Introduction
Gastric cancer (GC) is one of the most common forms
of cancer and is the second highest cause of cancer-associated
mortality worldwide (1). Due to
its aggressive growth and the lack of effective treatment options,
the disease has a high mortality rate, with a 5-year survival rate
of ~20% (2). It is of great
importance to identify novel biomarkers for an early diagnosis,
targeted treatment and prognosis evaluation in GC.
Mitogen-activated protein kinases (MAPKs) are a
family of conserved serine/threonine protein kinases, which are
essential in transmitting extracellular signals into the cytoplasm
(3). MAPKs are important in
modulating and regulating several crucial cellular processes,
including growth, migration, differentiation, apoptosis and
stress-associated responses. MAPK kinase kinase kinase isoform 4
(MAP4K4; also termed hepatocyte progenitor kinase-like/germinal
center kinase-like kinase) is involved in the regulation of cell
motility, rearrangement of the cytoskeleton and cell proliferation
(4–7). Previous studies revealed that MAP4K4
is overexpressed in numerous types of human cancer (5,8–10).
In addition, the overexpression of MAP4K4 is a prognostic marker
for stage II pancreatic ductal (8)
and lung (10) adenocarcinomas. In
particular, silencing of MAP4K4 by small interfering RNA inhibits
the invasion and migration of cancer cells from different anatomic
origins, including breast cancer, prostate cancer, ovarian cancer
and malignant melanoma (4). The
suppression of MAP4K4 protein expression in hepatocellular
carcinoma cells reduces cell proliferation, inhibits the cell cycle
progression and increases cell apoptosis (9). These results suggest an involvement
of MAP4K4 in cancer progression. However, little is known about the
expression pattern and biological functions of MAP4K4 in GC.
To investigate the roles of MAP4K4 in GC, the
protein was overexpressed in GC and normal tissue. The effects of
knocking down MAP4K4 on the proliferation, invasion and apoptosis
of GC cells were assessed, and a putative mechanism was also
investigated. The present study provided for the first time, to the
best of our knowledge, an assessment of the overexpression of
MAP4K4 in GC, and how this may be an effective therapeutic target
for this disease.
Materials and methods
Bioinformatics analysis
The Cancer Genome Atlas (TCGA) RNA sequencing
(RNA-Seq) information and corresponding clinical data, were
downloaded from the TCGA website (http://cancergenome.nih.gov), following approval of
this project by the consortium of Shanghai Jiao Tong University
Affiliated Sixth People's Hospital (Shanghai, China). RNA-Seq
analysis used data from 249 stomach cancer samples and 33 adjacent
normal tissues. To gain further insights into the biological
pathways involved in the pathogenesis of stomach cancer via the
MAP4K4 pathway, a gene set enrichment analysis (GSEA) was
performed. The gene sets demonstrating a false discovery rate of
0.25, a well-established cut-off for the identification of
biologically relevant genes, were considered enriched between the
classes under comparison.
Cancer specimens
Specimens of GC and paired non-cancerous tissues
were obtained from 25 patients, including 8 females and 17 males,
aged between 42 and 83 years (median age, 64 years). All tissues
were snap-frozen in liquid nitrogen immediately following
resection. Written informed consent was obtained from the
patients.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
The total RNA was extracted using TRIzol®
reagent (Invitrogen Life Technologies, Carlsbad, CA, USA),
according to the manufacturer's instructions. The complementary DNA
was synthesized using a cDNA synthesis kit (Thermo Fisher
Scientific Inc., Rockford, IL, USA). RT-qPCR analyses were
performed using SYBR Green (Takara Biotechnology Co., Ltd., Dalian,
China), and data collection was conducted using an ABI 7500
(Applied Biosystems Life Technologies, Foster City, CA, USA).
RT-qPCR was performed to detect the mRNA expression levels of the
genes, as indicated below. GAPDH was used an internal control for
normalization. The gene expression was calculated using the
2−ΔΔCt method (11).
The primers (Sangon Biotech Co., Ltd., Shanghai, China) used were
as follows: MAP4K4, forward: 5′-GATGAGGAGGACGACGATGTG-3′ and
reverse: 5′-GTCTGGCGGACGATTAGAGTG-3′; GAPDH, forward:
5′-CACCCACTCCTCCACCTTTG-3′ and reverse: 5′-CCACCACCCTGTTGCTGTAG-3′;
Notch2, forward: 5′-TGAGTGTCTGAAGGGTTATG-3′ and reverse:
5′-TGAAGCCTCCAATCTTATCC-3′; Notch3, forward: 5′-CATCCGAAACCGCTC
TAC-3′ and reverse: 5′-GTCTCCTCCTTGCTATCC-3′; Hesl, forward:
5′-CAGTTTGCTTTCCTCATTC-3′ and reverse: 5′-TCTCCCAGTATTCAAGTTC-3′.
The PCR cycling conditions were as follows: 95°C for 10 min,
followed by 40 cycles at 95°C for 15 sec and 60°C for 45 sec and a
final extension step of 95°C for 15 sec, 60°C for 1 min, 95°C for
15 sec and 60°C for 15 sec.
Cell lines
All culture media were supplemented with 10% fetal
bovine serum (FBS; Invitrogen Life Technologies), 100 mg/ml
penicillin G (Invitrogen Life Technologies) and 50 µg/ml
streptomycin (Invitrogen Life Technologies). The BGC-823, SGC-7901
and AGS GC cell lines, and MKN-28 cells (all obtained from the
Institute of Biochemistry and Cell Biology, Shanghai, China) were
cultured in RPMI-1640 medium (Invitrogen Life Technologies). The
MGC-803 and HEK-293T cells were cultured in Dulbecco's modified
Eagle's medium (DMEM; Invitrogen Life Technologies). All cells were
maintained at 37°C in 5% CO2.
RNA interference (RNAi) and construction
of stable cell lines
A total of three short hairpin (sh)RNAs (Sangon
Biotech Co., Ltd.) targeting nucleotide positions 5923–5945 (AAGATG
GAAATGGATGTTTCA; termed MAP4K4-Ri-1), 602–624
(AATACTCTCATCACAGAAACA; termed MAP4K4-Ri-2) and 3779–3801
(AACGCAATGACAAGGTGTTCT; termed MAP4K4-Ri-3) of human MAP4K4 mRNA
were cloned into a lentiviral vector (PLKO.1-EGFP; Sangon Biotech
Co., Ltd.). A non-specific scramble shRNA sequence was used as the
negative control (Sangon Biotech Co., Ltd.). The constructs were
subsequently transfected into HEK-293T cells with lentiviral
packaging vectors using Lipofectamine 2000 (Invitrogen Life
Technologies), according to the manufacturer's instructions. The
viruses were collected at 48 h following transfection, and were
used to infect the BGC-823 cells. All assays were performed 48 h
following infection.
Western blotting
The total cell lysates were extracted using
radioimmunoprecipitation buffer, containing 50 mmol/l Tris-HCl (pH
8.8),150 mmol/l NaCl, 1% Triton X-100, 0.1% SDS, 1% deoxycholic
acid sodium). The protein concentration was measured using a
bicinchoninic acid protein assay kit (Pierce Biotechnology, Inc.,
Rockford, IL, USA), and absorbance was measured using a micro-plate
reader (SM600 Labsystem; Shanghai Utrao Medical Instrument Co.,
Ltd., Shanghai, China). Equal quantities of cell lysates were
subjected to electrophoresis using 10 or 15% sodium dodecyl
sulfate-polyacrylamide gel electropho-resis and transferred to
polyvinylidene fluoride membranes (Sigma-Aldrich, St. Louis, MO,
USA), followed by blocking in fat-free milk overnight at 4°C. The
membranes were subsequently incubated with primary antibodies
overnight at 4°C, followed by incubation with horseradish
peroxidase-conjugated goat anti-rabbit/anti-mouse secondary
antibodies (cat nos. A0208 and A0216; dilution 1:1,000; Beyotime
Institute of Biotechnology, Haimen, China) for 1 h at 37°C, prior
to being washed three times with Tris-buffered saline containing
20% Tween (Amresco, Solon, OH, USA). Rabbit polyclonal antibodies
against Notch2 (cat. no. ab137665; 1:600), Notch3 (cat. no.
ab178948; 1:20,000) and MAP4K4 (cat. no. ab155583; 1:1,000) were
purchased from Abcam (Cambridge, MA, USA). Rabbit monoclonal
antibodies against GAPDH (cat. no. 5174; 1:1,500) and Hes1 (cat.
no. 11988; 1:1,000) were purchased from Cell Signaling Technology,
Inc. (Danvers, MA, USA). Rabbit polyclonal antibodies against Bcl-2
(cat. no. sc-492; 1:150) and Bax (cat. no. sc-493; 1:100) were
purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA,
USA). The blots were visualized using enhanced chemiluminescence
(Millipore, Billerica, MA, USA) and signals were quantified by
densitometry (Quantity One software version 4.62; Bio-Rad
Laboratories, Inc., Hercules, CA, USA).
Cell proliferation assay
The Cell Counting kit-8 (CCK-8; Dojindo
Laboratories, Kumamoto, Japan) was used to detect cell
proliferation. Briefly, BGC-823 cells were seeded into 96-well
plates at a density of 3×103 cells/well. The BGC-823
cells were subsequently infected with MAP4K4-RNAi virus or negative
control virus (Neg.) following culture overnight. At the indicated
time points, CCK-8 solution (10 µl in 100 µl DMEM)
was added to each well and incubated for 1 h at 37°C. Optical
density (OD) values at 450 nm were measured using a microplate
reader (SM600 Labsystem; Shanghai Utrao Medical Instrument Co.,
Ltd.
Cell cycle analysis
The cell cycle was assessed by flow cyto-metric
analysis using propidium iodide (PI; Sigma-Aldrich) staining on a
flow cytometer (BD Accuri C6, software version 1.0.264.21; BD
Biosciences, Franklin Lakes, NJ, USA). Briefly, the BGC-823 cells
were seeded into 6-well plates, infected with the indicated virus
and cultured for 48 h. The cells were collected and fixed in 70%
ethanol at −20°C overnight. The cells were subsequently washed in
phosphate-buffered saline (PBS) and resuspended in staining
solution, containing 20 µg/ml PI and 200 µg/ml RNase
A. All experiments were performed in triplicate and 30,000 cells
were analyzed per sample.
Annexin V/PI staining and flow cytometric
analysis
Annexin V/PI staining (BD Biosciences) and flow
cytometric analysis were performed, according to the manufacturer's
instructions. Briefly, the BGC-823 cells were seeded into 6-well
plates, infected with the indicated virus and cultured for 48 h.
The cells were subsequently collected by trypsinization (JRDUN
Biotechnology, Shanghai, China) and incubated with annexin
V-fluorescein isothiocyanate (FITC) and PI, prior to analysis by a
flow cytometry. The early apoptotic cells, which were stained with
FITC and emit green fluorescence, are represented in the lower
right quadrant of the fluorescence-activated cell sorting
histogram, and the late apoptotic cells, which were stained with
FITC and PI, emit red-green fluorescence and are represented in the
upper right quadrant of the histogram.
In vitro invasion assay
The upper well of the Transwell (Corning, Inc.,
Corning, NY, USA) was coated with Matrigel (BD Biosciences) at 37°C
in a 5% CO2 incubator for 1 h. The cells were
serum-starved for 24 h and subsequently, 5×104 cells in
500 µl serum-free DMEM were seeded into the upper well of
the transwell chamber. Cell culture medium, supplemented with 10%
FBS (750 µl), was added into the lower well of the chamber.
Following 48 h incubation, the cells in the upper well were removed
with a cotton swab. The cells, which migrated into the lower well,
were washed with PBS, fixed in 3.7% paraformaldehyde and stained
with 0.2% crystal violet (JRDUN Biotech). Images of the cells were
captured and cell numbers were counted under a microscope (CX41RF;
Olympus Corporation, Tokyo, Japan). The number of migrated cells
was expressed as the mean ± standard deviation.
Statistical analysis
The data are expressed as the mean ± standard
deviation. Statistical analyses were performed using GraphPad Prism
5 software (GraphPad Software, Inc., La Jolla, CA, USA). The
paired, two-tailed Student's t-test was used to analyze the
significance of the difference between groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
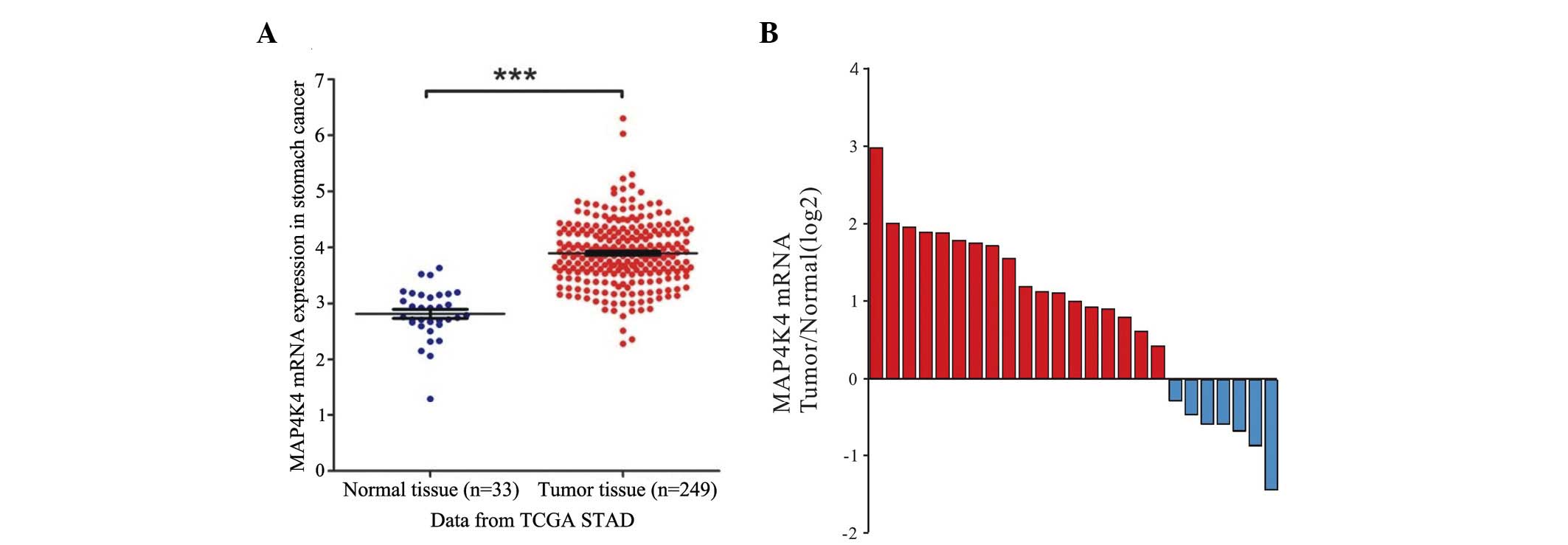
MAP4K4 mRNA is overexpressed in GC
To determine whether MAP4K4 was overexpressed in GC,
the mRNA expression of MAP4K4 in GC and adjacent tissues was
analyzed by bioinformatics. As shown in Fig. 1A, the mRNA expression levels of
MAP4K4 were significantly increased in GC tissues compared with the
adjacent tissues of patients from TCGA stomach adenocarcinoma
data-sets, which indicated that MAP4K4 may be an oncogene in
GC.
To further determine the expression of MAP4K4 in GC,
RT-qPCR analysis was performed on 25 pairs of GC and their matched
non-cancerous tissue samples. Overexpressed levels of MAP4K4 mRNA
were identified in 72% (18/25) of the assessed GC tissues (Fig. 1B). Statistical analysis revealed
that the MAP4K4 mRNA was significantly overexpressed in gastric
tumor tissues as compared with the normal tissues (P<0.001).
MAP4K4 is downregulated by RNAi in GC
cells
The protein expression of MAP4K4 was also detected
in five GC cell lines by western blotting. A high expression level
of MAP4K4 was observed in the BGC-823 cells (Fig. 2A); consequently, BGC-823 cells were
selected for the further analysis.
 | Figure 2MAP4K4 expression is suppressed by
RNAi in BGC-823 cells. (A) The protein expression of MAP4K4 in five
GC cell lines (MGC-803, BGC-823, SGC-7901, AGC and MKN-28) was
detected by western blotting. GAPDH was used as loading control.
The BGC-823 cells demonstrated the highest expression of MAP4K4,
and were subsequently selected for further analysis. (B) Western
blotting and (C) reverse transcription-quantitative polymerase
chain reaction demonstrated the efficiency of MAP4K4 knockdown.
**P<0.01, vs. WT group. WT, wild-type; Neg., negative
control; Ri-1, Ri-2 and Ri-3, MAP4K4-shRNA-1, -2 and -3; MAP4K4,
mitogen-activated protein kinase kinase kinase kinase 4. |
To investigate the function of MAP4K4 on GC, shRNA
plasmids were constructed for the downregulation of MAP4K4
expression. A total of three pairs of human MAP4K4 gene shRNA
sequences and a non-specific scrambled shRNA sequence, negative
control (Neg.), were cloned into a lentiviral plasmid. The
recombinant lentivirus was packaged into HEK-293T cells. BGC-823
cells were subsequently infected with MAP4K4-RNAi or Neg. virus.
The silencing effect of the MAP4K4-RNAi was confirmed by western
blotting (Fig. 2B) and RT-qPCR
(Fig. 2C). MAP4K4-Ri-3 was
identified as the most efficient infected cell type, and was used
for subsequent experiments.
Downregulation of MAP4K4 by RNAi inhibits
cell proliferation and induces G1 phase cell cycle arrest in GC
cells
To determine the role of MAP4K4 downregulation in
the proliferation of hepatocellular carcinoma cells, the
proliferation of BGC-823 cells infected with MAP4K4-Ri-3 was
analyzed using a CCK-8 assay. As shown in Fig. 3A, cell growth was markedly impaired
in the MAP4K4-Ri-3 virus-infected cells (MAP4K4-Ri-3) compared with
the wild-type cells (WT) and scrambled shRNA virus-infected cells
(Neg.). These data indicated a role for MAP4K4 in the proliferation
of GC cells.
Subsequently, the potential inhibitory effect of
MAP4K4 knockdown on cell cycle progression was investigated. As
shown in Fig. 3B, the suppression
of MAP4K4 overexpression resulted in a higher number of cells in
the G1 phase (64.9±2.9%) compared with the WT (46.0±2.8%) and Neg.
cells (44.9±1.7%). There was a concomitant reduction in the number
of cells in the S and G2-M phases. These data suggested that the
knockdown of MAP4K4 induced cell cycle arrest in G1 in GC cells,
which may be associated with the inhibition of cell proliferation
in MAP4K4-Ri-3 cells.
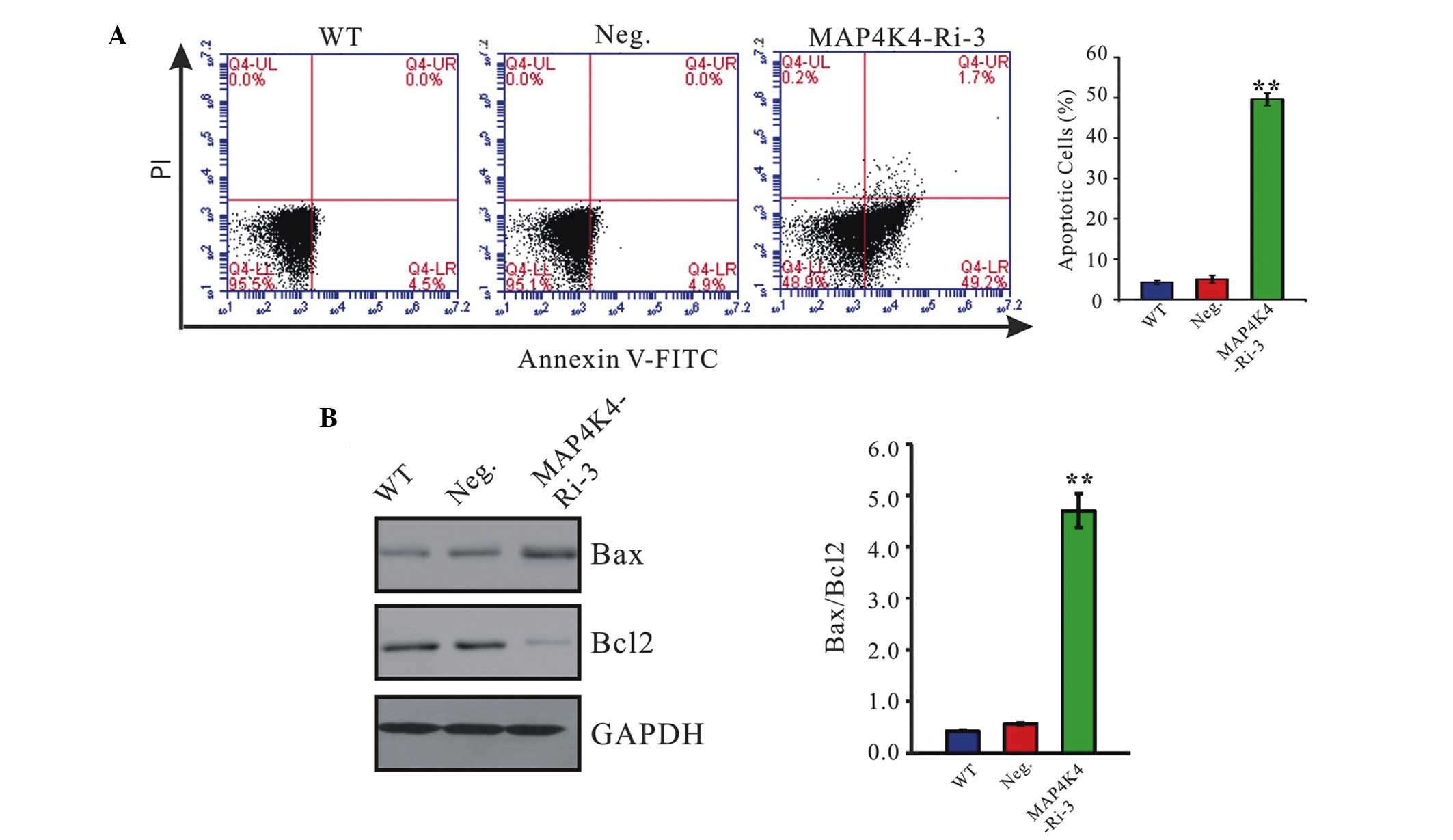
MAP4K4 knockdown induces cell apoptosis
by increasing the ratio of Bax/Bcl-2
To assess the effects of MAP4K4 on cell apoptosis,
annexin V/PI staining was performed (Fig. 4A). The ratio of cells undergoing
apoptosis was significantly increased to 49.6±1.5% in MAP4K4-Ri-3
cells compared with the WT (4.3±0.5%) and Neg. cells (5.0±1.0%).
These data suggested that MAP4K4 may fulfil an antiapoptotic role
in GC cells.
The proteins of the Bcl-2 family perform critical
roles in the regulation of apoptosis by functioning as promoters
(e.g. Bax) or as inhibitors (e.g. Bcl-2) of cell death processes
(12–14). Western blotting was performed to
detect the protein expression levels of Bcl-2 and Bax in the
MAP4K4-Ri-3 cells (Fig. 4B).
MAP4K4 knockdown resulted in a marked reduction in the level of the
antiapoptotic protein, Bcl-2, with a concomitant increase in the
level of proapoptotic protein Bax, compared with the control cells
(WT and Neg.). These data revealed that the suppression of MAP4K4
expression may increase the ratio of Bax/Bcl-2, which may
contribute to the increase in cell apoptosis.
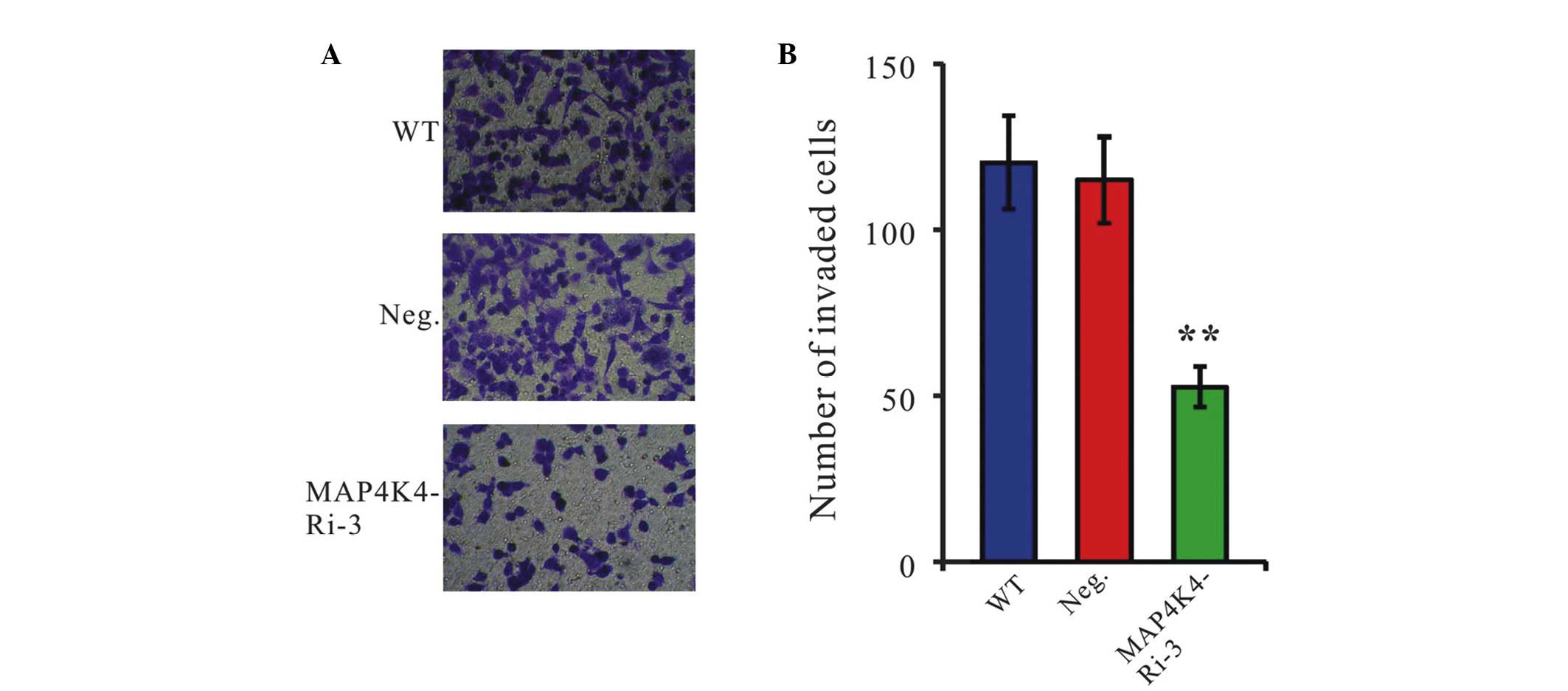
Downregulation of MAP4K4 by RNAi inhibits
the invasiveness of GC cells
To examine whether MAP4K4 affected the invasive
ability of GC cells, a Matrigel-coated membrane chamber invasion
assay was performed. As shown in Fig.
5, a markedly reduced invasive ability was observed in MAP4K4
knockdown cells compared with the control cells. The number of
invading MAP4K4-Ri-3 cells was 44% compared with the Neg. cells
(WT, 120±14; Neg., 115±13; MAP4K4-Ri-3, 53±6).
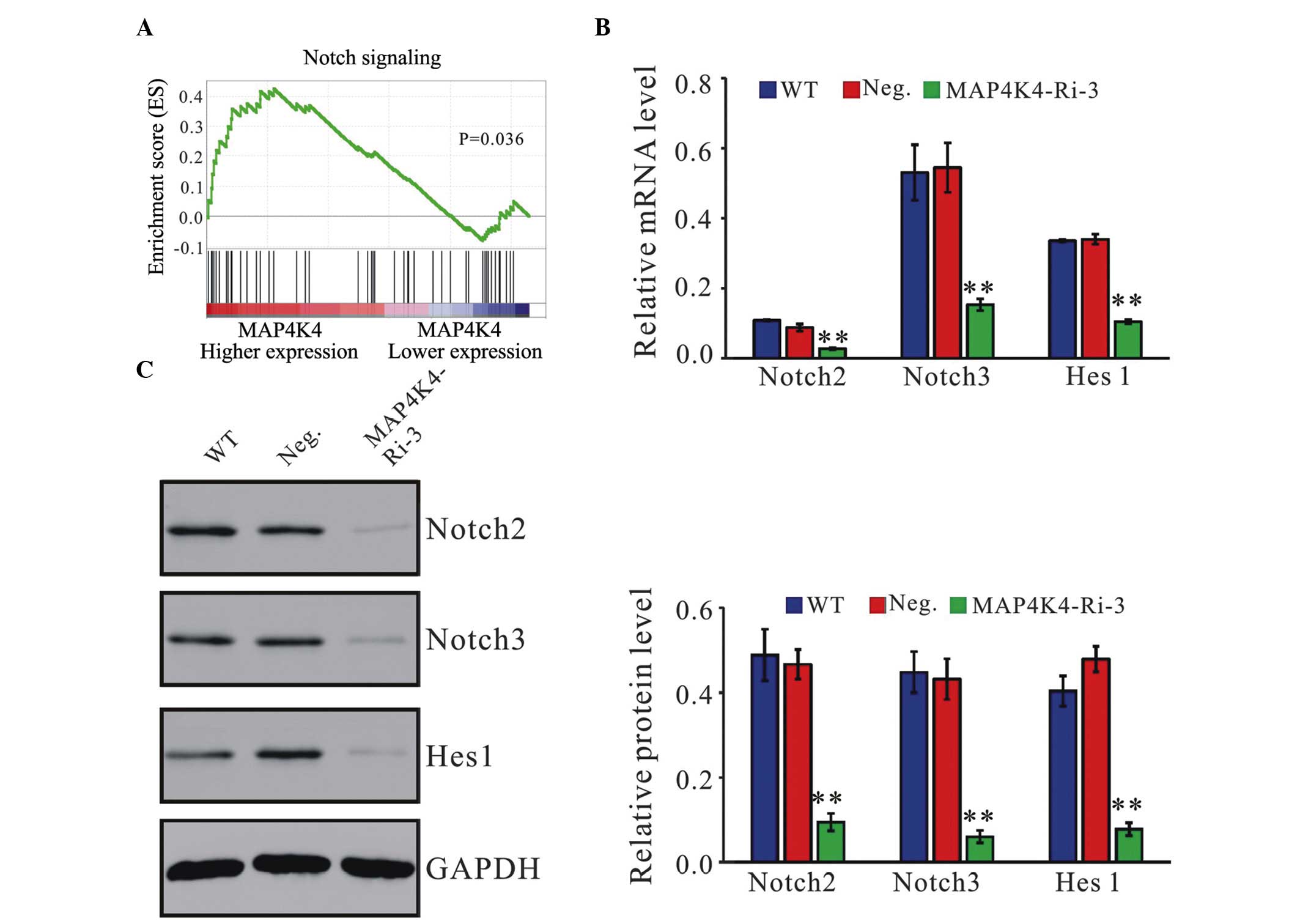
MAP4K4 positively correlates with the
Notch signaling pathway
To further investigate the biological pathways
involved in the pathogenesis of GC stratified by the median of
MAP4K4 expression level, a GSEA analysis was performed. The
enrichment plots of GSEA revealed that the MAP4K4 signature was
positively correlated with the Notch signaling pathway (Fig. 6A). The gene expression patterns of
important regulators in the Notch signaling pathway were determined
at the mRNA and protein expression levels. The mRNA (Fig. 6B) and protein levels (Fig. 6C) of Notch2, Notch3 and Hes1 were
markedly decreased following the downregulation of MAP4K4.
Discussion
MAPKs have an essential role in modulating the
regulation of several important cellular processes, including
growth, migration, differentiation, apoptosis and stress-associated
responses. The involvement of MAP4K4 in cancer has generated great
interest. High expression levels of MAP4K4 have been reported in
several cancer types (5,8–10).
The molecular mechanisms underlying the development and progression
of GC remain to be fully elucidated. Consistent with these previous
studies, the bioinformatics analysis in the present study, using
data from TCGA, indicated that MAP4K4 was highly expressed in GC
(Fig. 1A), which was further
confirmed by RT-qPCR (Fig.
1B).
In the present study, MAP4K4 was subsequently
knocked down in BGC-823 cells using RNAi (Fig. 2). The data revealed that
suppressing MAP4K4 expression markedly inhibited the proliferation
(Fig. 3) and the invasion
(Fig. 5) of BGC-823 cells, which
is consistent with previous reports in pancreatic cancer (15) and hepatocellular carcinoma
(9). These data further indicated
the importance of MAP4K4 in gastric cell carcinogenesis.
Cell cycle regulation is frequently abnormal in the
majority of the common malignancies, resulting in aberrant cell
proliferation (16,17). Silencing of MAP4K4 by RNAi markedly
induced G1 phase arrest of cell cycle progression (Fig. 3B), which indicated that the
inhibition of cell proliferation in GC cells is due to the
inhibition of cell cycle progression.
The G1 phase arrest of cell cycle progression
provides an opportunity for the cells to either undergo repair, or
to enter into apoptosis. The effects of knockdown of the MAP4K4
protein on the induction of apoptosis were subsequently determined
in BGC-823 cells. The flow cytometry data revealed that the
silencing of MAP4K4 resulted in a marked induction of apoptosis
(Fig. 4A). Apoptosis is of major
importance during the elimination of the mutated neoplastic and
hyperproliferating neoplastic cells, and therefore, is considered
as a protective mechanism against cancer progression (18). Due to its anti-apoptotic role in
GC, MAP4K4 may be a putative therapeutic target worthy of further
investigation.
The proteins of the Bcl-2 family either promote cell
survival (e.g. Bcl-2) or induce programmed cell death (e.g. Bax).
The ratio of Bax/Bcl-2 is critical for the induction of apoptosis,
and this ratio determines whether cells undergo apoptosis (14). The present study revealed that
MAP4K4 knockdown resulted in an increase in the expression of Bax
and a decrease in the expression of Bcl-2 (Fig. 4B), and the ratio of Bax/Bcl-2 was
increased (Fig. 4B), which
suggested that MAP4K4 exerted its antiapoptotic role by regulating
the ratio of Bax/Bcl-2.
The GSEA indicated that the overexpression of MAP4K4
correlates with the activation of the Notch signaling pathway
(Fig. 6A). The Notch pathway is an
evolutionarily conserved cell signaling mechanism, which is
involved in several cellular processes, including proliferation,
differentiation, apoptosis and stem cell maintenance (19). Dysregulation of Notch signaling was
reported in several cancer types (20–22).
It was reported that Notch2 (23,24)
and Notch3 (25) are important in
controlling the progression of GC. Hes1, a target gene of Notch
signaling activation, is considered to be critical for the
development of prostate cancer (26–28).
In the present study, MAP4K4 knockdown markedly decreased the mRNA
(Fig. 6B) and protein expression
levels (Fig. 6C) of Notch2, Notch3
and Hes1, which indicated an association between MAP4K4 function
and the regulation of Notch signaling in GC cells.
In conclusion, the results of the present study
indicated that MAP4K4 knockdown inhibited cell proliferation via
induction of the G1 phase arrest. Mechanistic evidence has been
provided to demonstrate that cell apoptosis induced by MAP4K4 RNAi
was mediated through an increase in the ratio of Bax/Bcl2.
Additionally, MAP4K4 was positively correlated with the Notch
signaling pathway in GC, and therefore may provide useful
information for the development of targeted therapeutic strategies
in the future.
References
|
1
|
Vogelaar IP, van der Post RS, Bisseling
TM, van Krieken JH, Ligtenberg MJ and Hoogerbrugge N: Familial
gastric cancer: Detection of a hereditary cause helps to understand
its etiology. Hered Cancer Clin Pract. 10:182012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Clegg LX, Ward E, Ries LA, Wu X,
Jamison PM, Wingo PA, Howe HL, Anderson RN and Edwards BK: Annual
report to the nation on the status of cancer, 1975–2001, with a
special feature regarding survival. Cancer. 101:3–27. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Turjanski AG, Vaqué JP and Gutkind JS: MAP
kinases and the control of nuclear events. Oncogene. 26:3240–3253.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Collins CS, Hong J, Sapinoso L, Zhou Y,
Liu Z, Micklash K, Schultz PG and Hampton GM: A small interfering
RNA screen for modulators of tumor cell motility identifies MAP4K4
as a promigratory kinase. Proc Natl Acad Sci USA. 103:3775–3780.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wright JH, Wang X, Manning G, LaMere BJ,
Le P, Zhu S, Khatry D, Flanagan PM, Buckley SD, Whyte DB, et al:
The STE20 kinase HGK is broadly expressed in human tumor cells and
can modulate cellular transformation, invasion and adhesion. Mol
Cell Biol. 23:2068–2082. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hu Y, Leo C, Yu S, Huang BC, Wang H, Shen
M, Luo Y, Daniel-Issakani S, Payan DG and Xu X: Identification and
functional characterization of a novel human misshapen/Nck
interacting kinase-related kinase, hMINK beta. J Biol Chem.
279:54387–54397. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zohn IE, Li Y, Skolnik EY, Anderson KV,
Han J and Niswander L: p38 and a p38-interacting protein are
critical for downregulation of E-cadherin during mouse
gastrulation. Cell. 125:957–969. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liang JJ and Wang H, Rashid A, Tan TH,
Hwang RF, Hamilton SR, Abbruzzese JL, Evans DB and Wang H:
Expression of MAP4K4 is associated with worse prognosis in patients
with stage II pancreatic ductal adenocarcinoma. Clin Cancer Res.
14:7043–7049. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu AW, Cai J, Zhao XL, Jiang TH, He TF,
Fu HQ, Zhu MH and Zhang SH: ShRNA-targeted MAP4K4 inhibits
hepatocellular carcinoma growth. Clin Cancer Res. 17:710–720. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Qiu MH, Qian YM, Zhao XL, Wang SM, Feng
XJ, Chen XF and Zhang SH: Expression and prognostic significance of
MAP4K4 in lung adenocarcinoma. Pathol Res Pract. 208:541–548. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun M, Xia R, Jin F, Xu T, Liu Z, De W and
Liu X: Downregulated long noncoding RNA MEG3 is associated with
poor prognosis and promotes cell proliferation in gastric cancer.
Tumour Biol. 35:1065–1073. 2014. View Article : Google Scholar
|
|
12
|
Adams JM and Cory S: Life-or-death
decisions by the Bcl-2 protein family. Trends Biochem Sci.
26:61–66. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hetz C: BCL-2 protein family. Essential
regulators of cell death. Preface Adv Exp Med Biol. 687:vii–viii.
2010.
|
|
14
|
Reed JC: Regulation of apoptosis by bcl-2
family proteins and its role in cancer and chemoresistance. Curr
Opin Oncol. 7:541–546. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao G, Wang B, Liu Y, Zhang JG, Deng SC,
Qin Q, Tian K, Li X, Zhu S, Niu Y, et al: miRNA-141, downregulated
in pancreatic cancer, inhibits cell proliferation and invasion by
directly targeting MAP4K4. Mol Cancer Ther. 12:2569–2580. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Evan GI and Vousden KH: Proliferation,
cell cycle and apoptosis in cancer. Nature. 411:342–348. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Molinari M: Cell cycle checkpoints and
their inactivation in human cancer. Cell Prolif. 33:261–274. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Koch U and Radtke F: Notch and cancer: A
double-edged sword. Cell Mol Life Sci. 64:2746–2762. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nickoloff BJ, Osborne BA and Miele L:
Notch signaling as a therapeutic target in cancer: A new approach
to the development of cell fate modifying agents. Oncogene.
22:6598–6608. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Westhoff B, Colaluca IN, D'Ario G,
Donzelli M, Tosoni D, Volorio S, Pelosi G, Spaggiari L, Mazzarol G,
Viale G, et al: Alterations of the Notch pathway in lung cancer.
Proc Natl Acad Sci USA. 106:22293–22298. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Stylianou S, Clarke RB and Brennan K:
Aberrant activation of notch signaling in human breast cancer.
Cancer Res. 66:1517–1525. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tseng YC, Tsai YH, Tseng MJ, Hsu KW, Yang
MC, Huang KH, Li AF, Chi CW, Hsieh RH, Ku HH and Yeh TS:
Notch2-induced COX-2 expression enhancing gastric cancer
progression. Mol Carcinog. 51:939–951. 2012. View Article : Google Scholar
|
|
24
|
Bauer L, Langer R, Becker K, Hapfelmeier
A, Ott K, Novotny A, Höfler H and Keller G: Expression profiling of
stem cell-related genes in neoadjuvant-treated gastric cancer: A
NOTCH2, GSK3B and β-catenin gene signature predicts survival. PLoS
One. 7:e445662012. View Article : Google Scholar
|
|
25
|
Kang H, An HJ, Song JY, Kim TH, Heo JH,
Ahn DH and Kim G: Notch3 and Jagged2 contribute to gastric cancer
development and to glandular differentiation associated with MUC2
and MUC5AC expression. Histopathology. 61:576–586. 2012.PubMed/NCBI
|
|
26
|
Lu JP, Zhang J, Kim K, Case TC, Matusik
RJ, Chen YH, Wolfe M, Nopparat J and Lu Q: Human homolog of
Drosophila Hairy and enhancer of split 1, Hes1, negatively
regulates δ-catenin (CTNND2) expression in cooperation with E2F1 in
prostate cancer. Mol Cancer. 9:3042010. View Article : Google Scholar
|
|
27
|
Leong KG and Gao WQ: The Notch pathway in
prostate development and cancer. Differentiation. 76:699–716. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Villaronga MA, Bevan CL and Belandia B:
Notch signaling: A potential therapeutic target in prostate cancer.
Curr Cancer Drug Targets. 8:566–580. 2008. View Article : Google Scholar : PubMed/NCBI
|