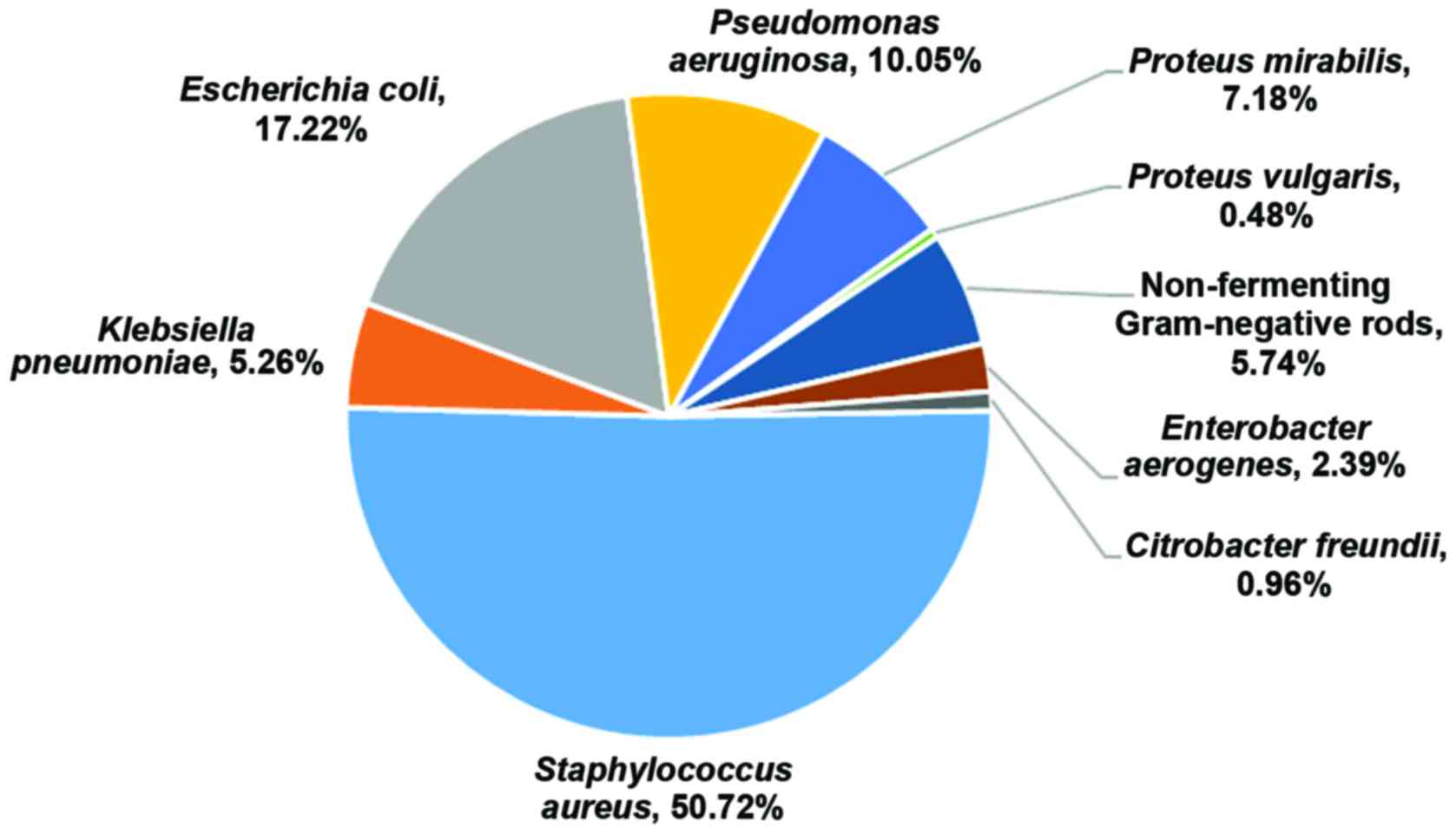
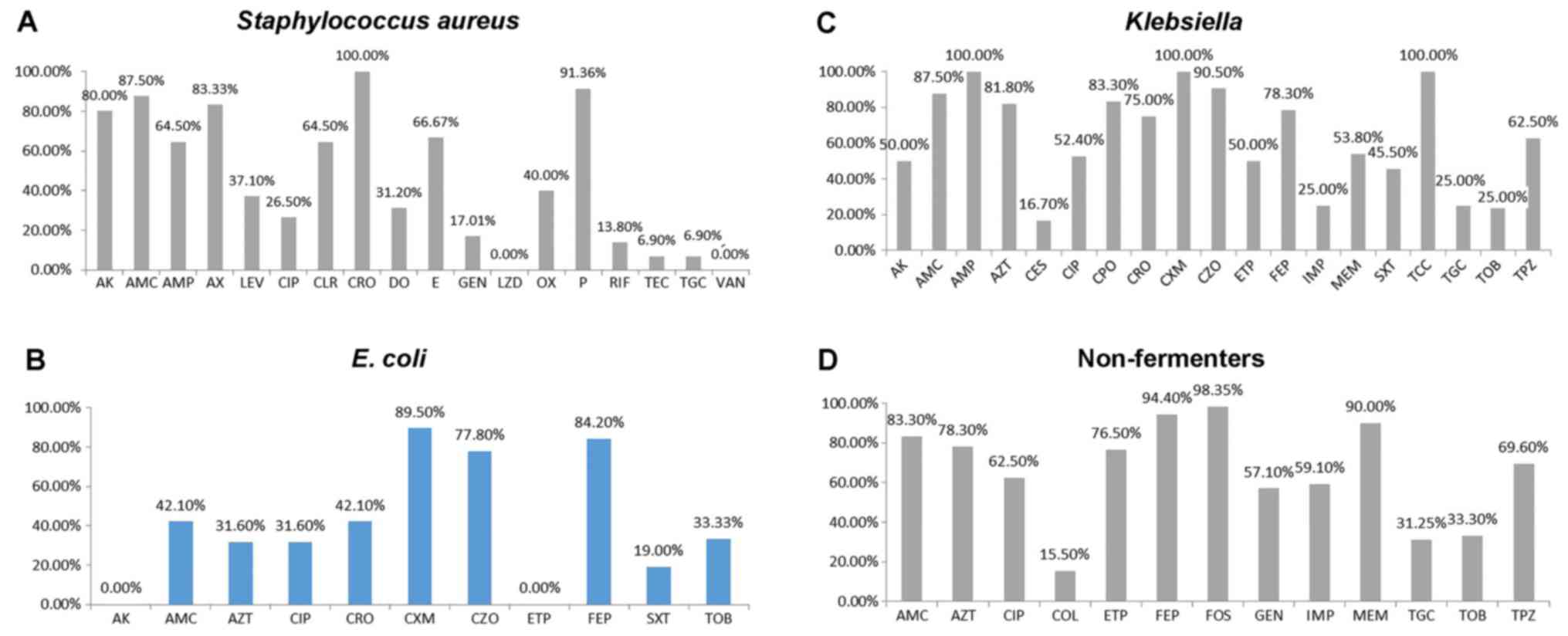
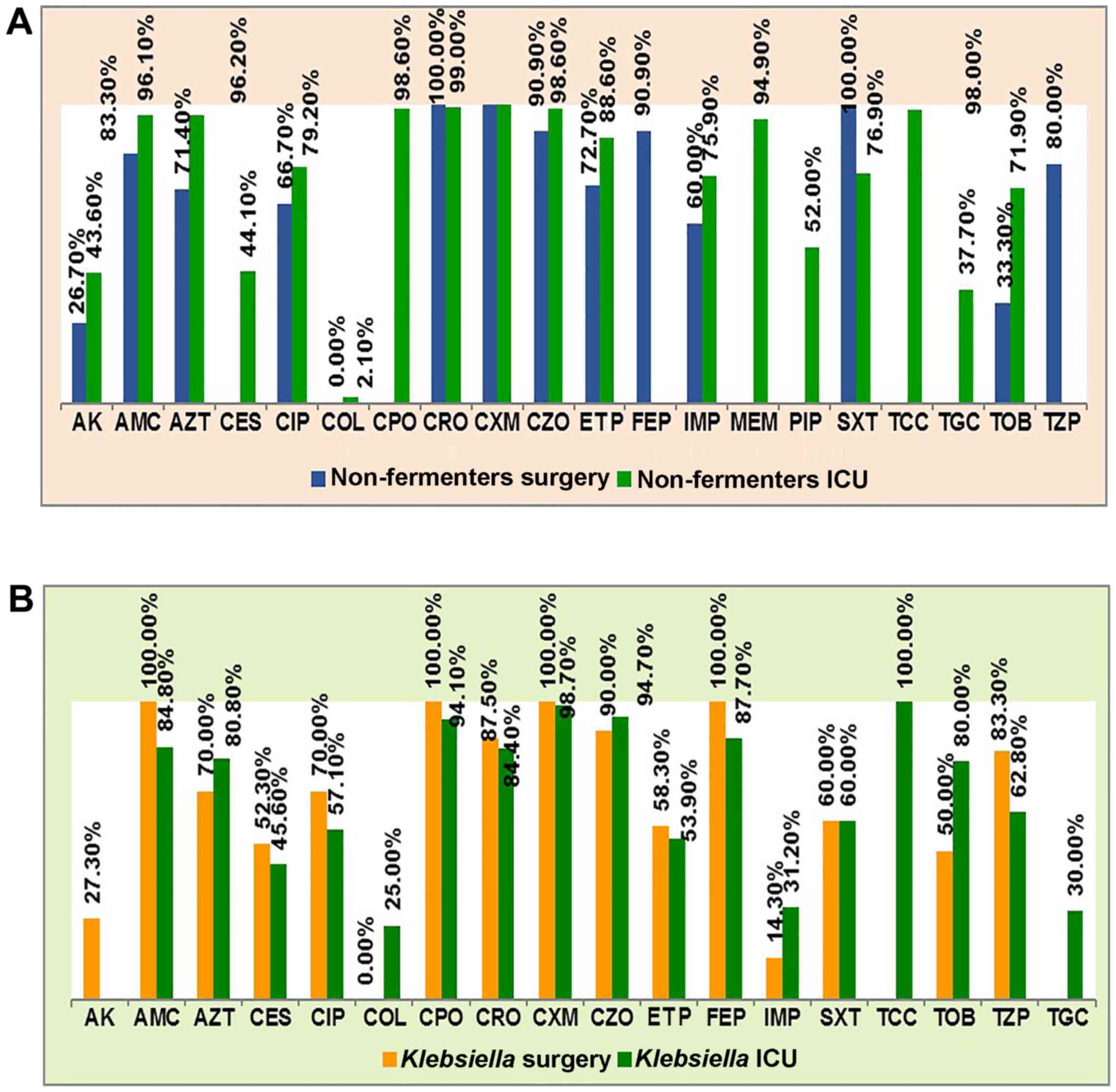
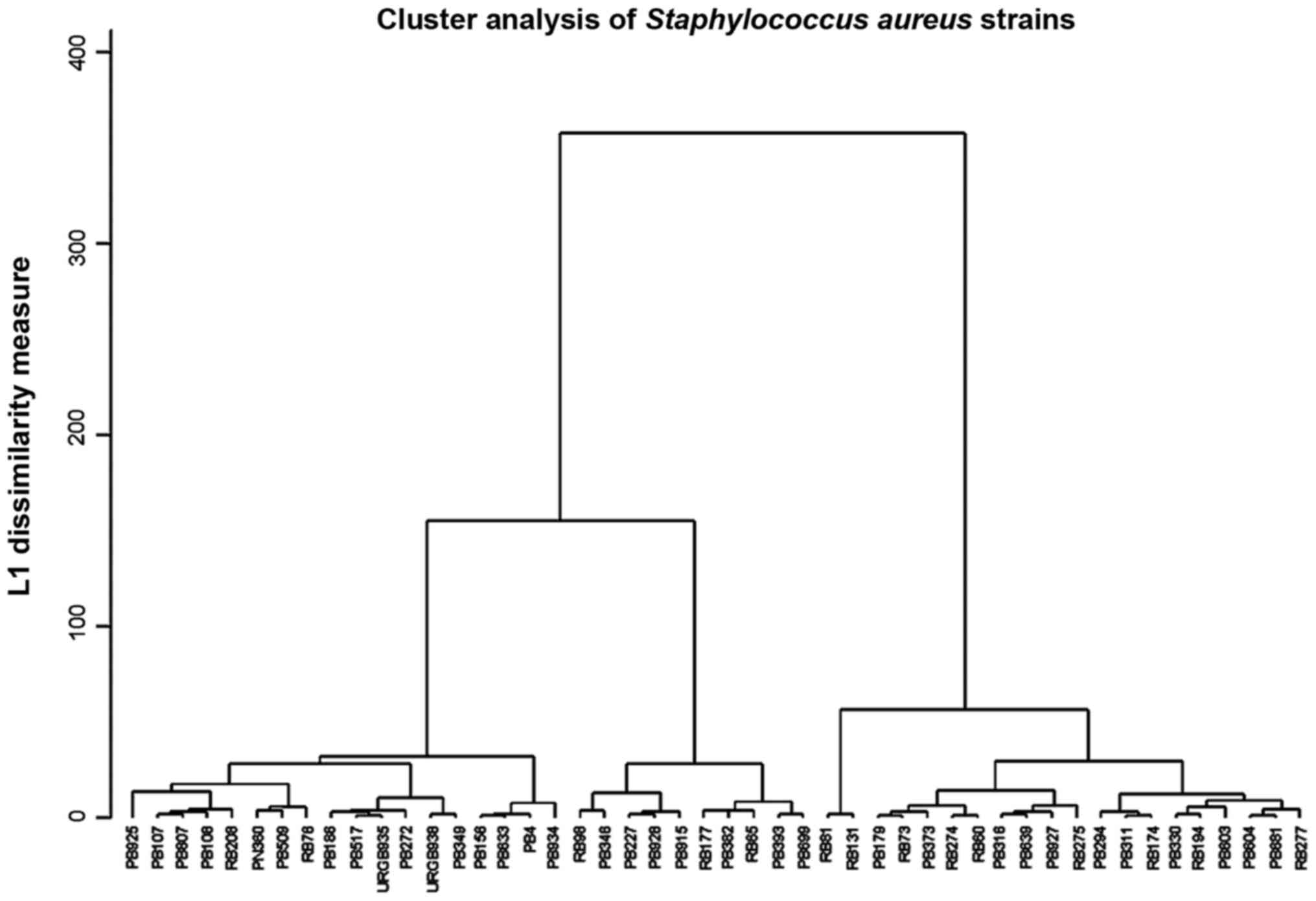
|
1
|
Staicus C, Calina D, Rosu L, Rosu AF and
Zlatian O: Involvement of microbial flora in aetiology of surgical
site infections. Eur J Hosp Pharm Sci Pract. 22:A592015.
|
|
2
|
Watanabe A, Kohnoe S, Shimabukuro R,
Yamanaka T, Iso Y, Baba H, Higashi H, Orita H, Emi Y, Takahashi I,
et al: Risk factors associated with surgical site infection in
upper and lower gastrointestinal surgery. Surg Today. 38:404–412.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Owens CD and Stoessel K: Surgical site
infections: Epidemiology, microbiology and prevention. J Hosp
Infect. 70 Suppl 2:3–10. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cristea OM, Zlatian OM, Dinescu SN,
Avramescu CS, Balasoiu M, Niculescu M and Calina DC: A comparative
study on antibiotic resistance of Klebsiella strains from surgical
and intensive care wards. Curr Heal Sci. 42:169–179. 2016.
|
|
5
|
Dulon M, Haamann F, Peters C, Schablon A
and Nienhaus A: MRSA prevalence in European healthcare settings: a
review. BMC Infect Dis. 11:1382011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
House of Commons Public Accounts
Committee, . Reducing Healthcare associated infection in hospitals
in England. Fifty-second Report of Session 2008–09. http://www.publications.parliament.uk/pa/cm200809/cmselect/cmpubacc/812/812.pdfNovember
10–2009.
|
|
7
|
Kassim A, Omuse G, Premji Z and Revathi G:
Comparison of Clinical Laboratory Standards Institute and European
Committee on Antimicrobial Susceptibility Testing guidelines for
the interpretation of antibiotic susceptibility at a University
teaching hospital in Nairobi, Kenya: a cross-sectional study. Ann
Clin Microbiol Antimicrob. 15:212016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li JY, Ma Y, Sun Z, Yao L, Zhang L, Hu C
and Jin S: Cluster analysis to detect the homogeneous strains of E.
coli isolated from clinical specimens. Chin J Microbiol Immunol.
23:384–387. 2003.
|
|
9
|
Xu J, Shi C, Song M, Xu X, Yang P, Paoli G
and Shi X: Phenotypic and genotypic antimicrobial resistance traits
of foodborne Staphylococcus aureus isolates from Shanghai. J Food
Sci. 79:M635–M642. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dzidic S and Bedeković V: Horizontal gene
transfer-emerging multidrug resistance in hospital bacteria. Acta
Pharmacol Sin. 24:519–526. 2003.PubMed/NCBI
|
|
11
|
Liapakis IE, Kottakis I, Tzatzarakis MN,
Tsatsakis AM, Pitiakoudis MS, Ypsilantis P, Light RW, Simopoulos CE
and Bouros DE: Penetration of newer quinolones in the empyema
fluid. Eur Respir J. 24:466–470. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kumar M, Dutta R, Saxena S and Singhal S:
Risk Factor Analysis in Clinical Isolates of ESBL and MBL
(Including NDM-1) Producing Escherichia coli and Klebsiella Species
in a Tertiary Care Hospital. J Clin Diagn Res. 9:DC08–DC13.
2015.PubMed/NCBI
|
|
13
|
Neonakis IK, Baritaki S, Georgiladakis A
and Spandidos DA: Analysis of the beta-lactam resistance phenotypes
of Escherichia coli. An 8-year survey conducted in Greece. Roum
Arch Microbiol Immunol. 67:10–13. 2008.PubMed/NCBI
|
|
14
|
Neonakis IK, Samonis G, Messaritakis H,
Baritaki S, Georgiladakis A, Maraki S and Spandidos DA: Resistance
status and evolution trends of Klebsiella pneumoniae isolates in a
university hospital in Greece: Ineffectiveness of carbapenems and
increasing resistance to colistin. Chemotherapy. 56:448–452. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sifakis S, Angelakis E, Makrigiannakis A,
Orfanoudaki I, Christakis-Hampsas M, Katonis P, Tsatsakis A and
Koumantakis E: Chemoprophylactic and bactericidal efficacy of 80 mg
gentamicin in a single and once-daily dosing. Arch Gynecol Obstet.
272:201–206. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Petrakis I, Gonianakis C, Vrachassotakis
N, Tsatsakis A, Vallilakis IS and Chalkiadakis G: Prospective study
of preincisional intraparietal single-dose ceftriaxone in reducing
postoperative wound infection in type I and II diabetic patients.
Acta Diabetol. 36:159–162. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Neonakis IK, Spandidos DA and Petinaki E:
Is minocycline a solution for multidrug-resistant Acinetobacter
baumannii? Future Microbiol. 9:299–305. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Youinou P, Garré M, Menez JF, Morin JF and
Masse R: Protein malnutrition and deficient cell-mediated immunity
in intensive care (author's transl). Nouv Presse Med. 10:3835–3837.
1981.(In French). PubMed/NCBI
|
|
19
|
Neonakis IK, Spandidos DA and Petinaki E:
Confronting multidrug-resistant Acinetobacter baumannii: A review.
Int J Antimicrob Agents. 37:102–109. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tanase A, Colita A, Ianosi G, Neagoe D,
Branisteanu DE, Calina D, Docea AO, Tsatsakis A and Ianosi SL: Rare
case of disseminated fusariosis in a young patient with graft vs.
host disease following an allogeneic transplant. Exp Ther Med.
12:2078–2082. 2016.PubMed/NCBI
|
|
21
|
Malini A, Deepa E, Gokul B and Prasad S:
Nonfermenting gram-negative bacilli infections in a tertiary care
hospital in kolar, karnataka. J Lab Physicians. 1:62–66. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Solomkin JS: Antibiotic resistance in
postoperative infections. Crit Care Med. 29 Suppl 4:N97–N99. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cockerill FR and Patel JB: M100-S25
Performance Standards for Antimicrobial Susceptibility Testing;
Twenty-Fifth Informational Supplement. Clin Lab Stand Inst.
35:44–49. 2015.
|
|
24
|
Schentag JJ, Hyatt JM, Carr JR, Paladino
JA, Birmingham MC, Zimmer GS and Cumbo TJ: Genesis of
methicillin-resistant Staphylococcus aureus (MRSA), how treatment
of MRSA infections has selected for vancomycin-resistant
Enterococcus faecium, and the importance of antibiotic management
and infection control. Clin Infect Dis. 26:1204–1214. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Manian FA, Meyer PL, Setzer J and Senkel
D: Surgical site infections associated with methicillin-resistant
Staphylococcus aureus: Do postoperative factors play a role? Clin
Infect Dis. 36:863–868. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Taiwo SS: Methicillin resistance in
Staphylococcus aureus: A review of the molecular epidemiology,
clinical significance and laboratory detection methods. West Afr J
Med. 28:281–290. 2009.PubMed/NCBI
|
|
27
|
Ianoşi S, Ianoşi G, Neagoe D, Ionescu O,
Zlatian O, Docea AO, Badiu C, Sifaki M, Tsoukalas D, Tsatsakis AM,
et al: Age-dependent endocrine disorders involved in the
pathogenesis of refractory acne in women. Mol Med Rep.
14:5501–5506. 2016.PubMed/NCBI
|
|
28
|
Thornsberry C and McDougal LK: Successful
use of broth microdilution in susceptibility tests for
methicillin-resistant (heteroresistant) staphylococci. J Clin
Microbiol. 18:1084–1091. 1983.PubMed/NCBI
|
|
29
|
Hiramatsu K, Kihara H and Yokota T:
Analysis of borderline-resistant strains of methicillin-resistant
Staphylococcus aureus using polymerase chain reaction. Microbiol
Immunol. 36:445–453. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Høiby N, Jarløv JO, Kemp M, Tvede M,
Bangsborg JM, Kjerulf A, Pers C and Hansen H: Excretion of
ciprofloxacin in sweat and multiresistant Staphylococcus
epidermidis. Lancet. 349:167–169. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Peterson LR, Quick JN, Jensen B, Homann S,
Johnson S, Tenquist J, Shanholtzer C, Petzel RA, Sinn L and Gerding
DN: Emergence of ciprofloxacin resistance in nosocomial
methicillin-resistant Staphylococcus aureus isolates. Resistance
during ciprofloxacin plus rifampin therapy for
methicillin-resistant S aureus colonization. Arch Intern Med.
150:2151–2155. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tarale P, Gawande S and Jambhulkar V:
Antibiotic susceptibility profile of bacilli isolated from the skin
of healthy humans. Braz J Microbiol. 46:1111–1118. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Berge AC, Atwill ER and Sischo WM:
Assessing antibiotic resistance in fecal Escherichia coli in young
calves using cluster analysis techniques. Prev Vet Med. 61:91–102.
2003. View Article : Google Scholar : PubMed/NCBI
|