Introduction
Oral squamous cell cancer (OSCC), characterized by
the development of malignant tumors in the mouth, is the eighth
most common type of cancer worldwide (1). In China, the incidence of oral cancer
is ~3–6/100,000 individuals; however, this number is increasing
every year. Smoking, alcohol abuse and betel quid-chewing serve
important roles in the etiology of oral cancer. At present,
although have been certain advances in surgery, radiotherapy and
chemotherapy for oral cancer, the mortality rate remains high
(2). Tumor metastasis is the
primary cause of mortality in patients with oral cancer (3). Therefore, an increased understanding
of the mechanisms underlying oral cancer metastasis is required to
develop novel therapeutic strategies.
microRNAs (miRNAs) suppress gene expression levels
via direct binding to the 3′ untranslated region (UTR) of target
mRNA, thereby regulating the expression levels of various oncogenes
and tumor suppressors involved in metastasis and tumor progression
(4–6). miRNA (miR)-375 may function as a
tumor suppressor, and there is increasing evidence to suggest that
it may be useful as a diagnostic biomarker and a therapeutic target
for cancer treatment. Winther et al (7) demonstrated that miR-375 served as a
prognostic biomarker in patients with primary esophageal squamous
cell carcinomas. In laryngeal squamous cell carcinoma, the
miR-21/miR-375 ratio was revealed as an independent prognostic
marker (8). In colorectal
carcinoma, Mao et al (9)
demonstrated that miR-375 was a potential therapeutic target for
the treatment of colorectal cancer, which reduces the proliferation
of conditionally reprogrammed cells via regulating the expression
levels of the Kruppel-like factor (KLF)4 protein. In tongue
squamous cell carcinoma, overexpression of miR-375 inhibited
specificity protein 1 expression levels, which resulted in
subsequent downregulation of cyclin D1. The same study suggested
that miR-375 may inhibit cell growth in tongue squamous cell
carcinoma (10). Song et al
(11) revealed that miR-375
modulated the radiosensitivity of high-risk human papilloma
virus-positive cervical cancer cells by targeting ubiquitin-protein
ligase E3A via the tumor suppressor p53 signaling pathway. In OSCC,
although miR-375 was demonstrated to be a potentially significant
early-stage biomarker and therapeutic target (12), its exact functions and underlying
mechanisms associated with cancer metastasis and invasion remain
unclear.
The present study investigated the role of miR-375
in the regulation of OSCC cell migration and invasion. These data
demonstrated that miR-375 was downregulated in highly metastatic
OSCC cell lines, and that its overexpression suppressed OSCC cell
migration and invasion. Mechanistic studies revealed that miR-375
may directly bind to the 3′-UTR of platelet-derived growth factor
subunit A (PDGF-A) and downregulate its expression levels, which
may suppress OSCC cell migration and invasion. The present study
provides important insight into miR-375-mediated regulation of OSCC
cell migration and metastasis.
Materials and methods
Cell lines and culture
The CAL-27, Tca8113, UM1 and UM2 OSCC cell lines
(China Center for Type Culture Collection, Wuhan, China) were
cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented
with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100
µg/ml streptomycin (Gibco; Thermo Fisher Scientific, Inc., Waltham,
MA, USA). All cells were incubated at 37°C in 5%
CO2.
Transfection with miR-375 mimic and
PDGF-A
miR-negative control (miR-NC,
5′-UCACAACCUCCUAGAAAGAGUAGA-3′) and miR-375 mimic
(5′-UUUGUUCGUUCGGCUCGCGUGA-3′) were purchased from GenePharma Co.,
Ltd. (Shanghai, China). The full-length PDGF-A (NM_002607.5) was
cloned and inserted into the pcDNA3.1 expression plasmid (Promega
Corporation, Madison, WI, USA). Cells were plated at 50% confluence
and transfected with 300 nM miR-NC, miR-375 mimic and/or 10 µM
PDGF-A or pcDNA3.1 empty vector using the Lipofectamine®
2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. Cells were harvested 24
or 48 h after transfection and subjected to further analysis.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis
Total RNA was extracted from cultured cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. miR-375 expression
levels were analyzed using the TaqMan Advanced miRNA assay (Applied
Biosystems; Thermo Fisher Scientific, Inc.) and U6 served as an
internal control. To evaluate PDGF-A mRNA expression levels,
RT-qPCR was performed using the PrimeScript RT Reagent kit and
SYBR® Premix Ex Taq reagent (Takara Biotechnology Co.,
Ltd., Dalian, China) according to the manufacturer's protocol.
GAPDH served as an internal control. RT-qPCR was performed on an
Applied Biosystems 7500 system (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The PDGF-A and GAPDH primer sequences used for
RT-qPCR are presented in Table I.
Gene expression levels were measured in triplicate, quantified
using the 2-ΔΔCq method, and normalized to the control
(13). The qPCR was conducted
using the following conditions: 95°C for 5 min followed by 40
cycles of 95°C for 15 sec and 60°C for 60 sec.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Primer name | Sequence (5′-3′) |
|---|
| PDGF-A-F |
CCCCTGCCCATTCGGAGGAAGAG |
| PDGF-A-R |
TTGGCCACCTTGACGCTGCGGTG |
| GAPDH-F |
ACACCCACTCCTCCACCTTT |
| GAPDH-R |
TTACTCCTTGGAGGCCATGT |
|
psiCHECK2-XhoI-F |
CCGctcgagcgGAGGAAGAGAAGCATCGAG |
|
psiCHECK2-NotI-R |
ATAAGAATgcggccgcTAAGGCTCTCAGGAAGGTTTCTGTA |
| psiCHECK2-mut-F |
TGTGTCCGAGAACACTCGGGATCGTTCGGAGACAGTGCACATTTGTTTAATGT |
| psiCHECK2-mut-R |
ACATTAAACAAATGTGCACTGTC
TCCGAACGATCCCGAGTGTTCTCGGACACA |
Transwell migration and invasion
assays
Cell migration and invasion were assessed using a
Transwell migration assay. For this, UM1 cells were harvested, and
5×104 cells in 200 µl DMEM containing 0.1% FBS were placed into the
upper chamber of an insert (8-µm pore size; BD Biosciences, San
Jose, CA, USA). The lower chamber was filled with 600 µl DMEM
containing 10% FBS. Migrating cells were imaged using a Leica light
microscope (Leica Microsystems GmbH, Wetzlar, Germany;
magnification, ×200) in five randomly selected fields per well, and
the mean count was calculated. For the invasion assays, 5×104 cells
were seeded into the upper chamber, which had been precoated with
Matrigel (BD Biosciences) and incubated for 24 h. Following the
removal of cells from the upper chamber with a cotton swab, the
cells in the lower chamber were fixed with 4% paraformaldehyde,
stained with 0.1% crystal violet solution in 20% ethanol, and
counted in five randomly-selected fields, using a phase contrast
light microscope at a magnification ×200 (Olympus Corporation,
Tokyo, Japan), following which the mean count was calculated. The
assays were performed in triplicate.
Western blotting
The different experimental groups of UM1 cells were
harvested and lysed using radioimmunoprecipitation assay buffer
(Takara Biotechnology Co., Ltd.) and the total protein
concentration was determined using the Bicinchoninic Acid Protein
Assay kit (Beyotime Institute of Biotechnology, Nantong, China).
Proteins (30 µg) were separated by 8% SDS-PAGE and transferred onto
polyvinylidene difluoride membranes. After blocked with 5% non-fat
milk in Tris-buffered saline containing 0.05% Tween-20 (TBST) for 1
h at 37°C, the membranes were incubated with a polyclonal rabbit
anti-human PDGF-A antibody (1:500; cat. no. ab125268; Abcam,
Cambridge, MA, USA) or a monoclonal rabbit anti-human β-actin
antibody (1:2,000; cat. no. ab115777; Abcam) for 1 h at 37°C.
Membranes were incubated with a horseradish peroxidase-conjugated
goat anti-rabbit IgG H&L secondary antibody (1:10,000; cat. no.
ab97080; Abcam) for 40 min and proteins were visualized using an
Enhanced Chemiluminescence reagent (Pierce; Thermo Fisher
Scientific, Inc.). The expression levels of the proteins of
interest were normalized against the expression levels of β-actin.
Quantification was performed using ImageJ version 6.0 (National
Institutes of Health, Bethesda, MD, USA).
Reporter vector construction and
luciferase reporter assay
miR-375 targets were predicted using the online
miRNA target prediction software TargetScan (www.targetscan.org) and miRanda (www.microrna.org/microrna/home.do). To
determine whether the 3′-UTR of PDGF-A mRNA was a direct target of
miR-375, the full-length wild-type 3′-UTR of PDGF-A and mutant
3′-UTR of PDGF-A were amplified and cloned into the psi-CHECK-2
vector (Promega Corporation, Madison, WI, USA). The primer
sequences used for the construction of the reporter vector are
presented in Table I. UM2 cells
were co-transfected with 100 ng plasmid and 200 nmol/l miR-375
mimic or miR-NC in 24-well plates. Cell lysates were harvested 48 h
after transfection. The firefly and Renilla luciferase fluorescence
intensities were measured using the Dual-Luciferase Reporter assay
system (Promega Corporation) and experiments were performed in
triplicate.
Statistical analysis
All statistical analyses were performed using SPSS
software version 19.0 (IBM SPSS, Armonk, NY, USA). Data are
presented as the mean ± standard deviation. A one-way analysis of
variance was used to compare the differences between miR-375
expression levels in different OSCC lines and followed by a least
significant difference post hoc test. A Student's t-test was used
to compare the differences prior to and following treatment.
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-375 is downregulated in highly
metastatic OSCC cell lines
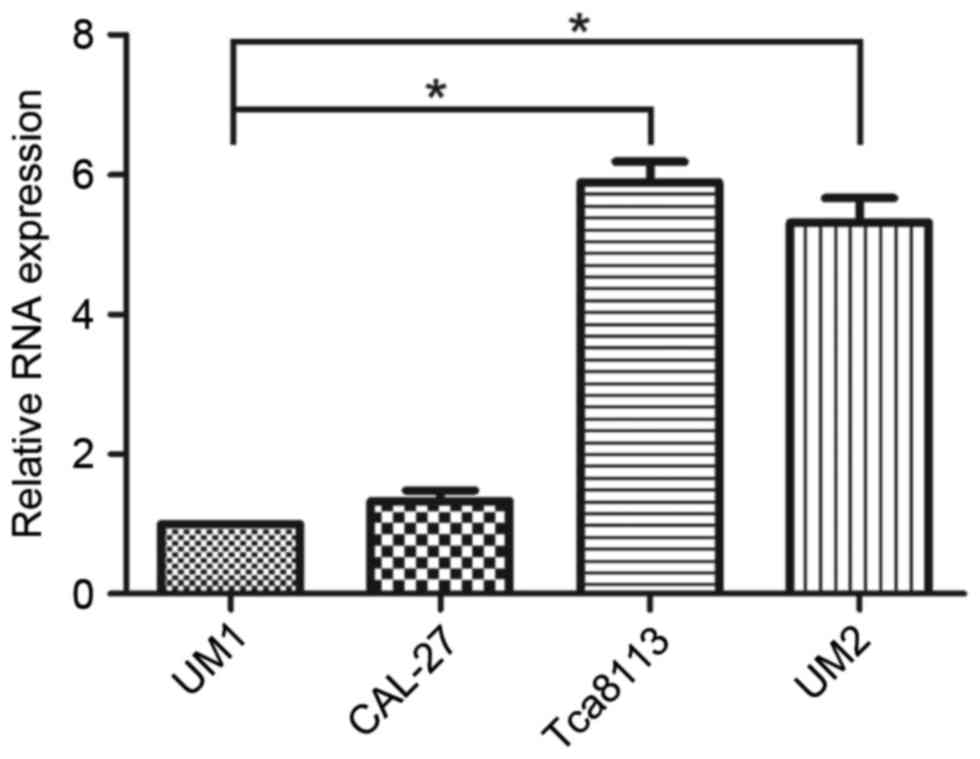
To assess the role of miR-375 in the regulation of
OSCC cell migration and invasion, miR-375 expression levels in UM2
and Tca8113 (less metastatic) and UM1 and CAL-27 (highly
metastatic) OSCC cell lines were determined using RT-qPCR. The
results demonstrated that expression levels of miR-375 were
significantly decreased in UM1 compared with Tca8113 (P<0.001)
and UM2 cell lines (P=0.003); however, no significant differences
were identified in the CAL-27 cell line (P=0.278; Fig. 1). The expression levels of miR-375
were reduced in the highly metastatic OSCC cell lines, particularly
in UM1, compared with the less metastatic ones. Based on these
results, the UM1 cell line was selected for further
experiments.
Overexpression of miR-375 suppresses
migration and invasion of UM1 cells
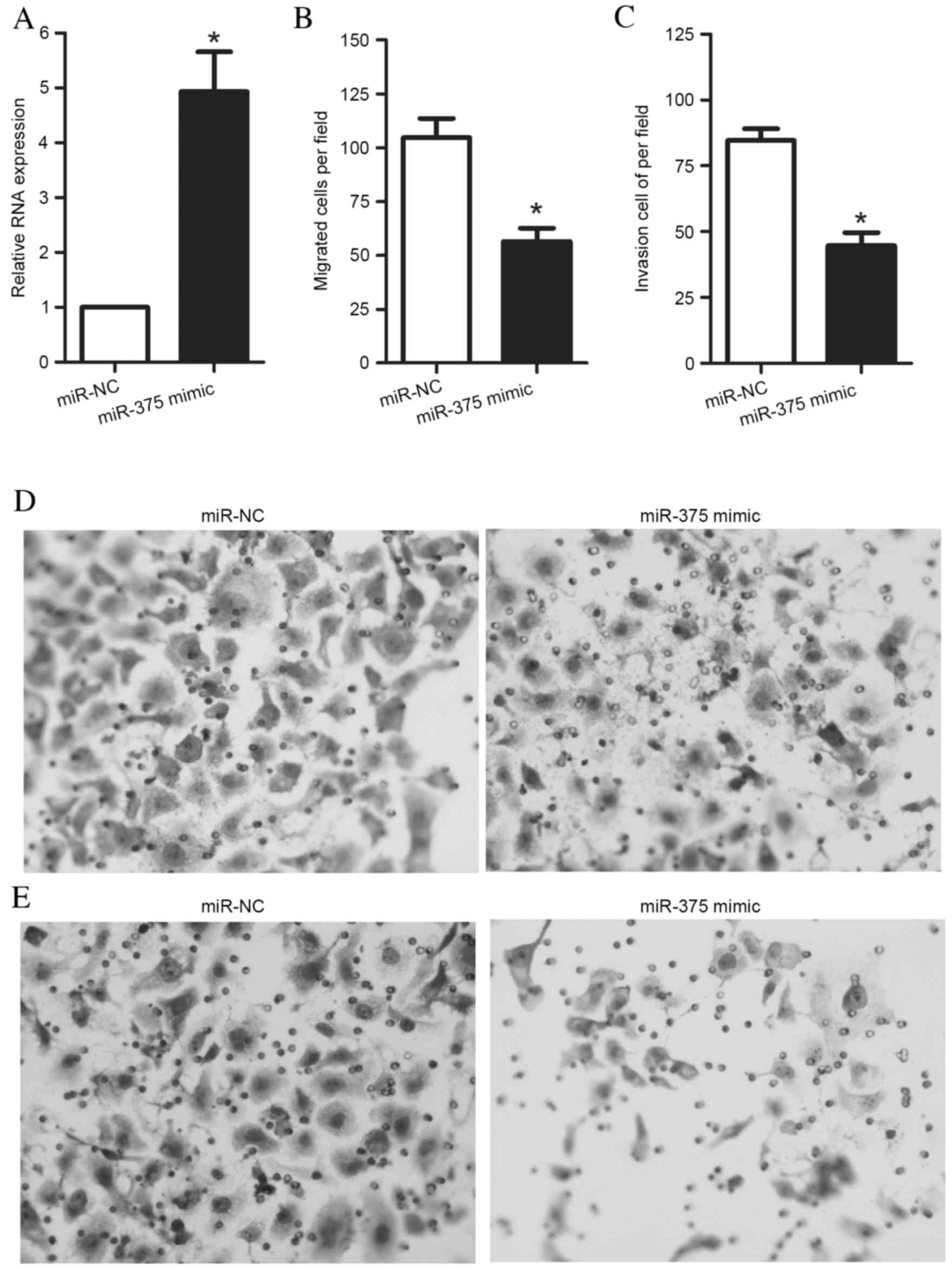
To assess the role of miR-375 in the regulation of
OSCC cell migration and invasion, an miR-375 mimic or an miR-NC was
transfected into UM1 cells. After 48 h of culture, UM1 cells were
harvested for RT-qPCR. The results demonstrated that transfection
of UM1 cells with an miR-375 mimic significantly increased miR-375
expression levels in UM1 cells (P=0.005) (Fig. 2A). Furthermore, following the
transfection of cells with the miR-375 mimic or miR-NC for 48 h,
Transwell assays were performed to measure UM1 cell migration and
invasion. The Transwell migration assay revealed that the ability
of the cells to migrate through the membrane into the lower chamber
was significantly inhibited in the miR-375 mimic-transfected cells
compared with miR-NC-transfected cells (P=0.002; Fig. 2B). The Transwell invasion assay
demonstrated that the ability of the cells to pass through a
Matrigel-coated membrane into the lower chamber was significantly
decreased in the miR-375 mimic-transfected cells compared with the
miR-NC-transfected cells (P<0.001; Fig. 2C). Representative images of
migrated and invaded cells are presented in Fig. 2D and E, respectively.
miR-375 regulates PDGF-A expression
levels via targeting of its 3′-UTR
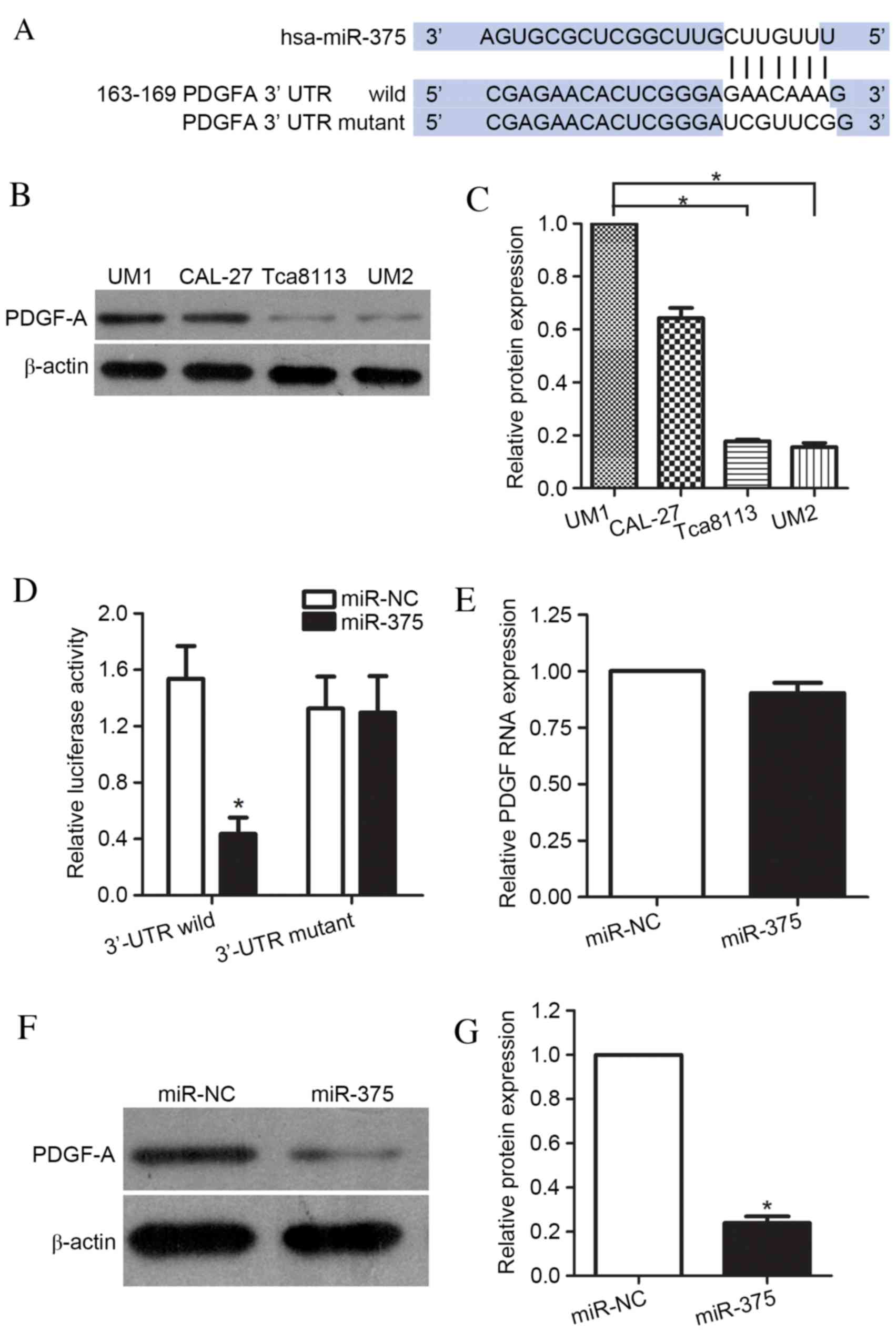
To further elucidate the mechanisms underlying
miR-375-mediated suppression of UM1 cell migration and invasion,
the targets of miR-375 were identified using the prediction
algorithms of TargetScan and miRanda. This analysis identified
PDGF-A as a potential target of miR-375, based on the putative
binding site at positions 163–169 of the PDGF-A 3′-UTR (Fig. 3A). To demonstrate the association
between miR-375 and PDGF-A, the protein expression levels of PDGF-A
in the UM1, CAL-27, UM2 and Tca8113 cell lines were determined. The
results confirmed that the UM1 and CAL-27 (highly metastatic) cell
lines had greater levels of PDGF-A protein compared with the
Tca8113 and UM2 (less metastatic) cell lines (P<0.001, Tca8113
and UM2 compared with UM1; P=0.003, P=0.009, respectively; Tca8113
and UM2 compared with CAL-27; Fig. 3B
and C). Luciferase reporter vectors, co-transfected into UM1
cells with miR-375 mimic or miR-NC, were used to further examine
whether PDGF-A was the target of miR-375. These contained wild-type
PDGF-A 3′-UTR or a mutant 3′UTR mutated at the predicted miR-375
binding site. A significant decrease in the luciferase activity of
the reporter was observed for the wild-type PDGF-A
3′-UTR-containing vector compared with miR-NC (P<0.001), whereas
this decrease did not occur when the target site was the mutated
PDGF-A 3′-UTR (P=0.158; Fig. 3D).
These results indicated that the sequence in the 163–169 bp region
of the PDGF-A 3′-UTR interacted with miR-375, leading to the
inhibition of PDGF-A expression levels in UM1 cells. Additionally,
the effects of miR-375 overexpression on PDGF-A mRNA and protein
levels were examined. As expected, overexpression of miR-375 did
not cause degradation of PDGF-A mRNA (P=0.096) (Fig. 3E); however, it did result in a
significant reduction in the protein expression levels of PDGF-A
(P=0.008; Fig. 3F and G). These
data indicated that miR-375 directly targeted the 3′-UTR of
PDGF-A.
PDGF-A is involved in the
miR-375-induced effects on migration and invasion of UM1 cells
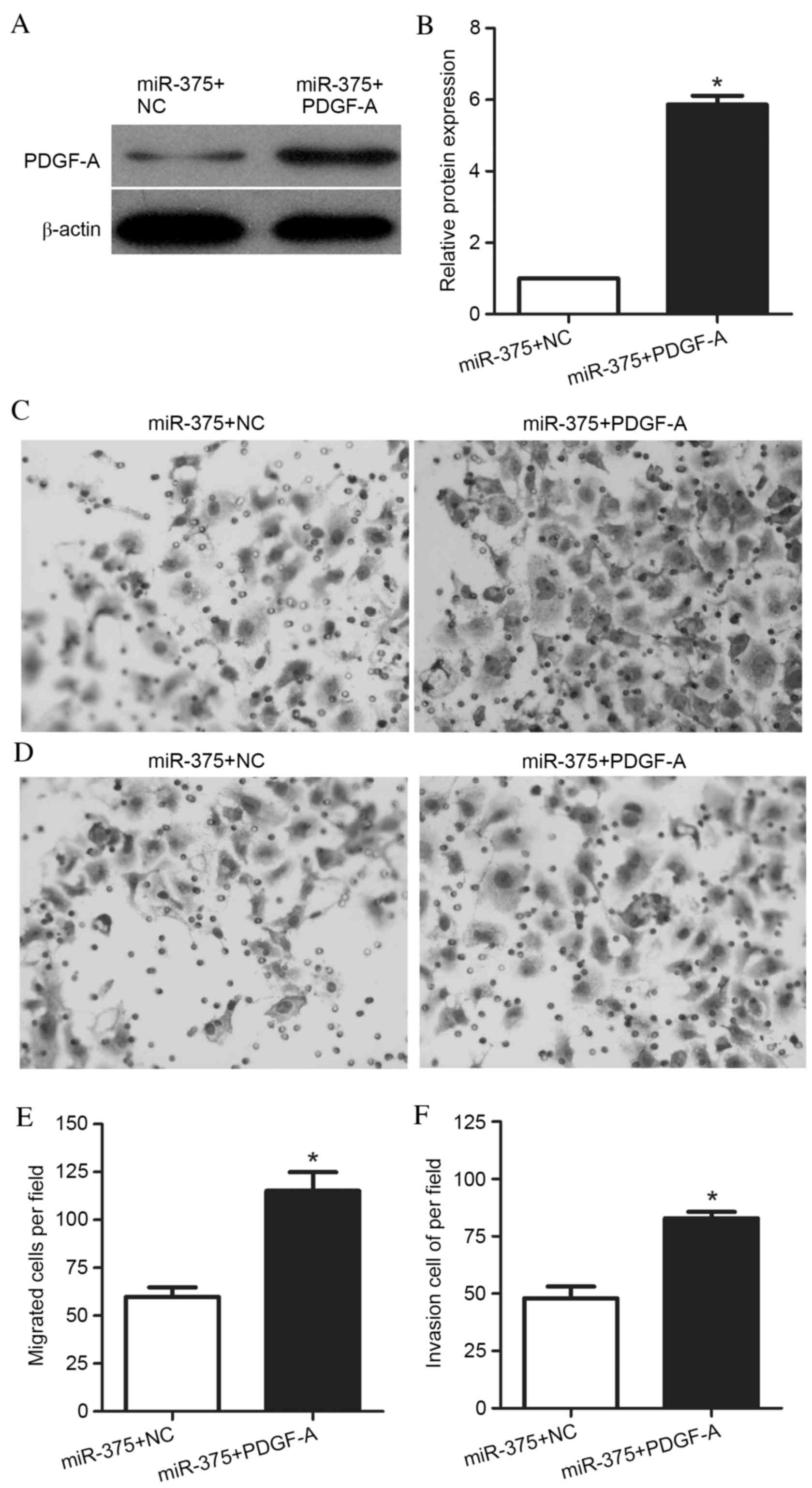
To determine whether miR-375 contributes to UM1
migration and invasion via PDGF-A, UM1 cells were transfected with
a PDGF-A expression vector for 48 h. PDGF-A protein expression
levels were increased in cells co-transfected with the PDGF-A
expression vector and miR-375 mimic, compared with cells
co-transfected with the NC vector and miR-375 mimic (P<0.001;
Fig. 4A and B). Transwell
migration and invasion assays were performed, and representative
images are presented in Fig. 4C and
D, respectively. The Transwell migration assay demonstrated
that the ability of these cells to migrate through the membrane
into the lower chamber was significantly enhanced in miR-375 mimic-
and PDGF-A-transfected cells compared with miR-375 mimic- and
NC-transfected cells (P=0.001; Fig.
4E). Additionally, the Transwell invasion assays revealed that
the ability of the cells to pass through the Matrigel-coated
membrane into the lower chamber was significantly increased in
miR-375 mimic- and PDGF-A-transfected cells compared with miR-375
mimic- and NC-transfected cells (P<0.001; Fig. 4F). These results suggested that
miR-375 affected the migration and invasion of UM1 cells via
regulation of its target gene, PDGF-A, and that overexpression of
PDGF-A was able to reverse the suppressive effect of miR-375 in UM1
cells.
Discussion
miRNAs have been reported to serve essential roles
in the progression of oral cancer (14–16).
Therefore, miRNAs may be promising novel targets for the
development of oral cancer treatments. miR-375, which is frequently
downregulated in oral cancer, is a prognostic biomarker for
early-stage OSCC patients (17).
Shi et al (12)
demonstrated that miR-375 was involved in the premalignant
progression of OSCC, via interactions with the transcription factor
KLF5, which modulates the expression levels of genes contributing
to proliferation and apoptosis. The present study aimed to
investigate the regulatory role of miR-375 in OSCC cell migration
and invasion. The results revealed that miR-375 was downregulated
in highly metastatic, compared with less metastatic, OSCC cell
lines. Additionally, overexpression of miR-375 suppressed the
migration and invasion of UM1 cells. These results indicated that
reduced expression levels of miR-375 may induce more aggressive
tumor behavior during oral cancer progression. These results are
similar to those reported by Siow et al (18), who demonstrated that low miR-375
expression levels were significantly associated with late-stage
disease, larger tumor size and a non-cohesive pattern of invasion
in OSCC.
To investigate the underlying mechanisms by which
miR-375 contributes to UM1 migration and invasion, software tools
were used to predict the target genes of miR-375. PDGF-A was of
particular interest due to its role in the migration and invasion
of cells during cancer development. PDGF was originally isolated
from blood platelets as a growth factor for cells of mesenchymal
origin, but the PDGF gene is additionally expressed by a
variety of cell types throughout development and in adult
vertebrates. PDGF-A serves a key role in the regulation of multiple
biological functions, including vascularization, cell migration and
invasion, and tumor development. Emerging evidence implicates
PDGF-A as a potential prognostic marker, independent of the
traditional pathologic parameters (19,20).
The promoter of this gene is a potential target for an efficient
and selective antineoplastic gene therapy in multiple cancer types,
including breast cancer and osteosarcoma (21,22);
however, the biological functions of PDGF-A in OSCC remain to be
determined. Identifying the association between miR-375 and PDGF-A,
which is essential for tumor progression, may provide novel
therapeutic targets. As demonstrated in the present study, the
protein expression levels of PDGF-A in the highly metastatic cell
lines was significantly greater compared with less metastatic cell
lines. The results revealed that the expression levels of PDGF-A
were negatively associated with the expression levels of miR-375.
Furthermore, these data indicated that miR-375 directly bound to
the 3′-UTR of PDGF-A mRNA and affected PDGF-A expression levels in
UM1 cells. Overexpression of PDGF-A significantly reversed the
suppressive effect of miR-375 on OSCC migration and invasion. The
reversal effect was full, as the migration and invasion ability of
miR-375 mimic- and PDGF-A-transfected cells was similar to that of
NC-transfected only cells. These results indicated that PDGF-A is a
potential tumor promoter that induces increased migration and
invasion in the UM1 OSCC cell line, consistent with the previously
described functions of this protein in other types of cancer
(22).
In conclusion, the present study demonstrated that
the expression levels of miR-375 were significantly reduced in
highly metastatic compared with less metastatic OSCC cell lines,
and that miR-375 inhibited cell migration and invasion by targeting
the tumor promoter PDGF-A in UM1 cells. Therefore, miR-375 may be a
promising therapeutic target for the treatment of oral cancer.
References
|
1
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gupta S, Kong W, Peng Y, Miao Q and
Mackillop WJ: Temporal trends in the incidence and survival of
cancers of the upper aerodigestive tract in Ontario and the United
States. Int J Cancer. 125:2159–2165. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wikner J, Gröbe A, Pantel K and Riethdorf
S: Squamous cell carcinoma of the oral cavity and circulating
tumour cells. World J Clin Oncol. 5:114–124. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP and Chen CZ: Micromanagers of
gene expression: The potentially widespread influence of metazoan
microRNAs. Nat Rev Genet. 5:396–400. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death and tumorigenesis. Br J Cancer.
94:776–780. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Winther M, Alsner J, Tramm T, Baeksgaard
L, Holtved E and Nordsmark M: Evaluation of miR-21 and miR-375 as
prognostic biomarkers in esophageal cancer. Acta Oncol.
54:1582–1591. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hu A, Huang JJ, Xu WH, Jin XJ, Li JP, Tang
YJ, Huang XF, Cui HJ, Sun GB, Li RL, et al: miR-21/miR-375 ratio is
an independent prognostic factor in patients with laryngeal
squamous cell carcinoma. Am J Cancer Res. 5:1775–1785.
2015.PubMed/NCBI
|
|
9
|
Mao Q, Quan T, Luo B, Guo X, Liu L and
Zheng Q: miR-375 targets KLF4 and impacts the proliferation of
colorectal carcinoma. Tumour Biol. 37:463–471. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jia L, Huang Y, Zheng Y, Lyu M, Zhang C,
Meng Z, Gan Y and Yu G: miR-375 inhibits cell growth and correlates
with clinical outcomes in tongue squamous cell carcinoma. Oncol
Rep. 33:2061–2071. 2015.PubMed/NCBI
|
|
11
|
Song L, Liu S, Zeng S, Zhang L and Li X:
miR-375 modulates radiosensitivity of HR-HPV-positive cervical
cancer cells by targeting UBE3A through the p53 pathway. Med Sci
Monit. 21:2210–2217. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shi W, Yang J, Li S, Shan X, Liu X, Hua H,
Zhao C, Feng Z, Cai Z, Zhang L, et al: Potential involvement of
miR-375 in the premalignant progression of oral squamous cell
carcinoma mediated via transcription factor KLF5. Oncotarget.
6:40172–40185. 2015.PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu X and Li Z: MicroRNA expression and its
implications for diagnosis and therapy of tongue squamous cell
carcinoma. J Cell Mol Med. 20:10–60. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xu H, Yang Y, Zhao H, Yang X, Luo Y, Ren
Y, Liu W and Li N: Serum miR-483-5p: A novel diagnostic and
prognostic biomarker for patients with oral squamous cell
carcinoma. Tumour Biol. 37:447–453. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Saad MA, Kuo SZ, Rahimy E, Zou AE,
Korrapati A, Rahimy M, Kim E, Zheng H, Yu MA, Wang-Rodriguez J and
Ongkeko WM: Alcohol-dysregulated miR-30a and miR-934 in head and
neck squamous cell carcinoma. Mol Cancer. 14:1812015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yoon AJ, Wang S, Shen J, Robine N,
Philipone E, Oster MW, Nam A and Santella RM: Prognostic value of
miR-375 and miR-214-3p in early stage oral squamous cell carcinoma.
Am J Transl Res. 6:580–592. 2014.PubMed/NCBI
|
|
18
|
Siow MY, Ng LP, Vincent-Chong VK,
Jamaludin M, Abraham MT, Rahman ZA Abdul, Kallarakkal TG, Yang YH,
Cheong SC and Zain RB: Dysregulation of miR-31 and miR-375
expression is associated with clinical outcomes in oral carcinoma.
Oral Dis. 20:345–351. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Katano M, Nakamura M, Fujimoto K, Miyazaki
K and Morisaki T: Prognostic value of platelet-derived growth
factor-A (PDGF-A) in gastric carcinoma. Ann Surg. 227:365–371.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jones AC, Antillon KS, Jenkins SM, Janos
SN, Overton HN, Shoshan DS, Fischer EG, Trujillo KA and Bisoffi M:
Prostate field cancerization: Deregulated expression of macrophage
inhibitory cytokine 1 (MIC-1) and platelet derived growth factor A
(PDGF-A) in tumor adjacent tissue. PLoS One. 10:e01193142015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mishra A, Ormerod AK, Cibull ML, Spear BT,
Kraner SD and Kaetzel DM: PDGF-A promoter and enhancer elements
provide efficient and selective antineoplastic gene therapy in
multiple cancer types. Cancer Gene Ther. 16:298–309. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yu G, Zhou A, Xue J, Huang C, Zhang X,
Kang SH, Chiu WT, Tan C, Xie K, Wang J and Huang S: FoxM1 promotes
breast tumorigenesis by activating PDGF-A and forming a positive
feedback loop with the PDGF/AKT signaling pathway. Oncotarget.
6:11281–11294. 2015. View Article : Google Scholar : PubMed/NCBI
|