Introduction
Stem cell treatment offers a promising approach in
providing an advanced and reliable therapeutic strategy for tissue
regeneration and disease therapy. Our previous studies, and those
of others, have demonstrated that mesenchymal stem cells derived
from dental tissues, including dental follicle stem cells (DFCs)
and dental pulp stem cells (DPCs), can be used to successfully
regenerate or repair bone, dentin and tooth roots (1–3).
DFCs are derived from the neural crest and give rise to the
periodontal ligament. DPCs are derived from the dental pulp and are
also considered to originate from the cranial neural crest. These
cells are multipotent and can differentiate into various cell
types, including osteoblasts, adipocytes and neurons (4–6). Of
note, these two types of cells can be readily isolated from
extracted third molars, which are usually otherwise discarded as
medical waste. Therefore, stem cell-derived dental tissues,
including DFCs and DPCs, represent an alternative source of stem
cells for tissue regeneration. However, the underlying mechanisms
controlling the fate of these stem cells remain to be fully
elucidated.
The kif3a protein is a member of the kinesin-2
family of motor proteins, which is associated with the
intraflagellar transport (IFT) system of primary cilia, is an
essential system for the maintenance of ciliogenesis and ciliary
function (7,8), and is key in the development of
several organs, including the bone, brain and the thyroid gland
(9–11). Previously, studies have provided
evidence that kif3a is involved in the maturation of osteoblasts
and osteoblastic differentiation of mesenchymal stem cells
(11,12). In particular, selective deletion of
kif3a in osteoblasts of kif3aOc-cKO mice impairs
postnatal bone formation through multiple pathways, including Wnt
signaling, similar to the phenotype exhibited following cilia
deletion by other IFT proteins (13), indicating that kif3a is involved in
regulating Wnt signaling in a cilia-associated manner (11). Furthermore, another IFT protein,
ofd1, is expressed in the dental mesenchyma during tooth
development and ofd1-deficient mice exhibited marked
disorganization of molar structure in a previous study (14), suggesting that kif3a may also be
associated with tooth development. Dental mesenchymal stem cells,
including DFCs and DPCs, can differentiate into osteoblasts and be
mineralized. Therefore, the present study hypothesized that kif3a
may be involved in the mechanism contributing to dental mesenchymal
stem cell osteoblastic differentiation.
In the present study, genetic approaches were used
to knock down the IFT protein, kif3a, for the suppression of
ciliogenesis, in human DFCs and DPCs and to provide insight into
whether kif3a and primary cilia have a direct function in
regulating the osteoblastic differentiation of hDFCs and hDPCs via
the Wnt signaling pathway. The present study may help elucidate the
involvement of kif3a and primary cilia in tooth mesenchymal
function and development.
Materials and methods
Cell culture
Normal impacted third molar tooth germs were
collected from patients (n=10; 16–18 years old) undergoing
orthodontic treatment in the West China Hospital of Stomatology
(Chengdu, China), following the provision of written informed
consent. All the experiments were performed in accordance with the
ethical protocol approved by the Ethics Committee of Sichuan
University (Chengdu, China). The DFCs and DPCs were isolated and
cultured, as previously reported (15,16).
In brief, the DFCs and DPCs were isolated from the dental follicle
and dental pulp, and incubated in α-minimum essential medium
(α-MEM; Gibco; Thermo Fisher scientific, Inc., Waltham, MA, USA),
supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
scientific, Inc.), 100 U/ml penicillin (Sigma-Aldrich, St. Louis,
MO, USA) and 100 mg/ml streptomycin (Sigma-Aldrich) in humid
conditions at 37°C of 5% CO2 in air. The medium was
renewed every three days.
Cell characteristics
hDFCs and hDPCs were seeded at a density of
3×103 into each well of a 24-well plate overnight,
respectively. The cells were then washed with phosphate-buffered
saline (PBS) three times, fixed with 4% cold polyoxymethylene
(Sigma-Aldrich) for 15 min and permeabilized by 0.5% Triton
(Sigma-Aldrich) for 10 min at room temperature. Following rinsing
three times with PBS, the cells were blocked with 1% bovine serum
albumin (Shanghai Zhangyun Chemical Co., Ltd., Shanghai, China)
diluted in PBS for 30 min at 37°C. The cells were then treated with
primary antibodies for 2 h in a humidified environment at 37°C.
Following being washed three times with PBS, the cells were treated
with secondary antibodies for 1 h in a humidified environment at
37°C. Following three rinses with PBS, the nuclei were
counterstained with 100 ng/ml DAPI blue (Beyotime Institute of
Biotechnology, Shanghai, China) for 3 min in the dark. The primary
antibodies used were as follows: Anti-vimentin (mouse IgG; 1:200;
OMA1-06001; Thermo Fisher Scientific, Inc.), anti-CK14 (mouse IgG;
1:200; MAB3232; EMD Millipore, Billerica, MA, USA) and anti-Stro-1
(monoclonal mouse IgG; 1:100; FAB1038F; R&D Systems,
Minneapolis, MN, USA). Secondary antibodies were conjugated to
Alexa FluoR 488 goat anti-mouse (A11001) or Alexa FluoR 555 goat
anti-mouse (1:500; A21422; Invitrogen; Thermo Fisher Scientific,
Inc.). Images were captured and analyzed under a confocal imaging
system (Olympus FV1000; Olympus Corporation, Tokyo, Japan).
Flow-cytometric analysis
To characterize the immunophenotype of the hDFCs and
hDPCs, flow cytometric analysis was used to measure the expression
of mesenchymal stem cell surface markers. Cells were trypsinized
(Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) and
incubated at 37°C with phycoerthyn (PE)-conjugated cluster of
differentiation (CD)29 integrin β1 (555443); fluorescein
isothiocyanate (FITC)-conjugated CD31 (555445), which is also known
as platelet-endothelial cell adhesion molecule-1; FITC-conjugated
CD34 (555821), which is also known as hematopoietic progenitor cell
antigen; FITC-conjugated CD44 (555478), which is also known as
hyaluronate/lymphocyte homing associated cell adhesion molecule;
PE-conjugated CD90 (Thy-1/Thy-1.1) (555596); and PE-conjugated
CD166 (559263), which is also known as activated leucocyte cell
adhesion molecule and is a mesenchymal stem cell marker. All
antibodies were purchased from BD Biosciences (Franklin Lakes, NJ,
USA). Flow cytometry was performed using a Beckman Coulter Cytomics
FC 500 MPL system (Beckman Coulter, Fullerton, CA, USA).
Lentiviral infection
According to previous studies, a total of
5×104 hDFCs and hDPCs were seeded into each well of a
6-well plate at 37°C overnight, respectively. At 50% cell density,
1 ml α-MEM (without FBS), containing 1.5 µl of
green-fluorescence protein-labeled kif3a short hairpin (sh) RNA
lentivirus (NeuronBiotech Co., Ltd., Shanghai, China) and 0.5
µl polybrene (Sigma-Aldrich), were added into each well.
Following incubation for 6 h at 37°C, the supernatant was removed
and replaced with 2 ml fresh α-MEM supplemented with 10% FBS. After
3 days at 37°C, the infected cells were observed and images were
captured under a fluorescence microscope (Leica Optical, Leica
Microsystems GmbH, Wetzlar, Germany) (17).
Osteoblastic induction
For osteoblastic induction, a total of
1×105 hDFCs and hDPCs at passage three were seeded into
each well of a 6-well plate overnight respectively. These cells
were then cultured at 37°C in osteoblastic inductive medium (α-MEM
supplemented with 10% FBS, 10 mmol/l β-glycerophosphate
(Sigma-Aldrich), 0.2 mmol/l ascorbic acid (Sigma-Aldrich), 10
nmol/l 1,25-dihydroxyvitamin D3 and 100 nmol/l dexamethasone
(Sigma-Aldrich) for 3–30 days, depending on the subsequent
experiments. The inductive medium was replaced every three
days.
Alizarin Red S staining
As previously described (18), following fixation with 4%
paraformaldehyde (Sigma-Aldrich) for 30 min at room temperature,
the cultures were incubated in 0.1% Alizarin Red solution
(Sigma-Aldrich) in Tris HCl (pH 8.3; 0.5–1 ml per well; Kelong
Chemical Co., Ltd., Chengdu, China) in a 6-well plate for 30 min at
room temperature.
Cellular immunofluorescent analysis
The control and kif3a-knockdown hDFCs and hDPCs were
seeded (3×103) into each well of a 24-well plate
overnight, respectively. The methods used were the same as
described above. The primary antibodies used were anti-acetylized
α-tubulin (mouse IgG; 1:500; ab24610; Abcam). Secondary antibodies
were conjugated to Alexa FluoR 555 (goat anti-mouse; 1:500;
Invitrogen; Thermo Fisher Scientific, Inc.). Images were captured
and analyzed under a confocal imaging system (Olympus FV1000;
Olympus Corporation).
RNA preparation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis
The control and kif3a-knockdown hDFCs and hDPCs were
harvested from each well of a 6-well plate following 3 days
osteoblastic induction in preparation for RNA isolation. Total RNA
was obtained using RNAiso™ Plus (Takara Bio Inc., Otsu, Japan).
cDNAs were synthesized by reverse transcription using the extracted
RNA with PrimeScript® Perfect Real Time reagent kit
(Takara Bio., Inc.). The relative expression levels of genes were
quantified using qPCR with SYBR® Premix Ex Taq™ (Perfect
Real Time; Takara Bio, Inc.) in a 10 µl total reaction
volume using an ABI Prism 7300 system (Applied Biosystems; Thermo
Fisher Scientific, Inc.). The qPCR conditions were as follows:
Initial cycle was 95°C for 30 sec, 40 cycles at 95°C for 5 sec and
60°C for 30 sec; the final cycle was 95°C for 15 sec, 60°C for 1
min and 95°C for 15 sec. Dissociation curves were used to confirm
primers specificity. D-glyceraldehyde-3-phosphate (GAPDH) was used
as an internal reference, and relative mRNA levels were quantified
using the 2−ΔΔCq method (19). The primer sequences used in the
present study for GAPDH, dentin sialophosphoprotein (DSPP), dentin
matrix protein 1 (DMP-1), osteocalcin (OCN), Runt-related
transcription factor 2 (Runx2), alkaline phosphatase (ALP),
osteopontin (OPN), axis inhibition protein 2 (Axin2) and β-catenin,
lymphoid enhancer-binding factor 1 (Lef1) are listed in Table I. All experiments were performed
three times.
 | Table IOligo-nucleotide primer sequences
used in reverse transcription-quantitative polymerase chain
reaction analysis. |
Table I
Oligo-nucleotide primer sequences
used in reverse transcription-quantitative polymerase chain
reaction analysis.
| Gene | Primer sequence
(5′–3′) | GenBank number |
|---|
| ALP | F:
TAAGGACATCGCCTACCAGCTC | NM_000478.4 |
| R:
TCTTCCAGGTGTCAACGAGGT | |
| DMP1 | F:
GTGAGTGAGTCCAGGGGAGATAA | NM_004407.3 |
| R:
TTTTGAGTGGGAGAGTGTGTGC | |
| DSPP | F:
CTGTTGGGAAGAGCCAAGATAAG | NM_014208.3 |
| R:
CCAAGATCATTCCATGTTGTCCT | |
| OCN | F:
CTCACACTCCTCGCCCTATTG | NM_199173.3 |
| R:
CTCCCAGCCATTGATACAGGTAG | |
| OPN | F:
CAGTTGTCCCCACAGTAGACAC | AB469789.1 |
| R:
GTGATGTCCTCGTCTGTAGCATC | |
| Runx2 | F:
CTTTACTTACACCCCGCCAGTC | NM_001024630.3 |
| R:
AGAGATATGGAGTGCTGCTGGTC | |
| Axin2 | F:
GACAGGAATCATTCGGCCAC | NM_004655 |
| R:
CCTTCAGCATCCTCCGGTAT | |
| β-catenin | F:
CTTACACCCACCATCCCACT | NM_001098209 |
| R:
CCTCCACAAATTGCTGCTGT | |
| Lef1 | F:
ACAGATCACCCCACCTCTTG | NM_016269.1 |
| R:
ATAGCTGGATGAGGGATGCC | |
| GAPDH | F:
CTTTGGTATCGTGGAAGGACTC | NM_002046.3 |
| R:
GTAGAGGCAGGGATGATGTTCT | |
Western blot analysis
Control and kif3a-knockdown hDFCs and hDPCs were
harvested from each well of a 6-well plate following 1 week of
osteoblastic induction prior to protein extraction. Cells were
rinsed three times in PBS prior to being lysed using cell lysis
buffer, containing 0.5% 100 mmol/l phenylmethanesulfonyl fluoride,
0.1% protease inhibitor and 0.5% phosphatase inhibitor, on ice.
Proteins in the supernatant were measured by modified Bradford
Protein Assay (Bio-Rad Laboratories, Inc., Hercules, CA, USA). An
equal quantity of total protein (50 mg) was separated by sodium
dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and
transferred onto polyvinylidene fluoride membranes (Invitrogen;
Thermo Fisher Scientific, Inc.). Following blocking with non-fat
milk in PBS with Tween 20 (PBST) for 1 h at room temperature, the
membranes were incubated with primary antibodies at 4°C overnight.
The membranes were subsequently washed three times with PBST for 10
min and incubated with secondary antibodies at room temperature for
1 h, respectively. Membranes were visualized with a
chemiluminescent horseradish peroxidase (HRP) substrate (EMD
Millipore) using a ChemiDoc XRS system (Bio-Rad Laboratories,
Inc.). The primary antibodies used for western blot analysis were
as follows: Anti-β-actin (1:500; ab8227), anti-ALP (1:500;
ab67228), anti-DMP1 (1:500; ab103203), anti-kif3a (1:500;
ab133587), anti-bone sialoprotein (BSP; 1:200; ab52128), anti-OPN
(1:1,000; ab8448), anti-Runx2 (1:500; ab76956), anti-phosphorylated
glycogen synthase kinase 3β (GSK3β; 1:1,000; ab32391; Abcam),
anti-Lef1 (1:500; ab85052) and anti-active β-catenin (1:4,000;
8814; Cell Signaling Technology, Inc., Danvers, MA, USA). The
secondary antibodies were HRP-conjugated AffiniPure goat anti-mouse
IgG (H+L; 1:10,000; ZB-2305) and HRP-conjugated AffiniPure goat
anti-rabbit IgG (H+L; 1:10,000, ZB-2301; ZSGB-Bio, Beijing, China).
All experiments were performed three times.
Statistical analysis
The data in the present study were collected from
three different samples of the same cell sample in triplicate.
Values are presented as the mean ± standard deviation. Statistical
significance was evaluated using one-way analysis of variance using
SPSS software (Version 10.0; SPSS, Inc., Chicago, IL, USA).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Characterization of hDFCs and hDPCs as
dental mesen- chymal stem/precursor cells
The present study identified the characterization of
hDFCs and hDPCs as dental mesenchymal stem cells using cellular
immunofluorescent detection. The two cell types were positive for
the vimentin and Stro-1 mesenchymal stem cells markers, but
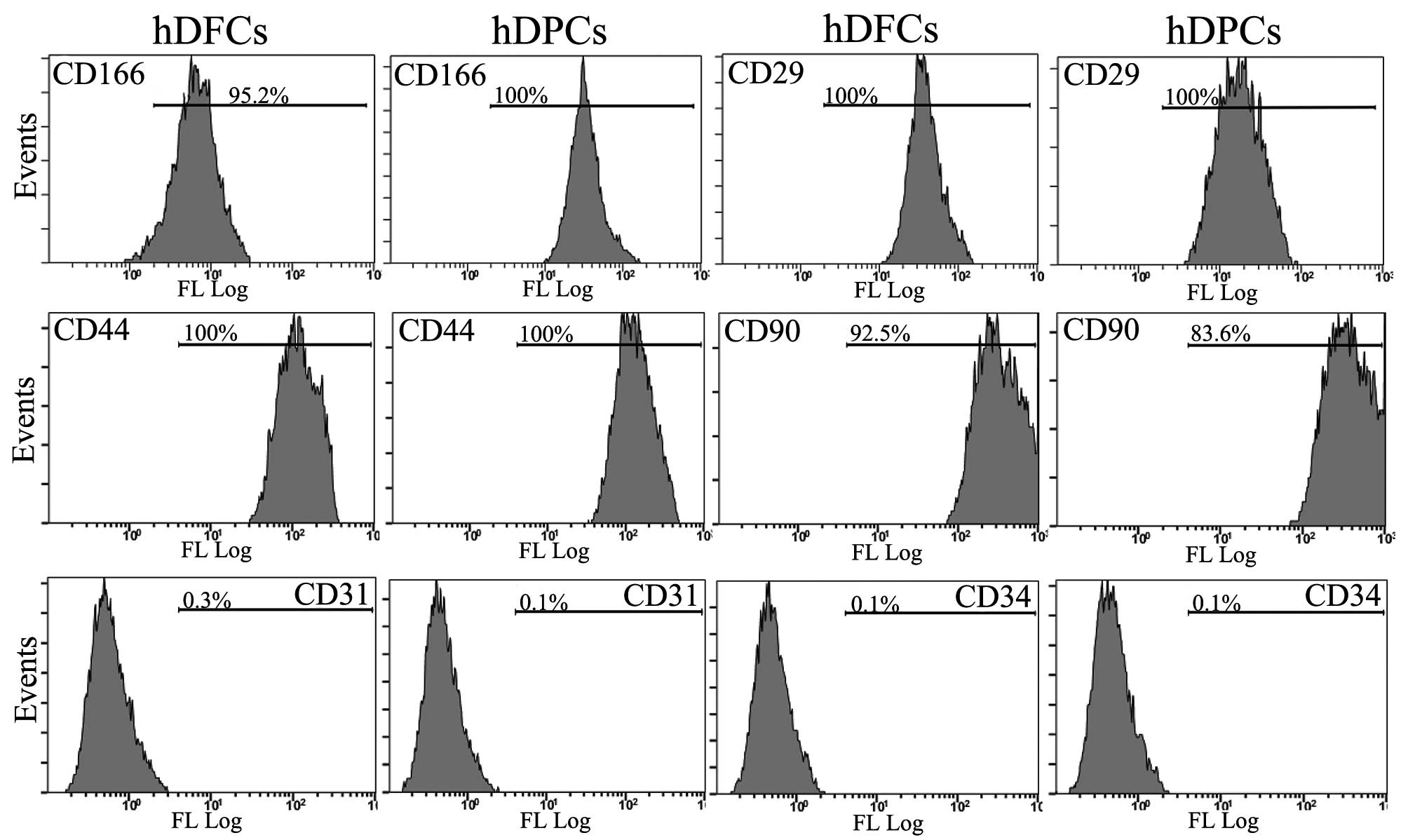
negative for the CK-14 epithelial cell marker (Fig. 1). The immunophenotypic
characterization was performed using flow cytometry. The DFCs and
DPCs were positive for the CD29, CD44, CD90 and CD166 mesenchymal
stem cell markers. However, the two types of cell were negative for
the leucocyte precursor markers, CD31 and CD34, suggesting a lack
of cells of hematopoietic and angiogenic lineages (Fig. 2). These data indicated that the DPC
and DFC cell lineage were pure mesenchymal cells and maintained
stem cell-like properties.
Knockdown of kif3a leads to loss of
primary cilia in hDFCs and hDPCs
The present study then investigated the existence of
primary cilia in the hDFCs and hDPCs. The primary cilia were
immunostained using acetylated α-tubulin antibody and observed
under a laser scanning confocal microscope, as described previously
(14) (Fig. 3A). The expression of kif3a was then
determined using western blot analysis, and the target bands were
detected with purified anti peptide antibodies in the two cell
cultures (Fig. 3B) Lentiviruses
expressing four different shRNAs specific for kif3a were
transfected into the cells, as described above, and the shRNA with
the most marked effect was selected. The efficiency of kif3a
suppression was determined using immunoblot analysis (Fig. 3C). Following kif3a knockdown, the
number of primary cilia were substantially reduced in the two cell
cultures (Fig. 3D–G).
Knockdown of kif3a suppresses
osteoblastic differentiation in hDFCs and hDPCs
To examine the association between the loss of
primary cilia caused by kif3a knockdown and osteoblastic
differentiation in hDFCs and hDPCs, the present study performed
Alizarin Red staining following treatment with osteoblastic
inductive medium. Following 10 days induction, the kif3a-knockdown
cells formed fewer mineralized nodules, compared with the cells in
the control group, in the hDFCs and hDPCs. This difference became
more significant following 30 days induction (Fig. 4A and B).
 | Figure 4Effects of kif3a knockdown on
osteoblastic differentiation in hDFCs and hDPCs. (A and B) Alizarin
Red staining for 10 and 30 days. (B) Staining viewed under
magnification (Scale bar=400 µm). Results of (C and D)
reverse transcription-quantitative polymerase chain reaction and (E
and F) western blot analyses following 6 days induction. Data are
representative of three independent experiments and are presented
as the mean ± standard deviation. *P<0.05 and
**P<0.01 vs. control. hDFCs, human dental follicle cells; hDPCs,
human dental pulp cells; CTR, control; kif3a KD, kif3a-knockdown;
ALP, alkaline phosphatase; DMP1, dentin matrix protein 1; BSP, bone
sialoprotein; OCN, osteocalcin; OPN, osteopontin; Runx2,
Runt-related transcription factor 2. |
The present study then assessed the expression
levels of osteoblastic genes using RT-qPCR (Fig. 4C and D), and protein expression
levels using western blot analysis (Fig. 4E). The results showed that the
osteoblastic genes and proteins, including ALP, BSP, DMP1, OCN and
Runx2, were significantly downregulated following kif3a knockdown
in the hDFC and hDPC cells.
Exogenous Wnt3a partially recovers
osteoblastic differen- tiation in hDFCs and hDPCs
To examine the mechanisms by which the loss of kif3a
causes alterations in hDFC and hDPC functions, the present study
examined whether the Wnt signaling pathway was involved in this
suppression, which has a close association with tooth development
(20,21). Following treatment with
osteoblastic medium and recombinant Wnt3a for 6 days, the mRNA and
protein expression levels of osteoblastic markers of hDPCs and
hDFCs, were assessed, respectively. Significant increases in the
mRNA (Fig. 5A and B) and protein
(Fig. 5C) expression levels of
osteoblastic markers were found in the Wnt3a-treated wild-type
cells, compared with the control cells. By contrast, there was no
significant increase in the kif3a-knockdown cells (Fig. 5A–C). In accordance with these
results, the kif3a-knockdown cells formed fewer mineralized
nodules, compared with the control group cells, in the hDFCs and
hDPCs following culture in osteoblastic medium for 14 days and
treatment with recombinant Wnt3a (Fig.
5D and E). This confirmed that the loss of kif3a suppressed
osteoblastic differentiation in the hDFCs and hDPCs by the
inhibition of Wnt signaling.
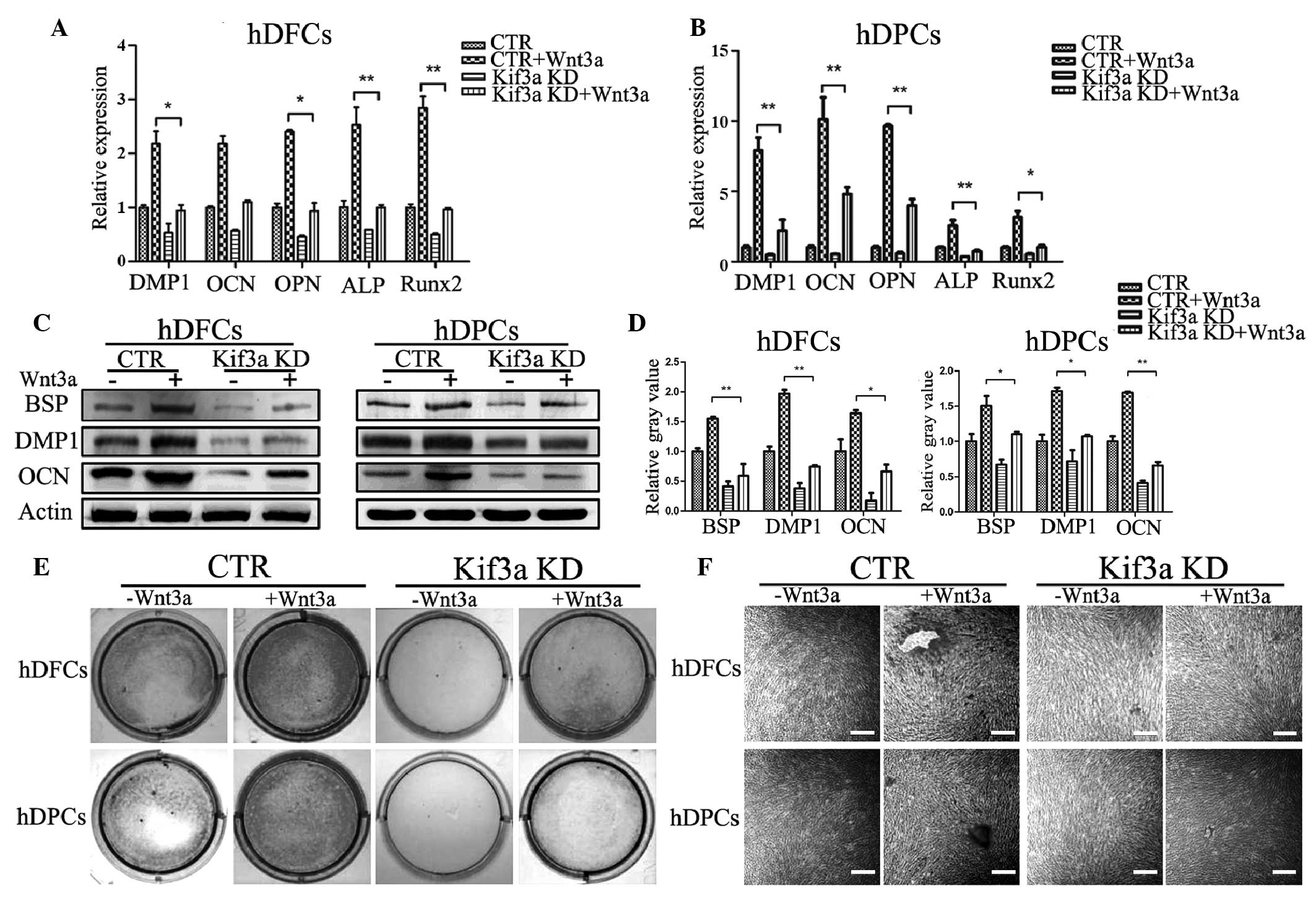 | Figure 5Osteoblastic differentiation of hDFCs
and hDPCs is rescued by exogenous Wnt3a. Effects on the mRNA levels
of osteoblastic markers of (A) hDPCs and (B) hDFCs following
stimulation with 100 ng/ml recombinant Wnt3a for 18 h were assessed
using reverse transcription-quantitative polymerase chain reaction
analysis. Data are presented as the mean ± standard deviation. (C)
Effects on protein levels were examined following 6 days
stimulation using Western blot analysis and were subsequently (D)
quantified. (E) After 14 days of treatment, culture plates were
analyzed for mineralization using Alizarin Red staining, and (F)
visualized at magnification (scale bar=200 µm).
*P<0.05; **P<0.01. hDFCs, human dental
follicle cells; hDPCs, human dental pulp cells; CTR, control; kif3a
KD, kif3a knockdown; ALP, alkaline phosphatase; DMP1, dentin matrix
protein 1; BSP, bone sialoprotein; OCN, osteocalcin; OPN,
osteopontin; Runx2, Runt-related transcription factor 2. |
Loss of kif3a causes abnormal Wnt
signaling in hDFCs and hDPCs
The present study further examined whether the loss
of primary cilia of the hDFCs and hDPCs following inhibition of
kif3a leads to interference in the canonical Wnt pathway. Using
RT-qPCR and western blot analyses, it was found that the expression
levels of canonical Wnt pathway factors, including Axin2, β-catenin
and Lef1, were significantly lower in the kif3a-knockdown cells,
compared with the control cells, at the transcriptional (Fig. 6A and B) and translational (Fig. 6C) levels following treatment in
osteoblastic medium for 6 days. However, the addition of 100 ng/ml
recombinant human Wnt3a protein to the osteoblastic hDFC and hDPC
cultures resulted in a significant increase in Wnt signaling
activity, at the mRNA (Fig. 6D and
E) and protein (Fig. 6F)
expression levels, in the control cells. By contrast, there was no
significant increase in the kif3a-knockdown cells (Fig. 6D–F), suggesting that the loss of
kif3a impaired the Wnt signaling response in the hDFCs and hDPCs.
This result confirmed that kif3a, at least part, regulated hDFC and
hDPC osteoblastic differentiation by the Wnt signaling pathway.
 | Figure 6Effects of kif3a knockdown on Wnt
signaling in hDFCs and hDPCs. There was a decrease of Wnt signaling
on (A and B) gene expression levels, revealed using reverse
transcription-quantitative polymerase chain reaction analysis, and
on (C) protein levels, revealed using Western blot analysis
following 6 days osteoblastic induction in hDFCs and hDPCs. (D and
E) mRNA and (F) protein expression levels of Wnt signaling
molecules were markedly higher in the control cells, compared with
the kif3a-knockdown cells following 100 ng/ml recombinant Wnt3a
stimulation for 18 h. Data are presented as the mean ± standard
deviation. *P<0.05 and **P<0.001, vs.
control. hDFCs, human dental follicle cells; hDPCs, human dental
pulp cells; CTR, control; kif3a KD, kif3a knockdown; Axin2, axis
inhibition protein 2; Lef1, lymphoid enhancer-binding factor 1;
p-GSK3β, phosphorylated glycogen synthase kinase 3β. |
Discussion
The kif3a protein is involved in embryogenesis
through a ciliary mechanism, which was initially reported in mice
with kif3a deficiency lacking cilia and exhibiting numerous
structural abnormalities (22). In
several stem/progenitor cell lines, kif3a is also reported to be
involved in the cell differentiation (23–25).
The present study showed that two dental-derived mesenchymal
stem/precursor cells, hDFCs and hDPCs, are cells with primary
cilia, rather than just odontoblasts, as described in previous
reports (14,26). The knockdown of kif3a in the hDFCs
and hDPCs resulted in a reduction in the numbers of primary cilia,
consistent with the known role of kif3a in primary cilia formation
(27,28).
Wnt signaling has been shown to be an important
regulatory pathway in the osteogenic differentiation of mesenchymal
stem cells (29–31). The persistent expression of
β-catenin in the dental mesenchyme results in the premature
differentiation of odontoblasts and differentiation of
cementoblasts, and induces immoderate dentin and cementum
generation in vivo (32).
In several tissues, kif3a has been reported to be involved in
regulating the Wnt pathway as an IFT motor subunit of primary cilia
(11,33,34).
The present study demonstrated that kif3a-knockdown
hDFCs and hDPCs caused abnormality of the Wnt signal pathway. Lef1
and active β-catenin were significantly downregulated following
kif3a knockdown. kif3a knockdown also attenuated the exogenous
Wnt3a-stimulated expression levels of Lef1 and active β-catenin in
the hDFCs and hDPCs (Fig. 6). In
the present study, the addition of recombinant Wnt3a protein
upregulated osteoblastic gene and protein expression levels, and
formed more mineralized nodules in vitro. This Wnt3a-induced
expression of osteoblastic markers was markedly reduced in the
kif3a-knockdown cells, in accordance with the effect of kif3a on
the Wnt signaling pathway, which indicated the importance of kif3a
and the integrity of primary cilia in the differentiation in hDFCs
and hDPCs in a Wnt-dependent manner.
Several lines of evidence support the hypothesis
that cilia and cilia-associated proteins regulate Wnt signaling in
a varied manner (11,33,35–37).
It is possible that the role of cilia and cilia-associated proteins
in the Wnt pathway is tissue or stage-specific. With bone
mesenchymal cell differentiation as an example, Chen et al
(38) and Silkstone et al
(39) found that β-catenin
signaling activation inhibited the differentiation of mesenchymal
stem cells (38). However, in
cells committed to the osteoblast lineage, osteoblast
differentiation was enhanced by β-catenin (39). In the present study, the disruption
of kif3a led to a reduction of Wnt signaling, consistent with the
role of kif3a in osteoblasts and prostate cancer cells demonstrated
in previous reports (11,33,40).
Further investigations are required to confirm the role of kif3a in
regulating cilia-dependent Wnt signaling in tooth development and
repair. Although further elucidation is required, analysis of the
data obtained in the present study demonstrated that kif3a was
essential in hDFC and hDPC differentiation though Wnt
signaling.
In conclusion, the present study provided novel
insight into the mechanisms of primary cilia as an indispensable
organelle regulating the osteoblastic differentiation of hDFCs and
hDPCs. The results suggested that the cilia-associated kinesin
protein, kif3a, may be a regulator of hDFC and hDPC osteoblastic
differentiation via the Wnt signaling pathway, and offer potential
as a therapeutic target in diseases associated with cilia. These
results not only enhance current understanding of tooth development
and diseases of tooth mineralization, but also indicate possible
strategies to regulate mineralization during tooth repair and
regeneration.
Acknowledgments
The present study was supported by the National
Natural Science Foundation of China (grant no. 81271119) and the
Basic Research Program of the Sichuan Province of China (grant no.
2013JY0019).
References
|
1
|
Yang B, Chen G, Li J, Zou Q, Xie D, Chen
Y, Wang H, Zheng X, Long J and Tang W: Tooth root regeneration
using dental follicle cell sheets in combination with a dentin
matrix-based scaffold. Biomaterials. 33:2449–2461. 2012. View Article : Google Scholar
|
|
2
|
Nie X, Tian W, Zhang Y, Chen X, Dong R,
Jiang M, Chen F and Jin Y: Induction of transforming growth
factor-beta 1 on dentine pulp cells in different culture patterns.
Cell Biol Int. 30:295–300. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jiao L, Xie L, Yang B, Yu M, Jiang Z, Feng
L, Guo W and Tian W: Cryopreserved dentin matrix as a scaffold
material for dentin-pulp tissue regeneration. Biomaterials.
35:4929–4939. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang C, Sun L, Li X, Xie L, Yu M, Feng L,
Jiang Z, Guo W and Tian W: The potential of dental stem cells
differentiating into neurogenic cell lineage after cultivation in
different modes in vitro. Cell Reprogram. 16:379–391. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang X, He F, Tan Y, Tian W and Qiu S:
Inhibition of Delta1 promotes differentiation of odontoblasts and
inhibits proliferation of human dental pulp stem cell in vitro.
Arch Oral Biol. 56:837–845. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Guo L, Li J, Qiao X, Yu M, Tang W, Wang H,
Guo W and Tian W: Comparison of odontogenic differentiation of
human dental follicle cells and human dental papilla cells. PLoS
One. 8:e623322013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kolpakova-Hart E, Jinnin M, Hou B, Fukai N
and Olsen BR: Kinesin-2 controls development and patterning of the
vertebrate skeleton by Hedgehog- and Gli3-dependent mechanisms. Dev
Biol. 309:273–284. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rosenbaum JL and Witman GB: Intraflagellar
transport. Nat Rev Mol Cell Biol. 3:813–825. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tong CK, Han YG, Shah JK, Obernier K,
Guinto CD and Alvarez-Buylla A: Primary cilia are required in a
unique subpopulation of neural progenitors. Proc Natl Acad Sci USA.
111:12438–12443. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
D'Amico E, Gayral S, Massart C, Van Sande
J, Reiter JF, Dumont JE, Robaye B and Schurmans S: Thyroid-specific
inactivation of KIF3A alters the TSH signaling pathway and leads to
hypothyroidism. J Mol Endocrinol. 50:375–387. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qiu N, Xiao Z, Cao L, Buechel MM, David V,
Roan E and Quarles LD: Disruption of Kif3a in osteoblasts results
in defective bone formation and osteopenia. J Cell Sci.
125:1945–1957. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Temiyasathit S, Tang WJ, Leucht P,
Anderson CT, Monica SD, Castillo AB, Helms JA, Stearns T and Jacobs
CR: Mechanosensing by the primary cilium: deletion of Kif3A reduces
bone formation due to loading. PLoS One. 7:e333682012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xiao ZS and Quarles LD: Role of the
polycytin-primary cilia complex in bone development and
mechanosensing. Ann N Y Acad Sci. 1192:410–421. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Thivichon-Prince B, Couble ML, Giamarchi
A, Delmas P, Franco B, Romio L, Struys T, Lambrichts I, Ressnikoff
D, Magloire H and Bleicher F: Primary cilia of odontoblasts:
Possible role in molar morphogenesis. J Dent Res. 88:910–915. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guo W, Chen L, Gong K, Ding B, Duan Y and
Jin Y: Heterogeneous dental follicle cells and the regeneration of
complex periodontal tissues. Tissue Eng Part A. 18:459–470. 2012.
View Article : Google Scholar :
|
|
16
|
Morsczeck C, Götz W, Schierholz J,
Zeilhofer F, Kühn U, Möhl C, Sippel C and Hoffmann KH: Isolation of
precursor cells (PCs) from human dental follicle of wisdom teeth.
Matrix Biol. 24:155–165. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang X, He H, Wu X, Hu J and Tan Y:
Promotion of dentin regeneration via CCN3 modulation on Notch and
BMP signaling pathways. Biomaterials. 35:2720–2729. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu J, He H, Tang C, Zhang G, Li Y, Wang R,
Shi J and Jin Y: Differentiation potential of STRO-1+ dental pulp
stem cells changes during cell passaging. BMC Cell Biol. 11:322010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2 (-Delta Delta C (T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
20
|
Chen J, Lan Y, Baek JA, Gao Y and Jiang R:
Wnt/beta-catenin signaling plays an essential role in activation of
odontogenic mesenchyme during early tooth development. Dev Biol.
334:174–185. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhu X, Zhao P, Liu Y, Zhang X, Fu J, Ivy
Yu HM, Qiu M, Chen Y, Hsu W and Zhang Z: Intra-epithelial
requirement of canonical Wnt signaling for tooth morphogenesis. J
Biol Chem. 288:12080–12089. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Marszalek JR, Ruiz-Lozano P, Roberts E,
Chien KR and Goldstein LS: Situs inversus and embryonic ciliary
morphogenesis defects in mouse mutants lacking the KIF3A subunit of
kinesin-II. Proc Natl Acad Sci USA. 96:5043–5048. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guadiana SM, Semple-Rowland S, Daroszewski
D, Madorsky I, Breunig JJ, Mykytyn K and Sarkisian MR: Arborization
of dendrites by developing neocortical neurons is dependent on
primary cilia and type 3 adenylyl cyclase. J Neurosci.
33:2626–2638. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhao C, Omori Y, Brodowska K, Kovach P and
Malicki J: Kinesin-2 family in vertebrate ciliogenesis. Proc Natl
Acad Sci USA. 109:2388–2393. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhu D, Shi S, Wang H and Liao K: Growth
arrest induces primary-cilium formation and sensitizes
IGF-1-receptor signaling during differentiation induction of 3T3-L1
preadipocytes. J Cell Sci. 122:2760–2768. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Couble ML, Farges JC, Bleicher F,
Perrat-Mabillon B, Boudeulle M and Magloire H: Odontoblast
differentiation of human dental pulp cells in explant cultures.
Calcif Tissue Int. 66:129–138. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Barakat MT, Humke EW and Scott MP: Kif3a
is necessary for initiation and maintenance of medulloblastoma.
Carcinogenesis. 34:1382–1392. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lin F, Hiesberger T, Cordes K, Sinclair
AM, Goldstein LS, Somlo S and Igarashi P: Kidney-specific
inactivation of the KIF3A subunit of kinesin-II inhibits renal
ciliogenesis and produces polycystic kidney disease. Proc Natl Acad
Sci USA. 100:5286–5291. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Delaine-Smith RM, Sittichokechaiwut A and
Reilly GC: Primary cilia respond to fluid shear stress and mediate
flow-induced calcium deposition in osteoblasts. FASEB J.
28:430–439. 2014. View Article : Google Scholar :
|
|
30
|
Holmen SL, Zylstra CR, Mukherjee A, Sigler
RE, Faugere MC, Bouxsein ML, Deng L, Clemens TL and Williams BO:
Essential role of beta-catenin in postnatal bone acquisition. J
Biol Chem. 280:21162–21168. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Leucht P, Monica SD, Temiyasathit S,
Lenton K, Manu A, Longaker MT, Jacobs CR, Spilker RL, Guo H,
Brunski JB and Helms JA: Primary cilia act as mechanosensors during
bone healing around an implant. Med Eng Phys. 35:392–402. 2013.
View Article : Google Scholar
|
|
32
|
Kim TH, Lee JY, Baek JA, Lee JC, Yang X,
Taketo MM, Jiang R and Cho ES: Constitutive stabilization of
ß-catenin in the dental mesenchyme leads to excessive dentin and
cementum formation. Biochem Biophys Res Commun. 412:549–555. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu Z, Rebowe RE, Wang Z, Li Y, Wang Z,
DePaolo JS, Guo J, Qian C and Liu W: KIF3a promotes proliferation
and invasion via Wnt signaling in advanced prostate cancer. Mol
Cancer Res. 12:491–503. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gerdes JM, Liu Y, Zaghloul NA, Leitch CC,
Lawson SS, Kato M, Beachy PA, Beales PL, DeMartino GN, Fisher S, et
al: Disruption of the basal body compromises proteasomal function
and perturbs intracellular Wnt response. Nat Genet. 39:1350–1360.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Han YG, Kim HJ, Dlugosz AA, Ellison DW,
Gilbertson RJ and Alvarez-Buylla A: Dual and opposing roles of
primary cilia in medulloblastoma development. Nat Med.
15:1062–1065. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kishimoto N, Cao Y, Park A and Sun Z:
Cystic kidney gene seahorse regulates cilia-mediated processes and
Wnt pathways. Dev Cell. 14:954–961. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lancaster MA, Louie CM, Silhavy JL,
Sintasath L, Decambre M, Nigam SK, Willert K and Gleeson JG:
Impaired Wnt-beta-catenin signaling disrupts adult renal
homeostasis and leads to cystic kidney ciliopathy. Nat Med.
15:1046–1054. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen Y, Whetstone HC, Lin AC, Nadesan P,
Wei Q, Poon R and Alman BA: Beta-catenin signaling plays a
disparate role in different phases of fracture repair: Implications
for therapy to improve bone healing. PLoS Med. 4:e2492007.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Silkstone D, Hong H and Alman BA:
Beta-catenin in the race to fracture repair: In it to Wnt. Nat Clin
Pract Rheumatol. 4:413–419. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Vuong LT, Mukhopadhyay B and Choi KW:
Kinesin-II recruits Armadillo and Dishevelled for Wingless
signaling in Drosophila. Development. 141:3222–3232. 2014.
View Article : Google Scholar : PubMed/NCBI
|