Introduction
Schizophrenia is a mental disease of unknown
etiology. The patients are characterized by experiencing
distortions of reality and disturbances of thought and language,
and tend to withdraw from social contact (1). Neurotransmitter disturbances are
considered an important factor leading to the development of
schizophrenia (2). The fact that
the functions of multiple known susceptibility genes seem to relate
them to the neurotransmitter system provides molecular evidence for
the validity of the neurotransmitter hypothesis.
In addition to the neurotransmitter system
abnormality hypothesis, a large amount of research focuses on
schizophrenia as a neurodevelopmental disease. Since the early
1980s, researchers have been documenting structural abnormalities
on the brains of schizophrenia patients shown on CAT images, and it
has been found that the weight of brains of patients is lighter on
average than those of non-affected individuals. Specialists
attribute this specific finding to abnormalities occurring during
early development (3).
With the progress in imaging science, studies now
further show that the abnormal cerebral development of
schizophrenic patients results in a decrease in volume of the
lateral cerebral ventricle, of the grey and white matter, and of
some special functional areas such as the hippocampus, and the
prefrontal cortex. Based on imaging findings and postmortem
examinations of brains, the number of neuron dendritic spines is
decreased significantly in schizophrenic patients compared to
non-affected individuals (4).
Pallidin is a protein widely expressed in various
mammalian tissues. The earliest research showed it to be highly
expressed in the brain and skeletal muscle (5). However, further studies have
confirmed it is also widely expressed in the central nervous system
(CNS) (6). Immunohistochemistry
shows pallidin is highly expressed in the glutamatergic neurons of
the hippocampus, including some large cone neurons in the
neocortex, stellate cells in the entorhinal cortex, and large
neurons in the cerebellar nuclei (7). The ACh neurons in the basal forebrain
and cranial motor nuclear cluster, the dopaminergic neurons and
GABA neurons in the substantia nigra, and the tryptamine neurons in
the brainstem also exhibit a certain expression level of the
pallidin protein. This widespread presence suggests its
participation in diverse neural networks (8). The studies focusing on the cellular
localization of pallidin have aided researchers trying to elucidate
the functions of this protein. Pallidin is a cytoplasmic protein,
and immunoelectron microscopy results, in the granule cell layers
of the mouse hippocampal area, have shown the presence of pallidin
on neural presynaptic and postsynaptic membranes, indicating that
the protein may participate in the regulation of synaptic activity.
Furthermore, cell biology research has found pallidin in the cell
nucleus, indicating that it may also exert nuclear functions
(9).
p38 is a protein with important roles in cell
apoptosis (10). However, a study
shows that it also plays other roles in cells, including as an
important transcription factor in the cell nucleus (11). p38 promotes the expression of
multiple genes and regulates various functions of the cells after
binding to a specific sequence on the genome. In recent years, a
large amount of research points to the role of p38 in neuronal
development, for instance as a result of its role in apoptosis, an
essential factor in neuronal development (12). Furthermore, studies on the PCI2
cell line have shown that the transcriptional activity of p38 has a
significant effect on the NGF-induced cell differentiation
(13). PCI2 cells expressing
exogenous p38 produce much shorter neurites than those that are
produced in cells expressing p38 under control by the NGF,
highlighting the importance of the interaction between these
factors. Finally, two effector molecules related to neurite growth,
namely small GTP enzyme Rab13 and F-actin binding protein coronin
1b, have also been found to be transcribed and are regulated by p38
during regenerating neurons (14).
The existing research shows that different enzymes
can post-translationally modify newly synthesized p38 protein. Such
modifications determine the downstream functionality of the
resulting protein. This probably explains the dual functions
attributed to p38 as an apoptosis-promoting and a
development-promoting mediator. Our experiments are based on the
hypothesis that both pallidin and p38 are somehow related to the
pathogenesis of schizophrenia at a neurodevelopmental level.
Materials and methods
Cell lines and experimental
animals
All of the mammalian cell lines used (HEK293A,
HCT116 and N2a) were purchased from the American Type Culture
Collection (ATCC, Manassas, VA, USA). For culture, cells were
immersed into MEM + 10% fetal bovine serum (FBS) + 1X antibiotic
culture medium, and placed in an incubator containing 5%
CO2 at 37°C. The Sandy mice (sdy/sdy) and wild mice (WT;
+/+) were purchased from the Jackson Laboratory (Sacramento, CA,
USA). All animal experiments were completed under the supervision
of the Animal Ethics Committee of Bingzhou People's Hospital
(Shandong, China). The mice were sacrificed by decollation after
the experiment and cerebral tissues were extracted for later
use.
Separation and culture of primary
neurons
Mouse cortical neurons were separated from the
cortical tissue of neonatal mice aged 0–1 days. The cortical tissue
was separated into fragments with tweezers in a 1X
phosphate-buffered saline (PBS) glass dish. The tissues were
incubated with 0.5% trypsin (Gibco-BRL Life Technologies Inc.,
Grand Island, NY, USA) for 20 min at 37°C, in an EP tube. The
dislodged cells were gently centrifuged at 3,900 × g, seeded in a
24-well microplate, and cultured with Dulbecco's modified Eagle's
medium (DMEM) + 10% FBS (Gibco-BRL Life Technologies Inc.)
containing glutamine (3 pg/ml). After 24 h, the solution was
changed to 1X NB/B27 containing glutamine (3 pg/ml) (15).
Polymerase chain reaction (PCR)
Mouse cerebral tissue (1 cm3) or
106 of the mammalian cells HEK293A, HCT116 and N2a were
harvested. RNA was extracted using the TRIzol method and
transcribed into cDNA. Each 20 µl PCR reaction solution including
DNA polymerase (1 µl), reverse transcriptase (0.35 µl), and
template RNA (5 µl) was well mixed and centrifuged at 3,900 × g for
10 sec. The amplification protocol program included a reverse
transcription step for 25 min at 50°C; a pre-denaturation step for
3 min at 95°C; and 5 cycles of denaturation at 95°C for 15 sec,
annealing for 30 sec at 50°C, and extension for 30 min at 72°C; and
finally 40 cycles of a denaturation step for 10 sec at 95°C, and an
annealing step for 40 sec at 55°C. The primers used in the
experiment are listed in Table
I.
 | Table I.The primers used in the
experiments. |
Table I.
The primers used in the
experiments.
| Primers | Sequence |
|---|
| Human p21-F |
5′-GCGACTGTGATGCGCTAAC-3′ |
| Human p21-R |
5′-GGGCTTCCTC订?GGAGAAG-3′ |
| Mouse p21-F |
5′-AGGAGCAAAGTGTGCCGTG-3′ |
| Mouse p21-R |
5′-GGAGTGArAGAAATCTGTCG-3′ |
| Human β-actin-F |
5′-GACCTGACTGACTACCT-3′ |
| Human β-actin-R |
5′-GACAGCGAGGCCAGGATGT-3′ |
| Mouse/rat
β-actin-F |
5′-GACCTGACTGACTACCTC-3′ |
| Mouse/rat
β-actin-R |
5′-GACAGCGAGGCCAGGATGC-3′ |
| Human coronin
1b-F |
5′-CAGAGCAAAGCCGGCATG-3′ |
| Human coronin I
b-R |
5′-GTCAGGGTGCAGGCTGTCC-3′ |
| Mouse coronin
1b-F |
5′-GTTGTGCGGCAGAGCAAATT-3′ |
| Mouse coronin
1b-R |
5′-CAAGCTGTCCAGACGGTAC-3′ |
| Rat coronin 1b-F |
5′-GTTGTGCGGCAGAGCAAAT-3′ |
| Rat coronin l
b-R |
5′-GTCAGGGTGCAAGCTGTCT-3′ |
| Human/mouse/rat
Rab13-F |
5′-ACGACCACCTCTTCAAGCC-3′ |
| Human/mouse/rat Rab
13-R |
5′-AAAGCCTCATCCACACTCA-3′ |
| Human/mouse
p38-F |
5′-TCTGGGACAGCCAAGTCTG-3′ |
| Human/mouse
p38-R |
5′-CTTCCAGTGTGATGATGGA-3′ |
| Rat p38-F |
5′-TCAGGGACAGCCAAGTCTG-3′ |
| Rat p38-R |
5′-CTTCCAGCGTGArGATGGT-3′ |
| Human pallidin-F |
5′-CTGGTGGACAGCGAGGTG-3′ |
| Human pallidin-R |
5′-CAGAGTTCAGGAAGACGT-3′ |
| Mouse pallidin-F |
5′-CTGGTGGACAGCGAGGT-3′ |
| Mouse pallidin-R |
5′-CTCGCCTCTCTGCGATCT-3′ |
| Mouse HDAC3-F |
5′-CTGGTGGACAGCGAGGT-3′ |
| Mouse HDAC3-R |
5′-CTCGCCTCTCTGCAATCT-3′ |
| Rat HDAC3-F |
5′-ATGAAGGACCAGAAGAGATG-3′ |
| Rat HDAC3-R |
5′-GTCCTCAGAGACACTGCG-3′ |
Agarose gel electrophoresis
Once the agarose gel solution cooled to about 65°C
it was mixed well, carefully poured into a glass plate, and allowed
to stand at room temperature until gel coagulation. The gel and the
glass plate were placed in the electrophoresis tank. A 1X TAE
electrophoretic buffer solution was added until the gel plate was
completely immersed. The DNA sample and loading buffer were mixed.
The gel plate was energized for electrophoresis at a voltage of
60–100 V immediately after loading. The electrophoresis was
terminated when the bromophenol blue reached a point ~1 cm from the
lower edge of the gel plate. The gels were stained for about 20 min
with ethidium bromide and rinsed for 10 min with clear water.
Observations were performed under an ultraviolet lamp (Gemini
Bio-Products, Woodland, CA, USA).
GST pull-down
The crude extract of the supernatant of the GST and
GST-Tag fusion proteins (Invitrogen, Carlsbad, CA, USA) were bound
to G4B. A crude extract of the supernatant of the second protein
bound to the GST-Tag fusion protein was added. The samples were
placed on a shaker platform for 1 h at 4°C, and then centrifuged
for 5 min at 500 × g at 4°C. The supernatants were discarded. A 2X
SDS gel loading buffer of appropriate amount was added to each
sample. They were boiled for 5 min at 100°C, subjected to sodium
dodecyl sulphate-polyacrylamide gel (SDS-PAGE), and Coomassie
Brilliant Blue stained, or detected by western blot analysis
(16).
Cell transfection (lipofection
method)
Six-well microplates were inoculated with
1×105-3×105 cells (HEK293A, HCT116 and N2a)
per well. For transfection, the cells were cultured in the
incubator overnight until the cell density reached 50–80%
confluence. The transfection procedure was done following the
manufacturer's instructions. Briefly, 1–4 µg plasmid DNA (Sangon
Biotech Co., Ltd., Shanghai, China) was used for transfections.
DMEM was used to prepare solutions containing plasmid DNA-liposome
compounds. The cells were washed with DMEM prior to the
transfections. Next, 200 µl of the DNA-liposome compound solution
were mixed with 0.8 ml DMEM and added to the cell incubation wells.
The cells were placed back in the incubator for 4–6 h. Finally, 1
ml of the culture solution at a serum concentration equivalent to 2
times the normal was added to the cells in each well. Transfections
were allowed to proceed for 18–24 h prior to changing the growth
media.
Co-immunoprecipitation
Collected lysis buffers of supernatants containing
lysed cell proteins were added to appropriate protein A or G gel
columns (Bio-Rad Laboratories, Inc., Hercules, CA, USA) coupled
with antibody. The protein extracts were incubated with the gel
while shaking the mixture for over 5 h at 4°C.
The columns were then centrifuged for 3–5 sec at the
highest speed at 4°C. The supernatant was carefully pipetted and
discarded. Washing buffer (1 µl) was added to the columns. The
washing proceeded for 20 min at 4°C. The washing steps were
repeated once. The columns were centrifuged for 3–5 sec at the
highest speed at 4°C. The supernatants were carefully pipetted out
and discarded. Finally, a 2X SDS gel loading buffer solution of
appropriate amount was added to the samples. The samples were
boiled for 5 min at 100°C and then subjected to the SDS-PAGE
analysis.
Western blot analysis
The pre-extracted mammalian cell proteins were
subject to SDS-PAGE. Next, the gels were placed in a sandwich with
transfer membrane in the electrical transfer tank (120 V 320 mA).
The transfer proceeded for 60–90 min. The membranes were placed in
a dish containing 25 ml blocking buffer. After 2 h on a shaker
platform at room temperature, the primary antibodies were added:
Anti-pallidin, HDAC3, his-Ndn, F-actin, GAPDH, and p21 (Jackson
ImmunoResearch Laboratories, Inc., West Grove, PA, USA). The
reactions were left incubating overnight at 4°C. The next day, each
membrane was washed 3 times for 5 min with TBST on a shaker at room
temperature. Appropriate secondary antibodies (Boster Inc., Wuhan,
China) were added. Incubations took place on a shaker for 1 h at
37°C. The membrane was washed 3 times × 5 min with TBST. A
developer was used to observe the results on a platform.
Immunocytochemistry
The mammalian cells (HEK293A, HCT116 and N2a) were
washed 3 times × 5 min with PBS after growing on glass slides. A
goat serum working solution was added carefully with a dropper. The
slides were blocked for 20 min at 37°C. The goat anti-pallidin
(1:500; cat. no. PA5-18160): rabbit polyclonal anti-HA-Ndn (1:500;
cat. no. MA1-012X), mouse polyclonal anti-EGFP (1:500; cat. no.
MA1953), and rabbit polyclonal anti-pal-EGFP (1:2,000; cat. no.
PA527854) were added. The cells were allowed to stand overnight at
4°C, and then were washed 3 times for 5 min with PBS the next day.
The specific secondary antibody polyclonal FITC (1:1,000; cat. no.
A-11090; Invitrogen Life Technologies) was added. The reactions
were incubated for 2 h at 37°C, stained with Hoechst for 5 min and
washed with PBS 3 times for 5 min each. The sections were placed on
clean glass slides, air dried, mounted with the anti-fluorescent
quenching mounting solution, and conserved away from light at 4°C
for microscopic examination.
RNA interference
The small RNA interference primers (Google
Biotechnology Co., Ltd., Shanghai, China) were designed as follows:
5′-AAGUGACAAGUCAAGAGAAGCAA-3′ (target to human pallidin mRNA);
5′-AAGUGAUAAGUCAAGAGAAGCAA-3′ (target to mouse pallidin mRNA);
5′-AAGCGACAAGUCAAAAGAAGCAA-3′ (target to rat pallidin mRNA); and
5′-CCACUUGAUGGAGAGUAUU-3′ (target to mouse p38 mRNA). The cell
densities were allowed to reach 40–60% confluence before
transfection. The following solutions were prepared: Solution A
with 2–10 µl Lipofectamine Oligofectamine (Invitrogen) transfection
reagent and 80 µl Opti-MEM, which was incubated for 5 min at room
temperature; and solution B with 4–8 µl of siRNA and 80 µl
Opti-MEM. Solution A and B were mixed well together. The mixtures
were allowed to stand for 30 min at room temperature. The cells
were washed with Opti-MEM. Then 0.8 µl 0pti-MEM were added to each
liposome mixture and the transfection mixture was added to the
cells. The cultures were incubated for 4–6 h. Then the culture
medium was replaced with a fresh one at 18–24 h after transfection.
The cells were harvested 4–72 h after transfection.
Cell differentiation experiment and
detection of the process of growth
N2a was cultured in complete medium (10% FBS + DMEM)
for 24 h. The plasmid or siRNA (as above) was transfected with
Lipofectamine 2000 or Oligo Lipofectamine for 6 h. Next, cells were
cultured with DMEM containing 2% FBS to induce cell
differentiation. The new processes on the cell membranes were
observed under a microscope. Approximately 200–250 cells were
counted in each experiment. The primary cortical neurons, separated
from the neonatal mice, were inoculated into a 24-well microplate.
The solution was replaced at 24 h. The cells were stained with Tuj
(Biosynthan GmbH, Berlin, Germany) to show the neuronal processes 5
days after being cultured with NB/B27.
Detection of living cells with the MTT
method
The plasmid at a determined concentration was used
for transfections at a cell confluence of 50–60%. The original
culture medium was pipetted out after 36–48 h. Cells were washed 2
times with 1X PBS. Five hundred microliters of MTT diluted with
phenol red-free MEM was added to obtain a final concentration of
0.5 mg/ml. The reactions proceeded for 2–3 h at 37°C. Then, the
excess MTT was absorbed, and 500 µl of 0.04 M acidulated
isopropanol was added. The cell culture dish was gently shaken back
and forth until the bluish violet crystal formazan was completely
dissolved in the acidulated isopropanol. The supernatant was
transferred to a 96-well ELISA plate (Thermo Fisher Scientific,
Inc., Beijing, China). The optical density was determined at 570 nm
(Thermo Fisher Scientific, Waltham, MA, USA).
Statistical analysis
The measurement data were expressed with mean ±
standard deviation. The significance of the average differences
between two groups of the data were tested with the t-test of two
independent samples. The measurement data were expressed as a
percentage. The Chi-square test was used for comparisons among
groups. The differences were considered statistically significant
when P<0.05. The SPSS 16.0 software was used to process the
statistical data.
Results
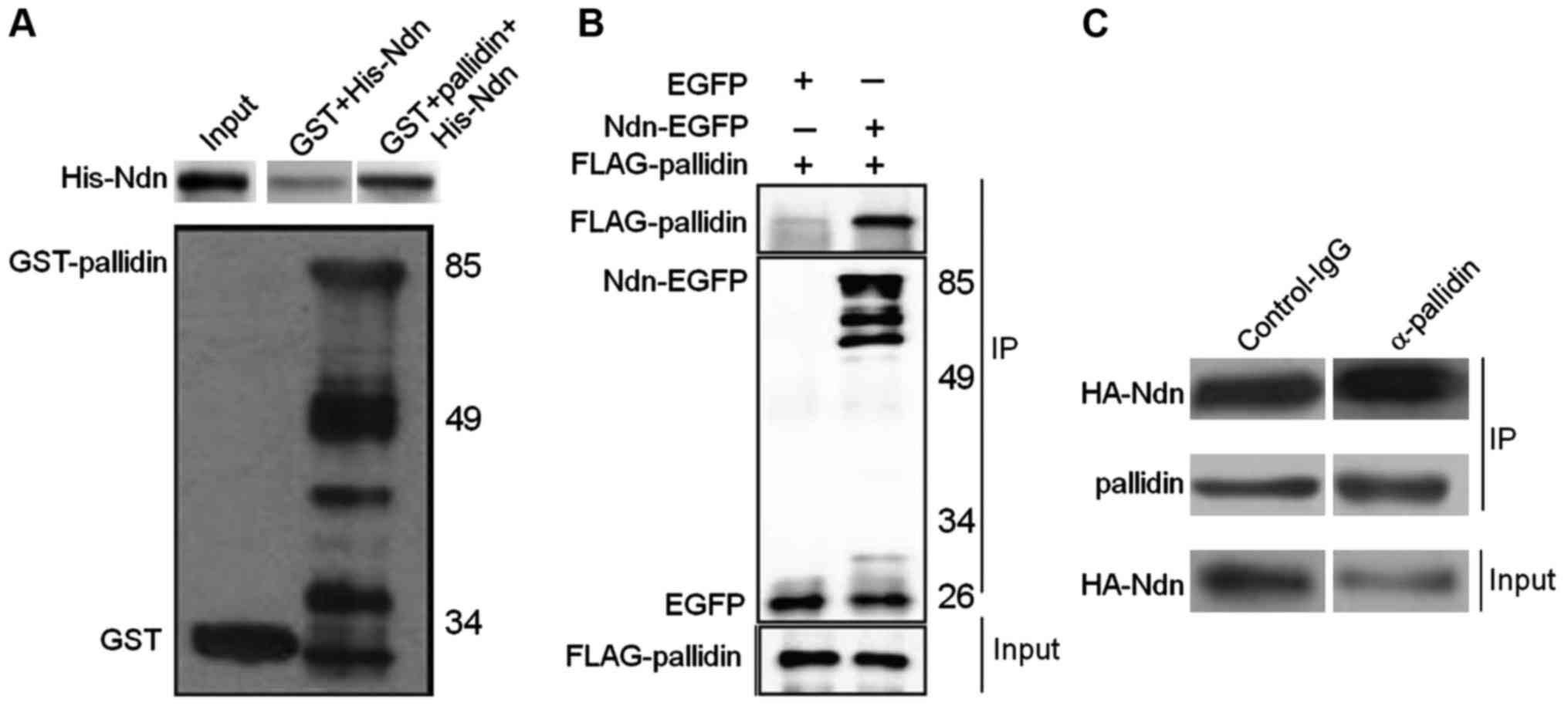
Identification of binding between
HDAC3 and pallidin
The experimental results showed that GST-pallidin
co-immunoprecipitates with His-Ndn in vitro, whereas, not
with the control GST alone. To further verify the binding between
pallidin and HDAC3 in the mammalian cell lines, FLAG-pallidin and
EGFP or Ndn-EGFP proteins were co-expressed in a human embryonic
renal cell line (HEK293A). Subsequently, a co-immunoprecipitation
experiment was conducted. The results indicated that FLAG-pallidin
also co-precipitates with Ndn-EGFP when Ndn-EGFP was bound to the
GFP antibody; as a negative control, EGFP alone was not able to
co-precipitate FLAG-pallidin (Fig.
1B). Next, a dysbindin antibody (which can also bind pallidin)
was used to precipitate the endogenous pallidin in a mouse neuroma
mother cell line (N2a) transfected with HA-Ndn. The results showed
that HA-Ndn and the endogenous pallidin co-precipitated, whereas
the control IgG antibody was not able to precipitate as much HA-Ndn
(Fig. 1C).
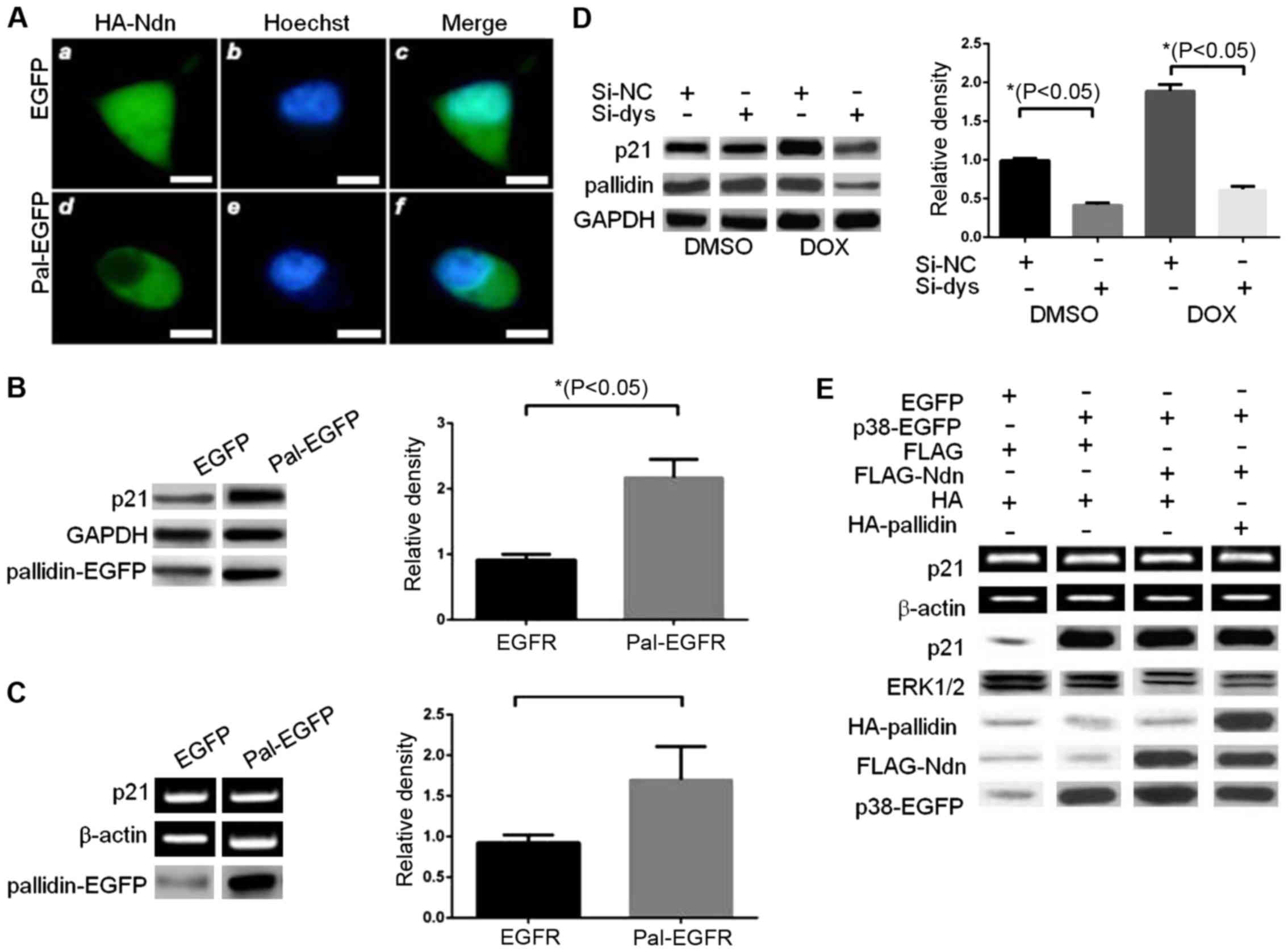
Pallidin changes localization and
influences the transcriptional activity of p38
Immunofluorescence results indicated that
overexpression of pallidin-EGFP caused a change in the cellular
distribution of HA-Ndn, from being present normally in both the
cytoplasm and nucleus to being present only in the cytoplasm
(Fig. 2A). During previous
experiments we observed that when exogenous p38 was overexpressed,
the activity of the reporter gene was markedly increased. By
contrast, the transcriptional activity of p38 was significantly
inhibited by HDAC3 in the cells co-transfected with HDAC3 and p38.
The transcriptional activity of endogenous HDAC3 increased
significantly when pallidin was overexpressed (data not shown).
Overexpression of pallidin-EGFP in the HCT116 p38 wild-type cell
line increased the endogenous p21 protein and mRNA levels (Fig. 2B and C). The small RNA interference
method was used to decrease the expression level of endogenous
pallidin and observe the effects on the expression level of
endogenous p21 in the HCT116 p38+/+ cell line. A
decrease in p21 was still significant even after the p38 activator
doxorubicin (DOX) was used to promote the transcriptional activity
of endogenous p38 (Fig. 2D).
Various different plasmids were transfected into HCT116
p38−/− cells as shown in Fig. 2E. It also indicated that pallidin
was able to reverse the decrease of the p21 protein and RNA levels
as a result of inhibitory effect of HDAC3. This coincided with the
previous result of the reporter gene. In conclusion, pallidin was
able to influence the transcriptional activity of the downstream
p38 by changing the intracellular localization of HDAC3.
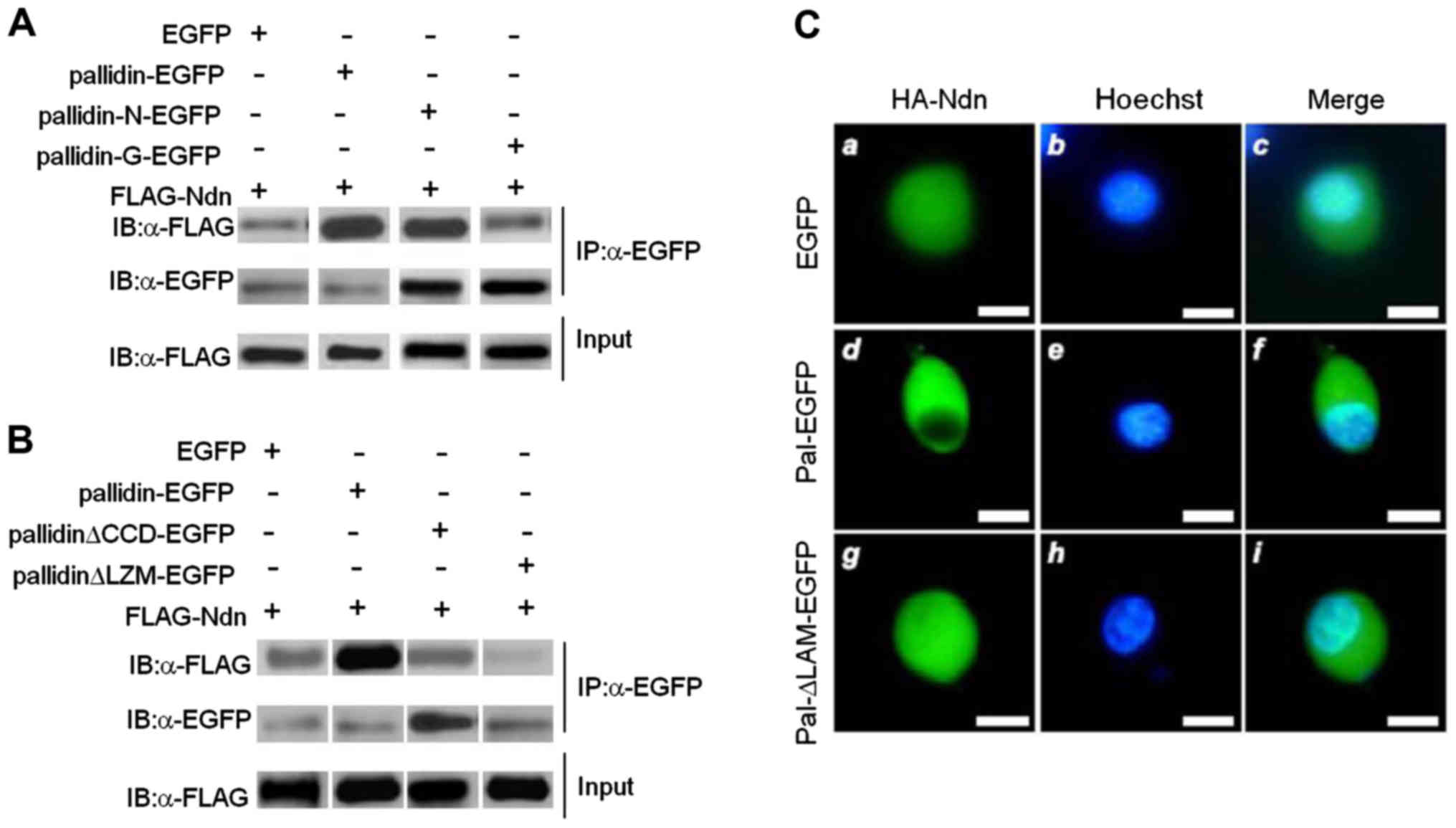
The 90–110 amino acid sequences of
pallidin are participating in HDAC3 regulation
In co-immunoprecipitation experiments both
pallidin-EGFP and pallidin-N-EGFP co-precipitated FLAG-HDAC3.
However, neither pallidin-C-EGFP nor the negative control EGFP,
were able to co-precipitate FLAG-HDAC3 (Fig. 3A). We established two pallidin
mutants with either the CCD or LZM domains deleted.
Co-immunoprecipitation assays were conducted after the two mutants
and the full-length pallidin were co-transfected with FLAG-HDAC3.
The result showed that both mutants lost their capacity to bind to
HDAC3 (Fig. 3B). The full-length
pallidin-EGFP enabled HA-HDAC3 to exhibit significant cytoplasmic
localization. On the contrary, the pallidin-ALZM-EGFP without its
leucine zipper was not able to influence the subcellular
localization of HA-HDAC3. HA-HDAC3 exhibited uniform distribution
in the cytoplasm and nucleus in the cells co-transfected with the
mutant (similar to those transfected with EGFP control) (Fig. 3C).
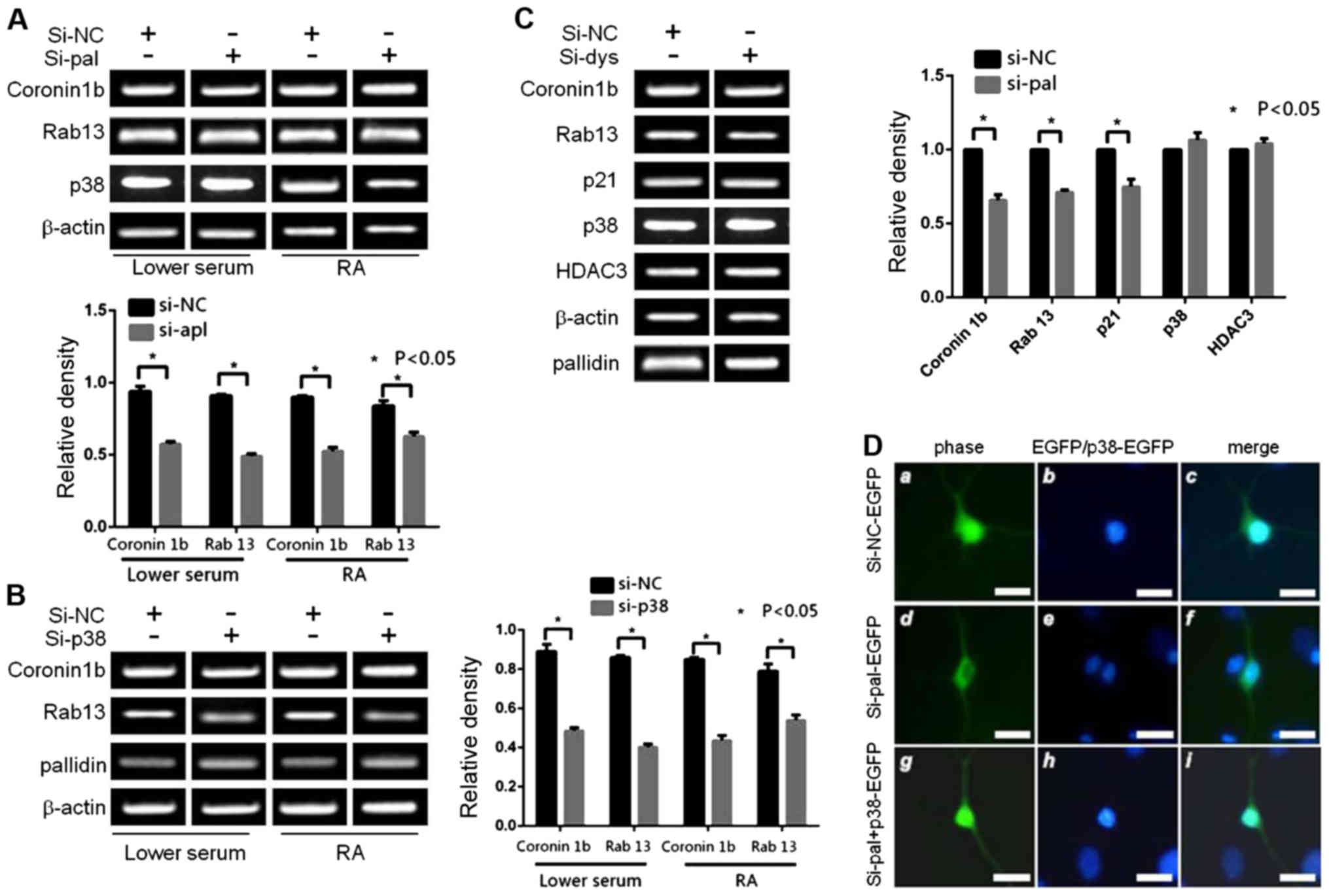
Pallidin influenced growth of the
differentiated cellular processes by regulating the downstream gene
product of p38
The levels of mRNA in the p38 downstream target
genes was detected with RT-PCR after the expression level of
endogenous p38 was knocked down. At the same time, a western blot
analysis demonstrated that the expression levels of the two effect
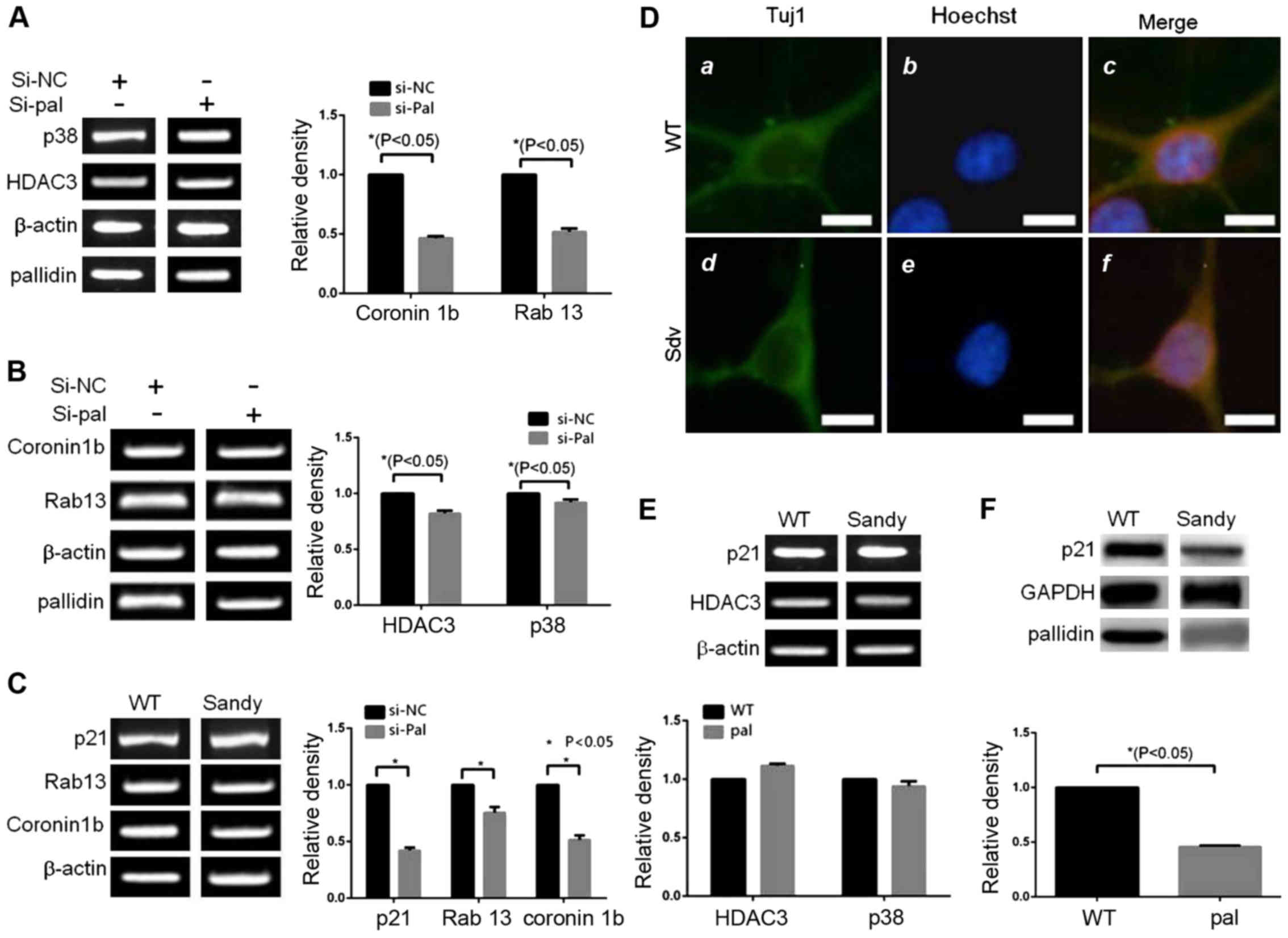
proteins coronin 1b and Rab13 decreased (Fig. 4A). The two mRNA molecules of
coronin 1b and Rab13 transcriptionally induced by p38 decreased
with the reduction in pallidin (Fig.
4B). Meanwhile, the expression levels of p38 and HDAC3 were not
influenced by pallidin (Fig. 4C).
Our results showed that the changes in pallidin can affect the
transcriptional activity of p38 during cell differentiation. The
decreased expression of endogenous p38 influenced the degree of N2a
cellular differentiation. The length of the nervous processes
(neurites) generated in the cells of the experimental group was
significantly shorter than the length of the processes in the
control group. Similar to the endogenous p38, the decrease of
endogenous pallidin also produced the same effect in the N2a cell
line (Fig. 4D).
The role of pallidin in regulating the
process growth
In the primary culture of rat cerebral cortex
neurons, when the expression level of pallidin decreased as a
result of small RNA interference, the expression levels of coronin
1b and Rab13 (the downstream transcription target genes of p38)
also decreased. The expression levels of p38 and HDAC3 did not
change (Fig. 5B). The detection of
the sandy mice without pallidin also indicated that the mRNA levels
of p21, coronin 1b, and Rab13 decreased significantly when compared
with those in the wild-type (WT) mice at the same age (Fig. 5C). The p21 protein level in the
hippocampal region of the sand mice decreased significantly when
compared with that in the wild mice (Fig. 5E). Similarly, there were no
significant differences in expression levels between p38 and HDAC3
(Fig. 5F). The length of the
processes of the cortical neurons of the sandy mice decreased
significantly at 5 days after the neurons were cultured in
vitro when compared with the processes in the WT mice
(decreased by ~20%, P=0.0068). No change in body size was observed
(Fig. 5D).
Discussion
In this study, we investigated the new functions of
the schizophrenia-related protein pallidin. Regulating the
transcriptional activity of p38 influences the neural system
development.
We have demonstrated that pallidin promotes the
transcriptional activity of the p38 inhibited by intranuclear HDAC3
by retaining HDAC3 in the cytoplasm. HDAC3 is a known transcription
inhibitor (17). It inhibits the
transcriptional activity by being bound to proteins such as E2F1
and p38, in the cell nucleus (5,8,18).
It regulates the functions of neurons by inhibiting important
intranuclear transcription factors (19,20).
This study shows that dysbindin-1 (a pallidin
homologue) can be directly bound to HDAC3 in vitro. The
results also show that overexpression of pallidin can change the
distribution ratio of HDAC3 in the cytoplasm/nucleus by retaining
HDAC3 within the cytoplasm. The inhibition of p38 transcription by
HDAC3 is reduced with lower amounts of HDAC3. Hence, pallidin can
change the transcriptional activity of p38 by regulating HDAC3. A
pallidin-ALZM mutant variant lacking 90–110 amino acid sequences
(21) demonstrates that pallidin
can change the HDAC3 location. The mutant cannot bind HDAC3 and
therefore, does not affect its location. Thus, we discuss a new
function of pallidin: It influences the transcriptional activity of
p38 by being bound to HDAC3 and changing the location of HDAC3.
In the knock-down experiments of the endogenous
pallidin, we observed the same result as when knocking down the
endogenous p38: The reduction significantly interferes with the
cellular differentiation. Overexpression of p38 in the pallidin
knocked down cells can restore the cellular differentiation
ability.
These results indicate that it is possible that
pallidin influences the growth of the nerve processes via p38. The
in vivo data also indicate that the genes related to neural
development downstream of p38, coronin 1b and Rab13 decreased in
the pallidin knocked down sandy mice. Moreover, the cortical
neurons of the sandy mice also show alterations in neuronal process
growth characteristics. Therefore, we believe that the endogenous
pallidin plays a role in maintaining the transcriptional activity
of p38 during neurodevelopment and guaranteeing the expression of
necessary genes. It has been reported that Rab13 promotes neuronal
development by binding and antagonizing the neuronal process growth
inhibiting factors (22–24). In addition, studies have indicated
that knockdown of coronin 1b can promote aggregation of F-actin in
leading edge and influence the morphology of the plate-shaped
filopodia on the cells (25,26).
Concentrations of both effector molecules, Rab13 and coronin 1b
decrease with overexpression of pallidin. Thus, the abnormal
arrangement of F-actin in the sandy mice may be associated with the
decrease of coronin 1b and Rab13.
Based on the results of our experiments, it seems
quite possible that pallidin influences the growth of the nerve
processes via p38, thereby participating in neuronal development.
An abnormally developed nervous system may also increase the risk
for schizophrenia. Given that pallidin has been linked to the
pathogenesis of schizophrenia, and we found pallidin to function
with p38 during neuronal development, our study argues for
schizophrenia as a neurodevelopmental disease.
References
|
1
|
Hermann GJ, Scavarda E, Weis AM, Saxton
DS, Thomas LL, Salesky R, Somhegyi H, Curtin TP, Barrett A, Foster
OK, et al: C. elegans BLOC-1 functions in trafficking to
lysosome-related gut granules. PLoS One. 7:e430432012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Larimore J, Ryder PV, Kim KY, Ambrose LA,
Chapleau C, Calfa G, Gross C, Bassell GJ, Pozzo-Miller L, Smith Y,
et al: MeCP2 regulates the synaptic expression of a
Dysbindin-BLOC-1 network component in mouse brain and human induced
pluripotent stem cell-derived neurons. PLoS One. 8:e650692013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Padeĭskaia EN, Kutchak SN and Polukhina
LM: Comparative activity of depot sulfanilamides in experimental
infection in mice caused by K1. pneumoniae. Antibiotiki.
25:193–198. 1980.(In Russian). PubMed/NCBI
|
|
4
|
Huard C, Martinez RV, Ross C, Johnson JW,
Zhong W, Hill AA, Kim R, Paulsen JE and Shih HH: Transcriptional
profiling of C2C12 myotubes in response to SHIP2 depletion and
insulin stimulation. Genomics. 89:270–279. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nazarian R, Starcevic M, Spencer MJ and
Dell'Angelica EC: Reinvestigation of the dysbindin subunit of
BLOC-1 (biogenesis of lysosome-related organelles complex-1) as a
dystrobrevin-binding protein. Biochem J. 395:587–598. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schmitz G and Schambeck CM: Molecular
defects in the ABCA1 pathway affect platelet function. Pathophysiol
Haemost Thromb. 35:166–174. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dotta L, Parolini S, Prandini A, Tabellini
G, Antolini M, Kingsmore SF and Badolato R: Clinical, laboratory
and molecular signs of immunodeficiency in patients with partial
oculo-cutaneous albinism. Orphanet J Rare Dis. 8:1682013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Larimore J, Zlatic SA, Gokhale A, Tornieri
K, Singleton KS, Mullin AP, Tang J, Talbot K and Faundez V:
Mutations in the BLOC-1 subunits dysbindin and muted generate
divergent and dosage-dependent phenotypes. J Biol Chem.
289:14291–14300. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cullinane AR, Curry JA, Carmona-Rivera C,
Summers CG, Ciccone C, Cardillo ND, Dorward H, Hess RA, White JG,
Adams D, et al: A BLOC-1 mutation screen reveals that PLDN is
mutated in Hermansky-Pudlak Syndrome type 9. Am J Hum Genet.
88:778–787. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Monfregola J, Napolitano G, D'Urso M,
Lappalainen P and Ursini MV: Functional characterization of
Wiskott-Aldrich syndrome protein and scar homolog (WASH), a
bi-modular nucleation-promoting factor able to interact with
biogenesis of lysosome-related organelle subunit 2 (BLOS2) and
gamma-tubulin. J Biol Chem. 285:16951–16957. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang P, Wang L, Wang S, Li S, Li Y and
Zhang L: Effects of calcium-sensing receptors on apoptosis in rat
hippocampus during hypoxia/reoxygenation through the ERK1/2
pathway. Int J Clin Exp Pathol. 8:10808–10815. 2015.PubMed/NCBI
|
|
12
|
Shao M, Tang ST, Liu B and Zhu HQ: Rac1
mediates HMGB1-induced hyperpermeability in pulmonary microvascular
endothelial cells via MAPK signal transduction. Mol Med Rep.
13:529–535. 2016.PubMed/NCBI
|
|
13
|
Sapkota M, Li L, Choi H, Gerwick WH and
Soh Y: Bromo-honaucin A inhibits osteoclastogenic differentiation
in RAW 264.7 cells via Akt and ERK signaling pathways. Eur J
Pharmacol. 769:100–109. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sabogal-Guáqueta AM, Osorio E and
Cardona-Gómez GP: Linalool reverses neuropathological and
behavioral impairments in old triple transgenic Alzheimer's mice.
Neuropharmacology. 102:111–120. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Delevoye C, Heiligenstein X, Ripoll L,
Gilles-Marsens F, Dennis MK, Linares RA, Derman L, Gokhale A, Morel
E, Faundez V, et al: BLOC-1 brings together the actin and
microtubule cytoskeletons to generate recycling endosomes. Curr
Biol. 26:1–13. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li W, Zhang Q, Oiso N, Novak EK, Gautam R,
O'Brien EP, Tinsley CL, Blake DJ, Spritz RA, Copeland NG, et al:
Hermansky-Pudlak syndrome type 7 (HPS-7) results from mutant
dysbindin, a member of the biogenesis of lysosome-related
organelles complex 1 (BLOC-1). Nat Genet. 35:84–89. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fu ZF, Li X and Dhingra V: Pathogenic
rabies virus alters host protein expression in the central nervous
system: implications for neuronal dysfunction. Dev Biol (Basel).
131:83–91. 2008.PubMed/NCBI
|
|
18
|
Spiegel S, Chiu A, James AS, Jentsch JD
and Karlsgodt KH: Recognition deficits in mice carrying mutations
of genes encoding BLOC-1 subunits pallidin or dysbindin. Genes
Brain Behav. 14:618–624. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Martina JA, Moriyama K and Bonifacino JS:
BLOC-3, a protein complex containing the Hermansky-Pudlak syndrome
gene products HPS1 and HPS4. J Biol Chem. 278:29376–29384. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Han MH, Hu Z, Chen CY, Chen Y, Gucek M, Li
Z and Markey SP: Dysbindin-associated proteome in the p2
synaptosome fraction of mouse brain. J Proteome Res. 13:4567–4580.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gwynn B, Martina JA, Bonifacino JS,
Sviderskaya EV, Lamoreux ML, Bennett DC, Moriyama K, Huizing M,
Helip-Wooley A, Gahl WA, et al: Reduced pigmentation (rp), a mouse
model of Hermansky-Pudlak syndrome, encodes a novel component of
the BLOC-1 complex. Blood. 104:3181–3189. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Valenti O and Grace AA: Antipsychotic
drug-induced increases in ventral tegmental area dopamine neuron
population activity via activation of the nucleus accumbens-ventral
pallidum pathway. Int J Neuropsychopharmacol. 13:845–860. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kouloumenta A, Mavroidis M and Capetanaki
Y: Proper perinuclear localization of the TRIM-like protein
myospryn requires its binding partner desmin. J Biol Chem.
282:35211–35221. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dhingra V, Li X, Liu Y and Fu ZF:
Proteomic profiling reveals that rabies virus infection results in
differential expression of host proteins involved in ion
homeostasis and synaptic physiology in the central nervous system.
J Neurovirol. 13:107–117. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Badolato R, Prandini A, Caracciolo S,
Colombo F, Tabellini G, Giacomelli M, Cantarini ME, Pession A, Bell
CJ, Dinwiddie DL, et al: Exome sequencing reveals a pallidin
mutation in a Hermansky-Pudlak-like primary immunodeficiency
syndrome. Blood. 119:3185–3187. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee HH, Nemecek D, Schindler C, Smith WJ,
Ghirlando R, Steven AC, Bonifacino JS and Hurley JH: Assembly and
architecture of biogenesis of lysosome-related organelles complex-1
(BLOC-1). J Biol Chem. 287:5882–5890. 2012. View Article : Google Scholar : PubMed/NCBI
|