Introduction
β-cell dysfunction is a key characteristic in the
pathogenesis of type 2 diabetes (1,2) and
hyperglycemia serves a direct role in pancreatic β-cell dysfunction
and death (3). However, the
mechanism underlying glucose-induced β cell apoptosis and
dysfunction remain to be fully understood. Deoxyribonuclease I
(DNase I) is a 38-kDa glycoprotein that is a type of
Ca2+/Mg2+-dependent non-restriction nuclease,
which can hydrolyze phosphodiester bonds in single and double
stranded DNA (4). Thus, endogenous
DNase I has been suggested as a candidate endonuclease facilitating
chromatin breakdown during apoptosis.
As a secretory endonuclease, DNase I is the
predominant nuclease observed in body fluids such as serum and
urine. In mammals, the pancreas exhibits the highest DNase I
activity (5), with ~60–65% total
serum DNase I secreted by the pancreas (6). Previously, two diseases, systemic
lupus erythematosus (SLE) and acute myocardial infarction (AMI),
have been demonstrated to be associated with alterations in serum
levels of DNase I (7,8). The association between DNase I and
type 2 diabetes remains to be fully elucidated.
Given the essential role of pancreatic tissue in the
development of metabolic syndrome, it is suggested that there may
be a functional significance of DNase I in the morbidity of type 2
diabetes (9). In the present
study, DNase I activity in patients with type 2 diabetes was
assessed, with the aim to explore the role of DNase I in high
glucose conditions in association with β cell apoptosis.
Materials and methods
Human serum collection
Individuals who were diagnosed with type 2 diabetes
[glycated hemoglobin (HbA1c) ≥6.5% or fasting blood glucose ≥7.0
mmol/l; 35–65 years old] were recruited at the China-Japan
Friendship Hospital (Beijing) between September and December, 2012
(Table I). Patients who were
pregnant, or presented with other acute or chronic complications,
including malignant tumors, SLE, AMI, uncontrolled hypertension or
other serious illnesses were excluded from the study. Healthy
volunteers with normal glucose, triglyceride and cholesterol levels
and with the absence of other diseases were enrolled in the study.
A consent form was signed by all subjects prior to enrollment.
Blood samples were taken from the antecubital vein without any
anticoagulant, and were centrifuged for 10 min at 1,500 × g at an
ambient temperature. Serum samples were aliquoted into 200
μl samples, which were stored at −80°C until required for
further experiments. The concentration of calcium, blood glucose
and HbA1c in the serum was measured using the Hitachi 7600
Auto-Biochemistry Instrument (Hitachi, Ltd., Tokyo, Japan) with the
reagents and methods provided by the manufacturer. This study was
approved by the ethics committee of the China-Japan Friendship
Hospital (Beijing, China).
 | Table IBaseline patient demographic and
background characteristics. |
Table I
Baseline patient demographic and
background characteristics.
| Characteristic | Healthy controls | Diabetes
patients |
|---|
| N (male/female) | 51 (26/25) | 66 (44/22) |
| Age (years) | 50.47±8.37 | 52.68±8.91 |
| FPG (mmol/l) | 5.07±0.37 | 8.07±3.07a |
| HbA1C (%) | 5.5±0.35 | 8.77±1.63b |
| Systolic pressure
(mmHg) | 118.47±17.49 | 130.45±13.22b |
| Diastolic pressure
(mmHg) | 77.93±12.25 | 79.68±7.71 |
| BMI
(kg/m2) | 23.24±2.91 | 26.83±3.73 |
| Cholesterol
(mmol/l) | 4.38±0.58 | 4.75±1.18 |
| Triglyceride
(mmol/l) | 1.11±0.43 | 1.81±0.97b |
| High density
lipoprotein (mmol/l) | 1.55±0.32 | 1.23±0.32b |
| Low density
lipoprotein (mmol/l) | 2.64±0.42 | 2.64±0.81 |
Human tissues
Surgically removed human pancreatic cancer tissue
specimens were obtained with approval from the China-Japan
Friendship Hospital Institutional Review Board (Beijing, China).
Human pancreatic tissues were collected from patients with
pancreatic cancer with a history of diabetes for at least 4 years
(3 male and one female) or without diabetes as control (2 male and
2 female). The patients were aged from 50 to 65. Written informed
consent was obtained from each patient. Surgically removed
pancreatic tissues from patients with pancreas cancer, with or
without diabetes, were used for the observation of DNase I.
Sections of tumor-free pancreas were immediately placed into
ice-cold fixative [(4% formaldehyde + 0.1% glutaraldehyde in
phosphate-buffered saline (PBS)] subsequent to resection, then were
processed for immunohistochemistry.
Measurement of DNase I activity
DNase I activity was measured using the radial
enzyme-diffusion method. Briefly, calf thymus DNA (Sigma-Aldrich,
St. Louis, MO, USA) was used as the substrate and was mixed with
SYBR Green I (Unique, Beijing, China) in DNase buffer (Worthington
Biochemical Corporation, Lakewood, NJ, USA). The end concentration
of substrate DNA in the mixture was 500 ng/ml. Regular (2%) agarose
(Biowest, HongKong, China) was melted using distilled water and
mixed with the solution. The mixed gel was then poured into the
96-well microplate. The serum samples (2 μl) were then
injected into the center of each well of the gel plate. A total of
2 μl DNase I (Worthington Biochemical Corporation) at
concentrations of 0.034–1.1 U/ml were additionally added into the
gel plate to calculate the standard curve. Each sample was
performed in duplicate. The gel plate was then incubated at 37°C in
a airtight black box for 12 h. The areas of hydrolyzed DNA were
assessed with a gel documentation and image analysis system
(ChampGel 5500; Beijing Sage Creation Science Co., Ltd., Beijing,
China) and were quantitated with Image-Pro Plus software, version
6.0 (Media Cybernetics, Inc., Rockville, MD, USA).
Immunohistochemistry for insulin,
glucagon and DNase I
Immunohistochemistry was conducted in 2
μm-thick paraffin (Sinopharm Chemical Reagent, Beijing,
China)-embedded pancreatic sections mounted in polylysine-coated
slides. Unstained sections were deparaffinized and rehydrated in
xylene (Sinopharm Chemical Reagent) and graded ethanol. Subsequent
to rinsing in distilled water, endogenous peroxidase was blocked
with 3% hydrogen peroxide (Sinopharm Chemical Reagent) for 30 min
to reduce nonspecific binding, then were incubated with primary
antibodies, mouse anti-insulin (1:5,000; ZM-0155; OriGene
Technologies, Inc., Beijing, China), rabbit anti-glucagon (1:5,000;
ZA-0119; OriGene Technologies) and rabbit anti-DNase I (1:1,000;
sc-30058; Santa Cruz Biotechnology, Inc., Dallas, TX, USA). The
primary antibodies were detected using the anti-mouse/rabbit
Poly-Horseradish-Peroxidase from the Immunohistochemistry
Ready-to-use Detection kit (GTVision™ III Detection
System/Mo&Rb; Gene Tech Biotechnology Co., Ltd., Shanghai,
China). Visualization was performed using diaminobenzidine
chromogen buffer. Sections were counterstained with hematoxylin
(Sinopharm Chemical Reagent). Digital morphometric analyses were
performed using the Leica DM5000 optical microscope with Leica Qwin
Plus analysis software DM5000 (Leica Microsystems, Inc., Buffalo
Grove, IL, USA).
INS-1 cell culture
The insulin-secreting cell line INS-1 (Cell Resource
Center of Peking Union Medical College, Beijing, China) was
cultured in RPMI-1640 (Hyclone, Logan, UT, USA), supplemented with
10% fetal calf serum (FCS; Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), 2 mmol/l L-glutamine, 10 mmol/l HEPES, 1 mmol/l
sodium pyruvate and 50 mmol/l 1,2-mercaptoethanol (all from
Sigma-Aldrich) in a humidified atmosphere (5% CO2,
37°C).
Small interfering RNA (siRNA)
transfection
DNase I siRNAs were synthesized by Shanghai
GenePharma Co., Ltd. (Shanghai, China). The siRNA sequences were as
follows: DNase I, 5′-GCCGCAAAAGCUACAAGGATT-3′ and
5′-UCCUUGUAGCUUUUGCGGCTT-3′; negative control siRNA,
5′-UUCUCCGAACGUGUCACGUTT-3′ and 5′-ACGUGACACGUUCGGAGAATT-3′. INS-1
cells were cultured in Opti-MEM (Invitrogen; Thermo Fisher
Scientific, Inc.) for 12 h and then transfected with the siRNAs and
Lipofectamine™ 2000 (Invitrogen; Thermo Fisher Scientific, Inc.).
The cells were then plated into wells and incubated at 37°C in a
CO2 incubator. A total of 6 h later, the medium was
replaced with RPMI-1640 containing either an 11.1 mM or 30 mM
concentration of glucose (Sinopharm Chemical Reagent). All
experiments using siRNA-transfected INS-1 cells were performed 48 h
subsequent to transfection unless otherwise stated.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA (2 μg) was converted to
first-strand cDNA using the RevertAid™ First Strand cDNA Synthesis
kit (Fermentas; Thermo Fisher Scientific, Inc.). RT-qPCR analysis
was used to measure the relative levels of DNase I, Bcl-2 and
caspase-3 mRNA expression. The amplification was performed on an
ABI 7500 thermocycler (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The nucleotide sequences of the primers were as
follows: DNase I, forward 5′-GGTCCGAGAGTTTGCGATTGT-3′ and reverse
5′-TGCAGCCAGCATTGAAATCTC-3′; caspase-3, forward
5′-AGCAGTTACAAAATGGATTAC-3′ and reverse
5′-ATCTCCATGACTTAGAATCAC-3′; Bcl-2, forward
5′-TGGTGGACAACATCGCTCTGT-3′ and reverse
5′-CCCAGGTATGCACCCAGAGTG-3′; β-actin, forward
5′-ATCGTGCGTGACATTAAGGAGAAG-3′ and reverse
5′-AGGAAGGAAGGCTGGAAGAGTG-3′. The PCR reaction conditions were as
follows: 94°C for 1 min followed 56°C for 30 sec and 72°C for 1 min
repeated for 40 cycles; and a final step at 72°C for 10 min to stop
the reaction. Levels of DNase I, caspase-3 and Bcl-2 mRNA were
subsequently normalized to β-actin mRNA levels.
Western blot analysis
Cells were lysed in lysis buffer [50 mM Tris, pH
7.5, 250 mM NaCl, 0.1% sodium dodecyl sulfate (SDS), 2 mM
dithiothreitol, 0.5% NP-40, 1 mM phenylmethanesulfonyl fluoride and
protease inhibitor cocktail] on ice for 30 min. Protein fractions
were collected by centrifugation at 15,000 × g at 4°C for 10 min
and then were subjected to 10% SDS-polyacrylimide gel (Sinopharm
Chemical Reagent) electrophoresis and were transferred to
polyvinylidene difluoride membranes (Merck Millipore, Billerica,
MA, USA). The membranes were blocked with 5% bovine serum albumin
(BSA) and incubated with the specific antibodies overnight. The
following antibodies were used: Rabbit anti-DNase I (1:500;
sc-30058; Santa Cruz Biotechnology, Inc.), mouse anti-caspase-3
(sc-56055; Santa Cruz, USA) and mouse β-actin (1:5,000; DKM9001;
Tianjin Sungene Biotech Co., Ltd., Tianjin, China) to examine the
concentrations of DNase I, caspase-3 and β-actin proteins in the
lysates, respectively. A goat anti-mouse (H+L: 115-035-003)and goat
anti-rabbit (H+L: 111-035-003) IgG horseradish peroxidase-labeled
secondary antibody (Jackson ImmunoResearch Laboratories, West
Grove, PA, USA). was added and visualized using a chemiluminescence
reagent (ECL; Engreen Biosystem, Ltd., Beijing, China).
Flow cytometry analysis
The cell apoptotic rate was detected by flow
cytometry. Briefly, INS-1 cells were plated in 6-well plates at a
density of 5×104 cells per well and were incubated at
37°C for 24 h. The media was then replaced, so that three groups of
cells were present: Cells exposed to constant normal glucose (11.1
mM), constant high glucose (30 mM) and high glucose with DNase I
knockout. Subsequent to incubation for an additional 48 h, the cell
apoptotic rate was detected by Annexin V staining [Fluorescein
Isothiocyanate (FITC) Annexin V Apoptosis Detection kit; BD
Biosciences, Franklin Lakes, NJ, USA]. Subsequently, cells were
treated with trypsin-ethylenediaminetetraacetic acid and were
centrifuged at 1,400 × g for 5 min at 4°C. Cells were washed with
PBS 2 times, then were resuspended with 100 μl binding
buffer. Supernatants were then added with 5 μl Annexin
V-FITC and 5 μl prop-idium iodide. Subsequent to incubation
for 15 min in the dark, 400 μl binding buffer was added, and
the cells were analyzed by the FACSCanto II flow cytometer (BD
Biosciences).
Cell proliferation assays
Cell viability was determined by the Cell Counting
Kit-8 assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan)
according to the manufacturer's protocol. Cells
(1.5×104) were grown as triplicates in 96-well plates
and allowed to adhere for 24 h. Cells were exposed to various DNase
I concentrations (1, 5, 10, 50 and 100 mU/μl), for 24 h,
then 10 μl tetrazolium substrate was added to each well of
the plate. Plates were incubated at 37°C for 1 h, then the optical
density was measured at 450 nm using a Bio-Rad 680 Enzyme-linked
Immunosorbent Assay Reader (Bio-Rad Laboratories, Inc., Hercules,
CA, USA). Cell viability was expressed as a percentage of control
cells (non-treated).
Terminal deoxynucleotidyl transferase
dUTP nick-end labeling (TUNEL) assay
For the TUNEL assay, INS-1 cells were seeded into
96-well plates (2×104 cells/well) and treated with or
without high glucose for 12 h. Subsequently, the cells were
incubated with 50 or 100 mM DNase I in RPMI-1640 containing 10% FCS
for 24 h. The cells were then washed with PBS (HyClone, Logan, UT,
USA), fixed in 4% paraformaldehyde (Sinopharm Chemical Reagent),
and permeabilized with 0.1% Triton X-100 (Sigma-Aldrich) in PBS/BSA
solution. The TUNEL assay was performed using the In Situ
Cell Death Detection kits (Roche Diagnostics, Indianapolis, IN,
USA).
Statistical analyses
SPSS software, version 11.0 (SPSS, Inc., Chicago,
IL, USA) was used for the statistical analysis. Statistical
analyses for parametric data and nonparametric data were performed
using the Kolmogorov-Smirnov test and the Mann-Whitney U test,
respectively. Spearman single regression analyses were used to
assess the correlation between healthy controls and patients with
diabetes. All of the data are presented as the mean ± standard
deviation. P<0.05 was considered to indicate a statistically
significant difference.
Results
Human DNase I activity
A total of 66 patients with diabetes were enrolled
in the current study. Of the participants, 22 were women (33.3%)
and 44 were men (66.7%) with a mean age of 52.68±8.91 (range, 35–65
years). Of the 52 healthy controls, 24 were women (46.1%) and 28
were men (53.8%), with a mean age of 50.47±8.37 (range, 35–65
years). Compared with the healthy controls, DNase I activity was
observed to tbe significantly elevated in patients with type 2
diabetes (0.2±0.06 U/ml vs. 0.27±0.12 U/ml; P<0.001; Fig. 1A). In addition, this increase was
identified to be positively correlated with serum Ca2+
concentration (r=0.387; P=0.002; Fig.
1B), however no correlation of DNase I activity with HbA1c
(r=0.06; P=0.641) or fasting glucose (r=0.132; P=0.297) was
observed. No significant difference of the DNase I activity was
identified between male and female patients with type 2 diabetes
(Fig. 1C).
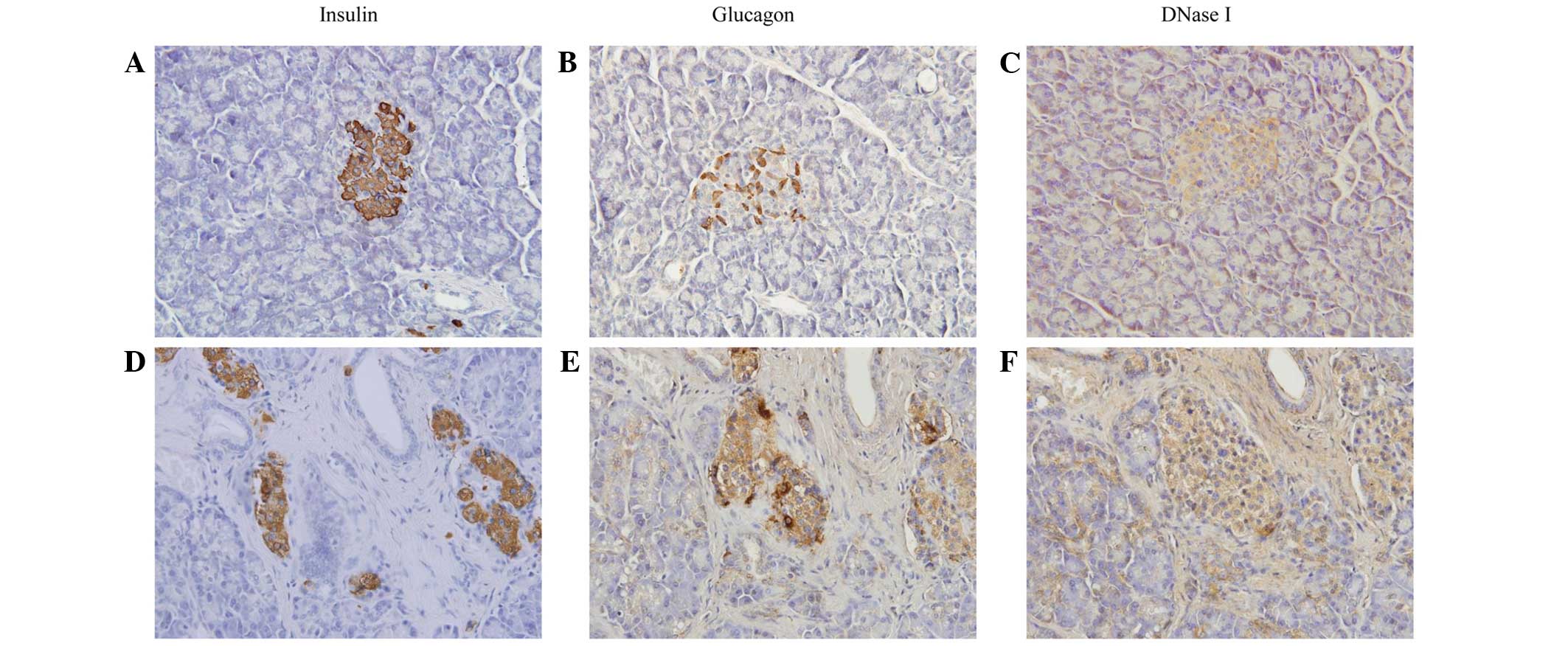
Immunohistochemistry results demonstrated that
islets in the diabetic patients exhibited an irregular morphology,
with poor insulin staining and increased glucagon staining. The
DNase I expression was marginally increased when compared with
those without diabetes (Fig.
2).
DNase I gene expression was increased in
high glucose-cultured cells
Compared with the normal group, DNase I was
significantly increased in the 30 mM glucose group, which is in
agreement with a previous study (10). High glucose additionally increased
the caspase-3 expression, whereas this increase was reversed by
DNase I siRNA transfection (Fig. 3B
and C). Notably, it was observed that when DNase I was knocked
down by DNase I siRNA, the caspase-3 level was observed to be
reduced when compared with the high glucose group. The specific
mechanism of this remains unclear. In the current study, it was
also identified that high levels of glucose resulted in cell
apoptosis. Subsequent to transfection with siRNA for 48 h, the rate
of cell apoptosis was examined, and it was identified hat the
apoptotic rate in the normal, high glucose and high glucose with
siRNA groups were 7.1%, 18.1% and 9.9%, respectively (Fig. 3D).
 | Figure 3DNase I knockdown can reduce the
apoptosis of cells cultured with high glucose. (A) Knockdown
efficiency examined by western blotting. Expression of DNase I,
Bcl-2 and caspase-3 in the three groups were examined by (B)
western blotting and (C) reverse transcription-quantitative
polymerase chain reaction. (D) Apoptotic rate examined by flow
cytometry [(a), normal; (b), high glucose; and (c), siRNA group].
Data are expressed as the mean ± standard deviation from three
independent experiments. *P<0.05 vs. the normal group
and #P<0.05 vs. the control group. DNase I,
deoxyribonuclease I; siRNA, small interfering RNA; NC, negative
control; N, normal; HG, high glucose; PI, propidium iodide; FITC,
fluorescein isothiocyanate. |
High glucose with 50 mU/μl DNase I leads
to cell apoptosis
To further explain the specific effects of DNase I
in cell proliferation, cells were cultured with high glucose and
DNase I siRNA. The cell proliferation results indicated that 50
mU/μl DNase I greatly suppressed the cell proliferation
(P<0.05; Fig. 4A). To more
accurately simulate the internal environment of patients with type
2 diabetes, the cells were cultured with high glucose and 50
mU/μl DNase I, and it was identified that this resulted in a
significant increase in caspase-3 levels (Fig. 4B and C). The TUNEL results
indicated that 50 mU/μl DNase I with high glucose resulted
in a marked increase in cell apoptosis, when compared with the high
glucose-treatment alone group. The flow cytometry results were in
agreement with those from TUNEL, indicating that high glucose
combined with 50 mU/μl DNase I resulted in clear cellular
apoptosis (34.6%), when compared with the normal and control groups
(6.9% and 17.3%, respectively) (Fig.
4D).
Discussion
Type 2 diabetes mellitus is a disease of high
prevalence, however its etiology remains to be fully elucidated.
Numerous factors including β cell apoptosis, oxidative stress and
inflammation have been suggested to contribute to the morbidity of
this disease. However, the specific mechanisms remain unclear. The
role of DNase I in SEL and AMI has been previously investigated,
however at present to the best of our knowledge, no correlative
studies on its association with type 2 diabetes have been conducted
(6,11,12).
In the current study, it was identified that DNase I levels were
increased in type 2 diabetes, and this increase in combination with
high glucose may aggravate β cell apoptosis. To the best of our
knowledge, the current study is the first to suggest that DNase I
may be involved in the pathophysiology of type 2 diabetes in the
development of metabolic syndrome.
DNase I is ubiquitously expressed in mammalian
tissues, particularly in the pancreas (13). It has been suggested to be highly
cytotoxic in mammalian cells when its activity and expression is
increased, as it can effectively digest single- and double-stranded
DNA (14). In addition, as a
'waste-management enzyme', it can trigger apoptosis (15,16).
Since its discovery in the bovine pancreas in 1905, correlations
have only been identified between DNase I and SLE and AMI (17). In SLE, the reduction of DNase I
levels were considered to be the predominant cause of antigen
accumulation, leading to autoimmunity (18). In AMI, the enzymatic activity was
identified to be elevated within 3 h and returned to basal levels
within 24 h, thus it is used for the early diagnosis of unstable
angina pectoris or non-ST-segment elevation myocardial infarction
(19,20). The associations between activity of
this enzyme and type 2 diabetes require elucidation.
In the present study, it was identified that high
glucose was able to induce an increase in DNase I expression, which
resulted in cell apoptosis. In order to confirm that apoptosis was
correlated with DNase I expression, siRNA was used to knock down
DNase I. It was observed that a reduction in DNase I expression
resulted in significant reductions in apoptotic rate and caspase-3
protein levels, even in the presence of high glucose. To further
investigate the role of DNase I in cell apoptosis, cells were
co-cultured with high glucose and 50 mU/μl DNase I to
simulate the conditions of type 2 diabetes. Notably, a significant
increase in apoptosis was observed in cells with 50 mU/μl
DNase I and high glucose, compared with high glucose-culture alone.
This suggested that DNase I may participate in β cell apoptosis in
type 2 diabetes.
However, the specific mechanisms of DNase I in type
2 diabetes remain to be fully elucidated, due to the complex
interactions among various factors. DNase I hypersensitive sites
(DHSs; short regions of chromatin that are highly sensitive to
DNase I digestion) in the cis-regulatory region on chromatin
has been previously suggested to be correlated with disease
(21,22). Human regulatory DNA encompasses a
variety of cis-regulatory elements, within which, the
cooperative binding of transcription factors creates focal
alterations in chromatin structure. Maurano et al (23) identified that disease-associated
variants in DHSs systematically alter the transcription factor
recognition sequences and allelic chromatin states, and form
regulatory networks. Stitzel et al (24) reported that DNase I hypersensitive
sites in the cis-regulatory elements of islet β-cells are
increased in the pancreatic islets, which may contribute to the
morbidity of type 2 diabetes. Thus, it is suggested that in type 2
diabetes, the increase of DNase I may affect the short regions of
chromatin cleavage and influence the secretion of insulin. This may
result in the development of type 2 diabetes.
In summary, the present study provided novel insight
into the central role of DNase I in high glucose-induced pancreatic
β-cell apoptosis. It was demonstrated that high glucose was able to
increase the expression of DNase I, and that this may aggravate
β-cell apoptosis. These observations may prove useful for the
development of novel new therapeutic strategies for the treatment
of type 2 diabetes mellitus.
Acknowledgments
The authors would like to thank Dr Shi Rui Li for
their technical assistance in conducting the experiments. The
current study was supported by grants from the National Natural
Scientific Foundation of China (grant nos. 81173422 and
81402814).
References
|
1
|
Butler AE, Janson J, Bonner-Weir S, Ritzel
R, Rizza RA and Butler PC: Beta-cell deficit and increased
beta-cell apoptosis in humans with type 2 diabetes. Diabetes.
52:102–110. 2003. View Article : Google Scholar
|
|
2
|
Mandrup-Poulsen T: Apoptotic signal
transduction pathways in diabetes. Biochem Pharmacol. 66:1433–1440.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ardestani A, Paroni F, Azizi Z, Kaur S,
Khobragade V, Yuan T, Frogne T, Tao W, Oberholzer J, Pattou F, et
al: MST1 is a key regulator of beta cell apoptosis and dysfunction
in diabetes. Nat Med. 20:385–397. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhou Z, Zhu C, Ren J and Dong S: A
graphene-based real-time fluorescent assay of deoxyribonuclease I
activity and inhibition. Anal Chim Acta. 740:88–92. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kaneko Y, Takeshita H, Mogi K, Nakajima T,
Yasuda T, Itoi M, Kuwano H and Kishi K: Molecular, biochemical and
immunological analyses of canine pancreatic DNase I. J Biochem.
134:711–718. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Funakoshi A, Wakasugi H and Ibayashi H:
Clinical investigation of serum deoxyribonuclease: II. Clinical
studies of serum deoxyribonuclease activity in pancreatic disease.
Gastroenterol Jpn. 14:436–440. 1979.PubMed/NCBI
|
|
7
|
Martinez-Valle F, Balada E, Ordi-Ros J,
Bujan-Rivas S, Sellas-Fernandez A and Vilardell-Tarres M: DNase 1
activity in patients with systemic lupus erythematosus:
Relationship with epidemiological, clinical, immunological and
therapeutical features. Lupus. 18:418–423. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kuribara J, Tada H, Kawai Y, Kawaguchi R,
Hoshizaki H, Arakawa K, Kitayama M, Kajinami K, Kurabayashi M,
Oshima S, et al: Levels of serum deoxyribonuclease I activity on
admission in patients with acute myocardial infarction can be
useful in predicting left ventricular enlargement due to
remodeling. J Cardiol. 53:196–203. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
St-Onge L, Wehr R and Gruss P: Pancreas
development and diabetes. Curr Opin Genet Dev. 9:295–300. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhu B, Gong Y, Chen P, Zhang H, Zhao T and
Li P: Increased DNase I activity in diabetes might be associated
with injury of pancreas. Mol Cell Biochem. 393:23–32. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tinazzi E, Puccetti A, Gerli R, Rigo A,
Migliorini P, Simeoni S, Beri R, Dolcino M, Martinelli N, Corrocher
R and Lunardi C: Serum DNase I, soluble Fas/FasL levels and cell
surface Fas expression in patients with SLE: A possible explanation
for the lack of efficacy of hrDNase I treatment. Int Immunol.
21:237–243. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Errami Y, Naura AS, Kim H, Ju J, Suzuki Y,
El-Bahrawy AH, Ghonim MA, Hemeida RA, Mansy MS, Zhang J, et al:
Apoptotic DNA fragmentation may be a cooperative activity between
caspase-activated deoxyribonuclease and the poly(ADP-ribose)
polymerase-regulated DNAS1L3, an endoplasm ic reticulum-localized
endonuclease that translocates to the nucleus during apoptosis. J
Biol Chem. 288:3460–3468. 2013. View Article : Google Scholar :
|
|
13
|
Shiokawa D and Tanuma S: Characterization
of human DNase I family endonucleases and activation of DNase gamma
during apoptosis. Biochemistry. 40:143–152. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rosner K, Kasprzak MF, Horenstein AC,
Thurston HL, Abrams J, Kerwin LY, Mehregan DA and Mehregan DR:
Engineering a waste management enzyme to overcome cancer resistance
to apoptosis: Adding DNase1 to the anti-cancer toolbox. Cancer Gene
Ther. 18:346–357. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Oliveri M, Daga A, Cantoni C, Lunardi C,
Millo R and Puccetti A: DNase I mediates internucleosomal DNA
degradation in human cells undergoing drug-induced apoptosis. Eur J
Immunol. 31:743–751. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hall AK: Molecular interactions between
G-actin, DNase I and the beta-thymosins in apoptosis: A hypothesis.
Med Hypotheses. 43:125–131. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Martínez Valle F, Balada E, Ordi-Ros J and
Vilardell-Tarres M: DNase 1 and systemic lupus erythematosus.
Autoimmun Rev. 7:359–363. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Martinez-Valle F, Balada E, Ordi-Ros J,
Bujan-Rivas S, Sellas-Fernandez A and Vilardell-Tarres M: DNase1
activity in systemic lupus erythematosus patients with and without
nephropathy. Rheumatol Int. 30:1601–1604. 2010. View Article : Google Scholar
|
|
19
|
Fujibayashi K, Kawai Y, Kitayama M, Akao
H, Ishida R, Motoyama A, Wakasa M, Arakawa K, Ueki M, Kajinami K
and Yasuda T: Serum deoxyribonuclease I activity can be a useful
diagnostic marker for the early diagnosis of unstable angina
pectoris or non-ST-segment elevation myocardial infarction. J
Cardiol. 59:258–265. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yasuda T, Iida R, Kawai Y, Nakajima T,
Kominato Y, Fujihara J and Takeshita H: Serum deoxyribonuclease I
can be used as a useful marker for diagnosis of death due to
ischemic heart disease. Leg Med (Tokyo). 11(Suppl 1): S213–S215.
2009. View Article : Google Scholar
|
|
21
|
Boyle AP, Davis S, Shulha HP, Meltzer P,
Margulies EH, Weng Z, Furey TS and Crawford GE: High-resolution
mapping and characterization of open chromatin across the genome.
Cell. 132:311–322. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Thurman RE, Rynes E, Humbert R, Vierstra
J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H,
Vernot B, et al: The accessible chromatin landscape of the human
genome. Nature. 489:75–82. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Maurano MT, Humbert R, Rynes E, Thurman
RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et
al: Systematic localization of common disease-associated variation
in regulatory DNA. Science. 337:1190–1195. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Stitzel ML, Sethupathy P, Pearson DS,
Chines PS, Song L, Erdos MR, Welch R, Parker SC, Boyle AP, Scott
LJ, et al: Global epigenomic analysis of primary human pancreatic
islets provides insights into type 2 diabetes susceptibility loci.
Cell Metab. 12:443–455. 2010. View Article : Google Scholar : PubMed/NCBI
|