega
Corp.) was used for reverse transcription. PCR was conducted using
an ABI7500 Real-Time PCR Instrument (Applied Biosystems Life
Technologies, Foster City, CA, USA). PCR conditions were as
follows: 95°C for 10 min, 95°C for 15 sec, 55°C for 15 sec, 72°C
for 20 sec, for 40 cycles. Each tube contained 20 µl PCR
reaction mixture. According to the comparative threshold method by
Livak and Schmittgen (10), Ct
values were read, and the expression quantity of the targeted
miR-21 gene was expressed using the 2−ΔΔCt formula, with
ΔΔCt=experimental group (Cttarget
gene-Cthousekeeping gene) - control group
(Cttarget gene-Cthousekeeping gene). Each
condition was repeated in three wells, and the experiment was
repeated three times.
Western blot analysis
The cells in the logarithmic growth phase and with
80% confluency were collected, and radio-immunoprecipitation assay
lysis buffer containing protease inhibitors (Beyotime Institute of
Biotechnology, Haimen, China) was used for conventional extraction
of total cellular protein. A bicinchoninic acid protein assay kit
(Beyotime Institute of Biotechnology) was used for protein
quantification. 5x loading buffer was added prior to sampling at
100°C for 5 min of boiling, and following cooling, the samples were
filled. 10% SDS-PAGE was performed, protein was then transferred to
a nitrocellulose membrane (Beyotime Institute of Biotechnology), 5%
skim milk was added, and membranes were agitated in a sealed
container for 1 h. The primary antibodies were then added: Rabbit
polyclonal SGOL1 (1:100; cat. no. R4392; Ucallm, Beijing Biocoen
Biotechnology Co., Ltd., Beijing, China) and mouse monoclonal GAPDH
(1:5,000; cat. no. A01020; Abbkine, Inc., Redlands, CA, USA), and
the membranes were incubated in a sealed container with agitation
at 4°C overnight. Phosphate-buffered saline (PBS) containing Tween
20 (PBST; Beyotime Institute of Biotechnology) was used for washing
the gels three times for 10 min each. Horseradish
peroxidase-labeled goat anti-mouse antibody (1:5,000; cat. no.
Ov02-03-02) and horseradish peroxidase-labeled goat anti-rabbit
antibody (1:2,000; cat. no. Ov03-03) (Luoyang Baitaike
Biotechnology Co., Ltd., Luoyang, China) were added and gels were
incubated at room temperature for 1 h. PBST was then used for
washing three times, gels were developed using enhanced
chemiluminescence and visualized using a Gel imager camera (Bio-Rad
Laboratories, Hercules, CA, USA). The target protein ratio was
calculated from the grayscale ratio (miR-21/GAPDH or
SPRY2/GAPDH).
Cell proliferation ability detected by
the MTT method
Cells in the logarithmic growth phase were collected
following transfection in each group. Single-cell suspensions were
prepared, the cell density was adjusted to 3×104/ml, and
cells were inoculated in collagen-coated 96-well plates with each
group set in three parallel wells. 0, 24, 48 or 72 h following
inoculation, 20 µl MTT solution (5 mg/ml; Beyotime Institute
of Biotechnology) was added to each well, cells were cultured for 4
h, and the supernatant was discarded. Dimethylsulfoxide (200
µl/well; Beyotime Institute of Biotechnology) was added and
plates were agitated to dissolve the formazan crystals over 10 min.
An ELISA plate reader was used to detect the absorbance (A) values
at 570 nm wavelength, with a blank well set as zero. The cell
growth curves were drawn, and the cell proliferation inhibition
rate was calculated from the optical density (OD) as: Inhibition
rate (%) = (1-ODexperimental g roup/ODcontrol
group) ×100%.
Flow cytometric analysis of the cell
cycle
The cells in each group were collected at 24, 48 and
72 h following transfection, and cold PBS was used to wash cells
three times. The cells were resuspended in 500 µl pre-cooled
binding buffer, and the concentration was adjusted to
5×106 ml. 100 µl of the cell suspension was added
to flow cytometry tubes and 5 µl Annexin V-fluorescein
isothiocyanate (Beyotime Institute of Biotechnology) was added.
Following mixing, samples were incubated at room temperature in the
dark for 15 min, and 5 min prior to the measurements, 5 µl
10 mg/l propidium iodide (PI) dye (Beyotime Institute of
Biotechnology) was added. Flow cytometry (FACScan; BD Biosciences,
Franklin Lakes, NJ, USA) was used to determine the cell cycle
distribution. Each sample was repeated three times. CellQuest FCS
3.0 software (BD Biosciences) was used for data analysis.
Detection of cell migration using wound
healing assay
Following transfection, the cells were collected and
seeded in 48-well plates (1×106/well). When the cells
merged into a monolayer, a sterile 200 µl pipette tip was
used to carefully cause a line-shaped scratch at the surface of the
cell layer. PBS was used for washing twice. An inverted microscope
(BX51; Olympus Corporation) was used to observe the non-suspended
or free cells at the borders of the scratches. The degree of
scratch healing was observed and images were captured in each group
(reflecting cell migration) at 0, 24 and 48 h. The cell migration
rate was calculated as: Mobility (%) = (1 – 48-h scratch
distance/initial distance) ×100%.
Transwell invasion assay
Cells were collected following transfection. A
Transwell invasion chamber (Beyotime Institute of Biotechnology;
polycarbonate pore membrane with pore size 8 µm) was placed
in a 24-well cell culture plate. Matrigel (15 µg/ml;
Beyotime Institute of Biotechnology) was placed on the surface of
the filter membrane of the Transwell chamber, and following
coagulation, RPMI 1640 serum-free medium (Beyotime Institute of
Biotechnology) (37°C) was used for hydration for 30 min.
0.25%trypsin (Beyotime Institute of Biotechnology) was used for
digestion of cells in the logarithmic growth phase. Following
suspension and dilution of serum-free medium, 1×105
cells were inoculated in each chamber containing a volume of 200
µl, and 600 µl complete medium was added to the lower
chamber. Following 24 h of incubation, the small chamber was
removed. A cotton swab was used to wipe the non-invaded cells from
the surface of the microporous membrane. The filter membrane was
then fixed with methanol for 20 min. Crystal violet (Shanghai
Qiaoxing Trading Corporation, Shanghai, China) staining was
performed for 10 min, and under an optical microscope
(magnification, x200), eight fields of view were randomly selected
to perform the averaged cell count. The above experiment was
repeated three times, with three wells per group.
Statistical analysis
Data were processed with SPSS 17.0 statistical
software (SPSS, Inc., Chicago, IL, USA). Continuous values are
expressed as the mean ± standard deviation (x±s), and differences
between two groups were tested using a small sample t test,
P<0.05 was considered to indicate a statistically significant
difference between values.
Results
PCR detection of miR-21 and SPRY2 gene
expression in MM cell lines
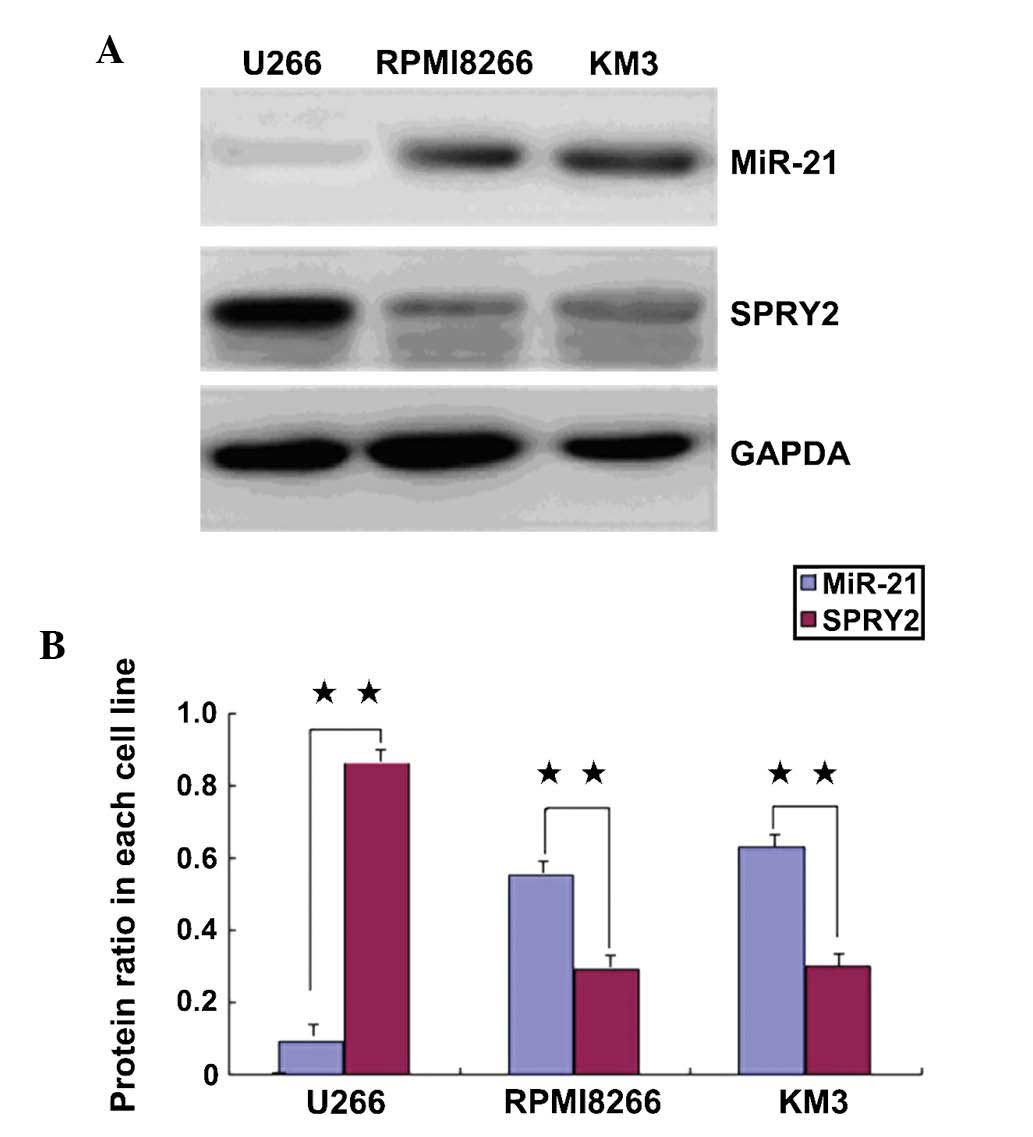
In the MM cell lines RPMI8226 and KM3, miR-21
expression was high and SPRY2 expression was low. In the U266 cell
line, miR-21 expression was low, and SPRY2 expression was high; the
differences were statistically significant (P<0.01) (Fig. 1).
Western blot analysis of miR-21 and SPRY2
protein expression in MM cells
Western blot analysis showed that endogenous miR-21
expression in the MM cell lines RPMI8226 and KM3 was high, while
SPRY2 expression was significantly lower. Conversely, in the U266
cell line, endogenous miR-21 expression was low and SPRY2
expression was significantly higher (Fig. 2A). The gray values of miR-21 and
SPRY2 protein in the respective cell lines showed statistically
significant differences (P<0.01) (Fig. 2B).
Inhibition of miR-21 expression following
infection with LV-anti-miR21 in U266 cells
RT-qPCR showed that following transfection of U266
cells, expression levels were as follows: miR-21 expression in the
U266/GFP group was higher than that in the U266/un group
(P>0.05); miR-21 expression in the U266/LV-anti-miR-21
lentiviral MOI 20 group and the MOI 40 group was significantly
lower than that in the U266/un group (P<0.05). miR-21 expression
in the U266/LV-anti-miR-21 lentivirus MOI 20 group was higher than
that in the MOI 40 group (P>0.05) (Fig. 3).
SPRY2 expression in MM cells following
transfection
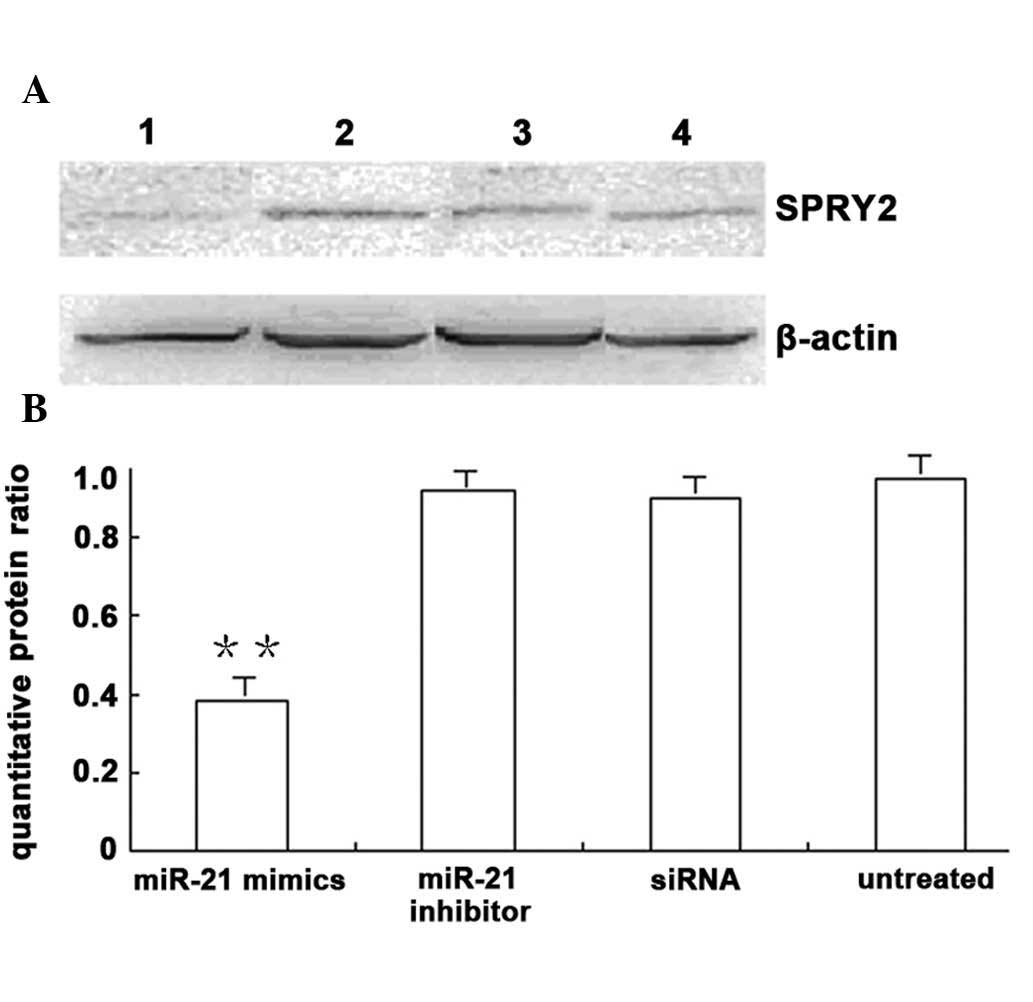
Western blot analysis showed that SPRY2 protein
expression in U266 cells in the transfected miR-21 mimics group was
significantly lower than that in the untreated group and the siRNA
negative control group (P<0.01) (Fig. 4).
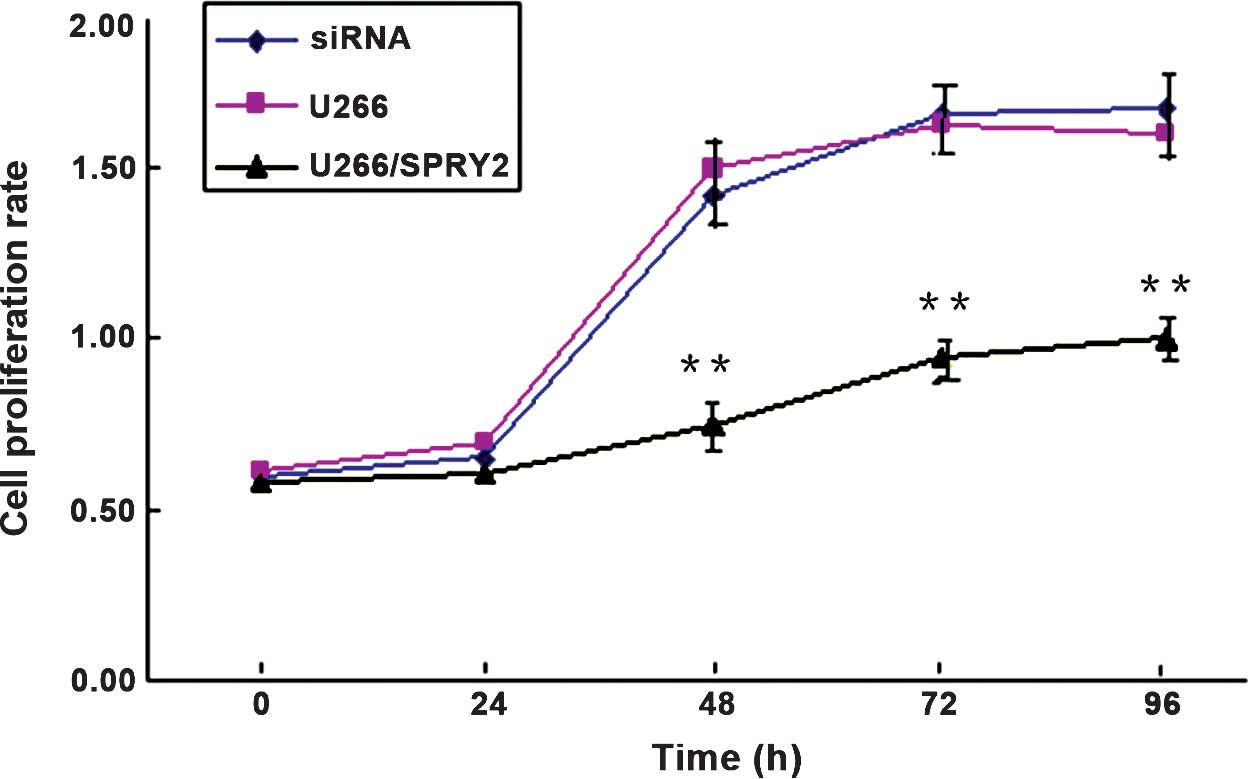
U266 cells overexpressing reduced SPRY2
proliferative capacity
The non-transfected group, the negative control
group and the SPRY2 plasmid-transfected U266 cells were cultured
for four days. An MTT assay showed that the growth of SPRY2
plasmid-transfected U266 cells was significantly decreased at 48,
72 and 96 h, and the proliferation rate decreased significantly
(P<0.01). No significant difference in cell growth was noted
between the untransfected group and the negative control group
(P>0.05) (Fig. 5).
Effect of miR-21 gene expression on
apoptosis of MM
Flow cytometry showed that 48 and 72 h following
transfection of U266 cells with miR-21 mimics, the apoptotic rates
were (24.7±1.97 and 38.6±1.56%, respectively) in the U266 group,
(27.3±1.72 and 37.3±1.59%, respectively) in the siRNA group and
(12.7±1.27 and 22.1±1.63%, respectively) in the U266/miR-21 group.
Compared with the two control groups, the apoptotic rate in the
U266/miR-21 group was significantly lower, and the cell population
in G0/G1 phase was significantly reduced
(P<0.05) (Fig. 6).
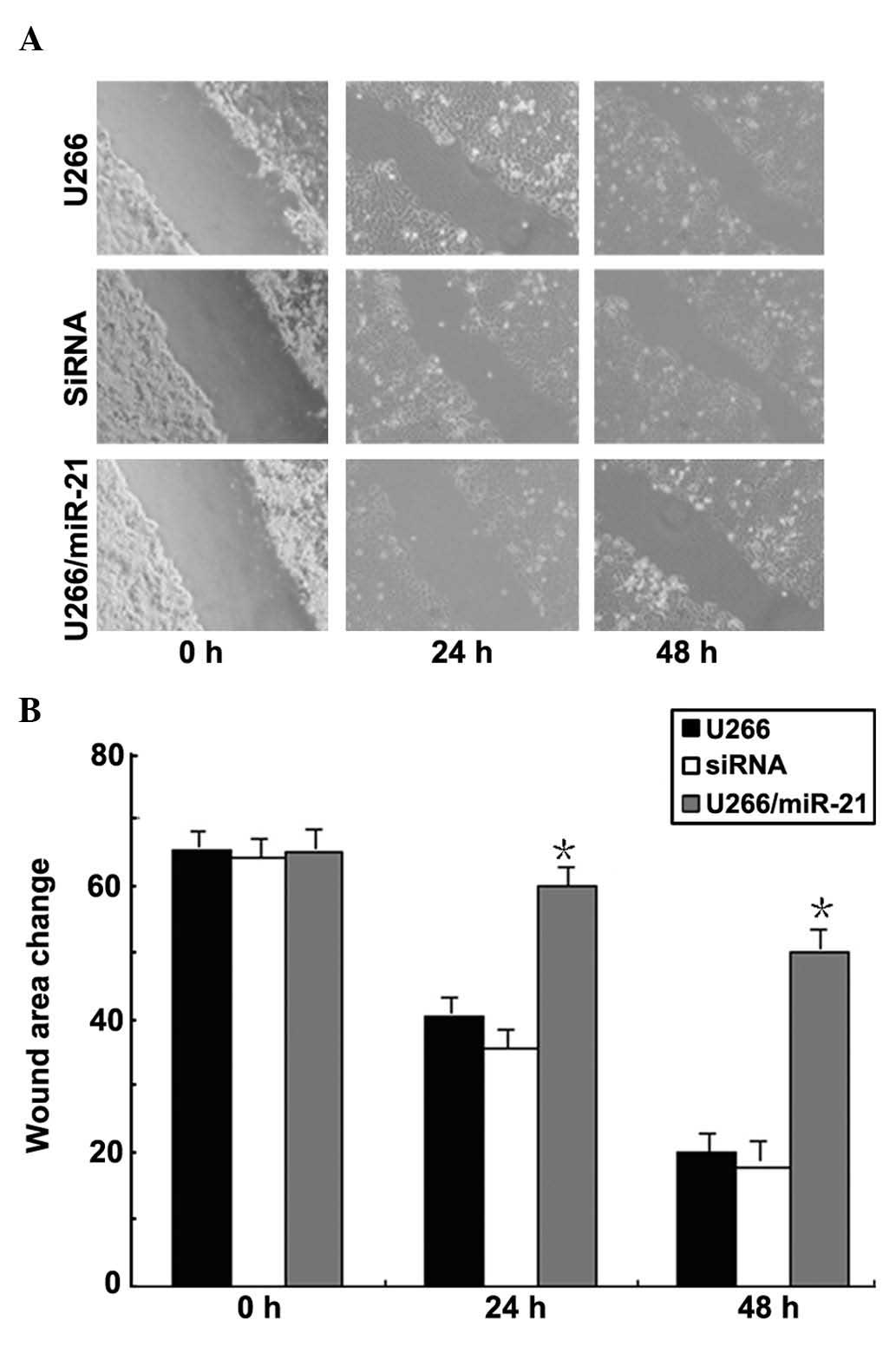
Effects of miR-21 gene expression on
migration of MM cells
Scratch test results showed that the edge of the
wound was neat subsequently following scratching. Following 24 and
48 h of incubation, the cell processes significantly increased and
migrated to the damaged area, whose size gradually decreased. The
ability of transfected cells to migrate into the wound area
significantly decreased when compared with that of the
non-transfected and negative control groups (P<0.05) (Fig. 7).
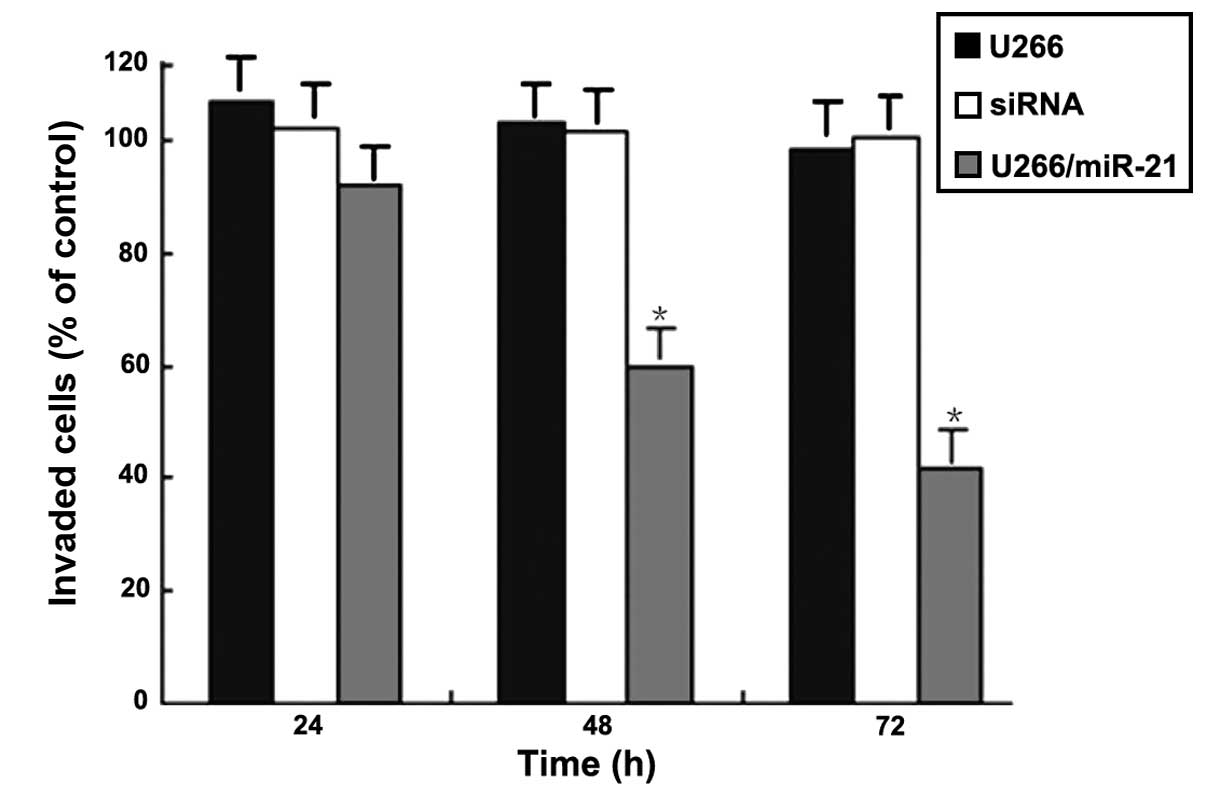
Effects of miR-21 gene expression on the
invasion of the MM cells
The Transwell invasion assay showed changes in U266
cell invasion at 24, 48 and 72 h following transfection with miR-21
mimics. At the same time-point, the differences in numbers of U266
cells which transgressed through the Matrigel-covered polyphosphate
membrane were not statistically significant between the two control
groups (P>0.05). At 48 and 72 h, the number of U266 cells which
passed through the Matrigel-covered polyphosphate membrane in the
transfected group significantly decreased as compared with that in
the non-transfected and negative control groups (P<0.05)
(Fig. 8).
Discussion
An ideal tumor marker should be easily detectable
using non-invasive methods. The use of miR-21 as a molecular marker
has been the focus of numerous studies (11–13).
It is abnormally expressed in a variety of malignant tumors and has
a pivotal regulatory role in the development of tumors (12–14).
Studies showed that the overexpression of miR-21 was associated
with proliferation, metastasis and prognosis of MM (15), non-Hodgkin’s lymphoma, leukemia
(16) and various non-hematologic
solid tumors (17). miR-21 was
shown to be able to regulate SPRY2 expression (5), and the structure of SPRY proteins was
shown to be rich in C-terminal of cysteine, highly evolutionarily
conserved (6), and its target
region was positioned in the activated cell membrane, with a strong
variation in the N-terminal region (7). Several studies showed that SPRY2 gene
expression was downregulated and inhibited in prostate cancer,
breast cancer, malignant glioma and other tumor types, leading to
uncontrolled and overactivated mitogen-activated protein
kinase/extracellular-regulated kinase (MAPK/ERK) signaling in tumor
cells (7,18–20).
Therefore, SPRY2 was considered an oncogene involved in MAPK/ERK
signaling. In the present study, RT-qPCR and western blot analyses
showed that in the MM cell lines with high endogenous miR-21
expression (RPMI8226 and KM3), SPRY2 expression was low.
Conversely, in the U266 cell line exhibiting low endogenous miR-21
expression, SPRY2 expression was higher. The gray values of miR-21
and SPRY2 protein in the respectve cell lines showed significant
differences (P<0.01). This illustrated that miR-21 may be
negatively correlated with SPRY2 in MM cells. To the best of our
knowledge, the effects of miR-21-mediated downregulation of SPRY2
gene expression on the proliferation and invasion of MM cells as
well as the underlying molecular mechanism have not been previously
reported. For this reason, in the present study, the LV-anti-miR-21
vector was constructed, and MM cell lines rediced expression levels
of SPRY2 were successfully established. In LV-anti-miR-21-infected
U266 cells, miR-21 expression was significantly inhibited
(P<0.05) and SPRY2 protein expression was significantly
increased (P<0.01). This further confirmed that miR-21 is able
to regulate the expression of SPRY2.
The results indicated that high levels of miR-21 and
downregulation of SPRY2 may inhibit cell proliferation, migration
and invasion, and promote apoptosis. It can be concluded that
miR-21 is able to downregulate SPRY2 expression; it is therefore
indicated that in the development of MM, low miR-21 levels lead to
the promotion of cell proliferation, invasion and metastasis, and
the inhibition of apoptosis. miR-21 is therefore a potential
biological target, which may be upregulated to increase apoptosis
signaling, and therefore may be used to treat and prevent the
generation of tumors.
The main pathogenesis of cancer involves
deregulation of the cell cycle, leading to unlimited regulation of
cell growth. Apoptosis is induced following insults originating
from intracellular processes of the external environment. It is the
active cell suicide process controlled by apoptotic proteins, which
participate in various parallel signaling pathways and/or
activation cascades (21). A
previous study showed that SPRY2 inhibits MAPK/ERK activation as
well as interleukin-6-stimulated MM cell growth (22). The ability of SPRY2 to inhibit
MAPK/ERK signaling pathway activation suggested that SPRY2
functions as a tumor suppressor gene in MM cells. The MTT assay in
the present study showed that SPRY2 overexpression decreased the
proliferation of U266 cells (P<0.01). Flow cytometric analysis
showed that 48 and 72 h after transfection, the apoptotic rate in
the U266/miR-21 group was significantly decreased, and the
G0/G1-phase population was significantly
reduced, suggesting that transfected miR-21 mimics can enhance the
tolerance of U266 cells to apoptosis. These results indicated that
miR-21 downregulated SPRY2 gene expression and promoted the
proliferation and migration functions of MM cells in vitro.
These results provided a molecular mechanisms underlying the
occurrence and development of MM.
The molecular mechanisms of invasion and metastasis,
which are malignant processes in cancer tissues, are complex
multi-step processes. The occurrence and development of metastases
is based on complex interactions between tumor cells and the host,
and the underlying mechanisms have remained to be fully elucidated,
as a large number of genes and proteins are involved (23–25).
Exploring the mechanism of invasion and metastasis of MM at the
molecular level, to determine the prognosis and prolonged survival
time as well as to improve the survival rate has been the focus of
studies on MM. In the present study, a wound healing assay showed
that the migration ability of the cells significantly decreased
following miR-21 transfection/SPRY2 downregulation (P<0.05). A
Transwell invasion assay demonstrated that the number of U266 cells
which transgressed through a Matrigel-covered polyphosphate
membrane significantly decreased. These results indicated that the
miR-21-mediated downregulation of SPRY2 expression inhibited the
migration and invasion of MM cells and may therefore have a
beneficial effect on MM. Further studies are required, using in
vivo experiments including tumor occurrence in nude mice and
transfection by intravenous injection. The present study provided
an experimental basis for further mechanistic studies on MM cell
migration and invasion.
In conclusion, RNA interference technology was used
to establish a stably SPRY2-silenced MM cell line. The results
suggested that miR-21 downregulated SPRY2 gene expression and
decreased cell proliferation and invasion of MM cells in
vitro. The present study provided experimental evidence and a
theoretical basis for the development of clinical treatments of MM,
and its prospects and potential clinical application deserve
further study.
References
|
1
|
Hatzimichael E, Dasoula A, Benetatos L,
Syed N, Dranitsaris G, Crook T and Bourantas K: Study of specific
genetic and epigenetic variables in multiple myeloma. Leuk
Lymphoma. 51:2270–2274. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Anderson KC: Multiple myeloma: How far
have we come? Mayo Clin Proc. 78:15–17. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pichiorri F, Suh SS, Ladetto M, Kuehl M,
Palumbo T, Drandi D, Taccioli C, Zanesi N, Alder H, Hagan JP,
Munker R, Volinia S, Boccadoro M, Garzon R, Palumbo A, Aqeilan RI
and Croce CM: MicroRNAs regulate critical genes associated with
multiple myeloma pathogenesis. Proc Natl Acad Sci USA.
105:12885–12890. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Murphy T, Hori S, Sewell J and
Gnanapragasam VJ: Expression and functional role of negative
signalling regulators in tumour development and progression. Int J
Cancer. 127:2491–2499. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Edwin F, Anderson K, Ying C and Patel TB:
Intermolecular interactions of Sprouty proteins and their
implications in development and disease. Mol Pharmacol. 76:679–691.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Feng YH, Wu CL, Shiau AL, Lee JC, Chang
JG, Lu PJ, Tung CL, Feng LY, Huang WT and Tsao CJ:
MicroRNA-21-mediated regulation of Sprouty2 protein expression
enhances the cytotoxic effect of 5-fluorouracil and metformin in
colon cancer cells. Int J Mol Med. 29:920–926. 2012.PubMed/NCBI
|
|
7
|
Lao DH, Chandramouli S, Yusoff P, Fong CW,
Saw TY, Tai LP, Yu CY, Leong HF and Guy GR: A Src homology
3-binding sequence on the C terminus of Sprouty2 is necessary for
inhibition of the Ras/ERK pathway downstream of fibroblast growth
factor receptor stimulation. J Biol Chem. 281:29993–30000. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sayed D, Rane S, Lypowy J, He M, Chen IY,
Vashistha H, Yan L, Malhotra A, Vatner D and Abdellatif M:
MicroRNA-21 targets Sprouty2 and promotes cellular out growths. Mol
Biol Cell. 19:3272–3282. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee
DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao
KQ, Livak KJ and Guegler KJ: Real-time quantification of microRNAs
by stem-loop RT-PCR. Nucleic Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C (T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar
|
|
11
|
Moriyama T, Ohuchida K, Mizumoto K, et al:
MicroRNA-21 modulates biological functions of pancreatic cancer
cells including their proliferation, invasion, and chemoresistance.
Mol Cancer Ther. 8:1067–1074. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Neri A, Giordano A, Munshi NC, Anderson
KC, Tagliaferri P and Tassone P: Targeting miR-21 inhibits in vitro
and in vivo multiple myeloma cell growth. Clin Cancer Res.
19:209–106. 2013.
|
|
13
|
Löffler D, Brocke-Heidrich K, Pfeifer G,
Stocsits C, Hackermüller J, Kretzschmar AK, Burger R, Gramatzki M,
Blumert C, Bauer K, Cvi jic H, Ullmann AK, Stadler PF and Horn F:
Interleukin-6 dependent survival of multiple myeloma cells involves
the Stat3-mediated induction of microRNA-21 through a highly
conserved enhancer. Blood. 110:1330–1333. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hu HY, Li KP, Wang XJ, Liu Y, Lu ZG, Dong
RH, Guo HB and Zhang MX: Set9, NF-kappaB and microRNA-21 mediate
berberine-induced apoptosis of human multiple myeloma cells. Acta
Pharmacol Sin. 34:157–166. 2013. View Article : Google Scholar
|
|
15
|
Liu C, Yu J, Yu S, Lavker RM, Cai L, Liu
W, Yang K, He X and Chen S: MicroRNA-21 acts as an oncomir through
multiple targets in human hepatocellular carcinoma. J Hepatol.
53:98–107. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu J, Wu C, Che X, Wang L, Yu D, Zhang T,
Huang L, Li H, Tan W, Wang C and Lin D: Circulating microRNAs,
miR-21, miR-122 and miR-223, in patients with hepatocellular
carcinoma or chronic hepatitis. Mol Carcinog. 50:136–142. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pichiorri F, Suh SS, Ladetto M, Kuehl M,
Palumbo T, Drandi D, Taccioli C, Zanesi N, Alder H, Hagan JP,
Munker R, Volinia S, Boccadoro M, Garzon R, Palumbo A, Aqeilan RI
and Croce CM: MicroRNAs regulate critical genes associated with
multiple myeloma pathogenesis. Proc Natl Acad Sci USA.
105:12885–12890. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cabrita MA and Christofori G: Sprouty
proteins, masterminds of receptor tyrosine kinase signaling.
Angiogenesis. 11:53–62. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lito P, Mets BD, Kleff S, O’Reilly S,
Maher VM and McCormick JJ: Evidence that sprouty 2 is necessary for
sarcoma formation by H-Ras oncogene-transformed human fibroblasts.
J Biol Chem. 283:2002–2009. 2008. View Article : Google Scholar
|
|
20
|
Asangani IA, Rasheed SA, Nikolova DA,
Leupold JH, Colburn NH, Post S and Allgayer H: MicroRNA-21 (miR-21)
post-transcriptionally downregulates tumor suppressor Pdcd4 and
stimulates invasion, intravasation and metastasis in colorectal
cancer. Oncogene. 27:2128–2136. 2008. View Article : Google Scholar
|
|
21
|
Yedjou CG and Tchounwou PB: In-vitor
cytotoxic and genotoxic effects of arsenic trioxide on human
leukemia (HL-60) cells using the MTT and alkaline single cell
gelelectrophoresis (Comet) assays. Mol Cell Biochem. 301:123–130.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Feng YH, Wu CL, Tsao CJ, Chang JG, Lu PJ,
Yeh KT, Uen YH, Lee JC and Shiau AL: Deregulated expression of
sprouty2 and microRNA-21 in human colon cancer: Correlation with
the clinical stage of the disease. Cancer Biol Ther. 11:111–121.
2011. View Article : Google Scholar
|
|
23
|
Wang X, Li C, Ju S, Wang Y, Wang H and
Zhong R: Myeloma cell adhesion to bone marrow stromal cells confers
drug resistance by microRNA-21 up-regulation. Leuk Lymphoma.
52:1991–1998. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xiong Q, Zhong Q, Zhang J, Yang M, Li C,
Zheng P, Bi LJ and Ge F: Identification of novel miR-21 target
proteins in multiplemyeloma cells by quantitative proteomics. J
Proteome Res. 11:2078–2090. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shah AA, Leidinger P, Blin N and Meese E:
miRNA: small molecules as potential novel biomarkers in cancer.
Curr Med Chem. 17:4427–4432. 2010. View Article : Google Scholar : PubMed/NCBI
|