Introduction
Interactions between intestinal epithelial cells and
intraepithelial lymphocytes (IELs) serve an important role in the
growth and maintenance of the intestinal epithelium (1–3).
Keratinocyte growth factor (KGF) is a member of the fibroblast
growth factor (FGF) family. It is thought that KGF, which is
expressed by gamma delta (γδ) intraepithelial lymphocytes (IEL) in
the mucosal layer (1), has an
important involvement in promoting epithelial cell growth in a
paracrine manner after it interacts with the KGF receptor (KGFR)
(4,5). Previously, the authors reported that
exogenous KGF may protect the small intestine from
ischemia-reperfusion injury (I/R) and radiation-induced intestinal
damage by promoting intestinal epithelial cell proliferation
(2,6). In addition, previous studies have
reported that KGFR is abundantly expressed in the gastrointestinal
tract, suggesting that the gut can both synthesize and respond to
KGF (7–9).
Previously, the aryl hydrocarbon receptor (AhR) has
been reported to be a significant factor in KGF-induced
regenerative growth in a zebrafish model (10). Furthermore, AhR expression may be
regulated by FGF in murine 3T3 fibroblasts (11), which indicates that endogenous AhR
might participate in FGF-mediated signaling.
AhR is a DNA binding protein that belongs to the
basic region-helix-loop-helix superfamily (4) and is expressed by most cells.
Unliganded AhR in the cytoplasmic compartment can form a stable
complex with HSP90 (12). However,
when the ligand for AhR activates this transcription factor, the
AhR-ligand complex translocates to the nucleus and subsequently
binds with the AhR nuclear translocator at dioxin-responsive
elements, leading to the transactivation of several genes that
encode phase I and II xenobiotic metabolizing enzymes, such as
cytochrome P450s (5,7,13).
For several decades, the toxic effects mediated by AhR have been
extensively studied (13–15). However, increasing evidence
indicates that AhR may serve an important role in the regulation of
receptor-mediated signaling. For example, Lee et al
(16) reported that Notch1 is a
downstream target of AhR based on microarray analysis of
Rorγt+ cells from wild type and AhR-deficient mice. In
addition, Qiu et al (17)
suggested that the expression of IL-7Rα was reduced by AhR
ablation, and Kiss et al (18) reported that the expression of cKit
was markedly lower in AhR−/− innate lymphoid cells
(ILCs). Based on these findings, it was hypothesized that
endogenous AhR may affect the signals mediated by KGF through the
regulation of KGFR expression.
Materials and methods
Animals
C57BL/6 wild type (12 male; weight, 18–22 g; age,
6–8 weeks) and C57BL/6 AhR+/− mice (2 female and 1 male;
weight, 18–22 g; age, 6–8 weeks) were purchased from the
Experimental Animal Center at Daping Hospital of the Third Military
Medical University (Chongqing, China). Animals were bred and
maintained under specific pathogen-free conditions in a
temperature-controlled room (20±2°C) with circadian light-dark
cycles and free access to standard rodent chow and water. All
animal experiments were performed in accordance with the National
Institutes of Health Guidelines for the Care and Use of Laboratory
Animals (Bethesda, MA, USA). All animal protocols used in the
current study were evaluated and approved by the Ethics Committee
of Xinqiao Hospital, Third Military Medical University (Chongqing,
China). Mice were bred in a scheme using
AhR+/−×AhR+/− breeders to generate
AhR−/− mice. Mice were randomly divided into the
following four groups: Control (n=6), KGF (n=6), KGF +
AhR−/− (n=6) and AhR−/− (n=6) groups.
Recombinant human KGF was intraperitoneally administered at 5
mg/kg/day for 5 days preoperatively.
Materials
KGF was purchased from Sino Biological Inc. (cat.
no. 10210-H07E-50; Beijing, China). Anti-GAPDH antibody (cat. no.
AB-P-R001) was obtained from Goodhere Biotechnology (Hangzhou,
China). Anti-E2F1 (cat. no. 12171-1-AP) and anti-PCNA (cat. no.
10205-2-AP) antibodies were purchased from Wuhan Sanying
Biotechnology (Wuhan, China). The anti-KGFR (cat. no. sc-6930 HRP)
antibody was purchased from Santa Cruz Biotechnology, Inc. (Dallas,
TX, USA) and the Anti-AhR (cat. no. ab166611) antibody was
purchased from Abcam (Cambridge, MA, USA).
LoVo cell culture
Dulbecco's modified Eagle's medium (DMEM)/Ham's F-12
medium (ratio, 1:1; cat. no. SH30023.01B; Thermo Fisher Scientific,
Inc. Waltham, MA, USA) containing 10% FBS (cat. no. SH30023.01B;
Thermo Fisher Scientific, Inc.) was used to culture LoVo cells
purchased from American Type Culture Collection (Manassas, VA,
USA). Additionally, 100 IU/ml penicillin and 100 mg/ml streptomycin
(cat. no. C0222; Beyotime Institute of Biotechnology, Haimen,
China) were added to the medium. Cells were incubated at 37°C in a
5% CO2 atmosphere. The medium was changed every 2 days.
LoVo cells were sub-cultured following partial digestion with 0.25%
trypsin and 0.53 mM EDTA.
Knockdown of AhR and E2F1 mRNA
transcripts by siRNA
Following culturing LoVo cells until reaching 30–40%
confluency in 6-well plates, they were transfected with siRNA
(Guangzhou RiboBio Co., Ltd., Guangzhou, China) at a concentration
of 50 nmol/well using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) in antibiotic-free and
serum-free Opti-MEM medium according to the manufacturer's
instructions. A random control siRNA (si-NC) was used as a negative
control. Following 4 h, the medium was replaced with normal LoVo
cell medium, and the cells were cultured prior to use in
experiments. The siRNA sequences used to target AhR were as
follows: Sense, 5′-GGAACACCUACAUCUAGAAdTdT-3′ and antisense,
3′-dGdTCCUUGUGGAUGUAGAUCUU-5′. The sequences use to target E2F1
were as follows: Sense, 5′-GGAACACCUACAUCUAGAAdTdT-3′ and
antisense, 3′-dGdTCCUUGUGGAUGUAGAUCUU-5′.
Cell proliferation assays
Cell proliferation assays were conducted based on
cell counting. LoVo cells were seeded in 96-well plates in a volume
of 100 µl/well and cultured in a 1:1 mixture of DMEM:Ham's F-12
medium containing 10% FBS for 24 h. The cells were transfected
using AhR-siAhR. Trypsin was used to collect the cells, and a
hemocytometer and trypan blue (cat. no. T8154; Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) were used to count the number of viable
LoVo cells. Numbers of cells per well were also assessed using flow
cytometry analysis.
Flow cytometry analysis
Cells were plated in 6-well culture dishes in growth
medium overnight prior to siRNA transfection. Following serum
starvation for 24 h, cells were stimulated with KGF (100 ng/ml) for
24 h. Cells were then dissociated with 800 µl trypsin per well at
37°C for 3–5 min, and following adding 500 µl DMEM/Ham's F-12
medium containing 10% FBS per well, cells were centrifuged at 1,000
× g at 4°C for 5 min. The supernatant was discarded and the
cell pellet was washed with ice-cold PBS thrice with centrifugation
at 1,000 × g at 4°C for 5 min after each wash. Cells were
then fixed in 80% ethanol at 4°C overnight. Following 2 more washes
with PBS, cells were treated with 0.1% Triton X-100, 5 mg/ml
propidium iodide and 50 mg/ml ribonuclease A in PBS at 37°C for 15
min in the dark. Finally, 2×105 cells were analyzed by
flow cytometry (MoFlo Astrios EQ; Beckman Coulter, Inc. Brea, CA,
USA). The results were analyzed using FlowJo (version 9.0, FlowJo
LLC, Asland, OR, USA).
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated using TRIzol reagent (Takara
Biotechnology Co., Ltd., Dalian, China) and was then reverse
transcribed into cDNA using a PrimeScript™ RT reagent kit with gDNA
Eraser (cat. no. RR047A; Takara Biotechnology Co., Ltd.). The
primers used in the present study were as follows: KGFR
(Invitrogen; Thermo Fisher Scientific, Inc.) forward,
5′-CGCGGATCCGCCGCCGGTGTTAACACCACGTACGGTCATCATCTGACAC-3′ and
reverse,5′-CGGAATTCACCATGCAGAGTGAAAGGATCGCCATCCTGGGAAGACTCC-3′;
β-actin (Invitrogen; Thermo Fisher Scientific, Inc.) forward,
5′-CCACGAAACTACCTTCAACTCC-3′ and reverse,
5′-GTGATCTCCTTCTGCATCCTG-3′). qPCR was performed using the Fast
SYBR® Green Master Mix (cat. no. 4385612; Thermo Fisher
Scientific, Inc.), in a Rotor-Gene Q system (Qiagen GmbH, Hilden,
Germany) and with the following conditions: 94°C for 10 min, then
45 cycles of 30 sec at 94°C, 30 sec at 60°C, and 45 sec at 72°C.
Relative fold changes in mRNA expression were calculated using the
formula 2−ΔΔCq (19).
Western blotting
Total cell lysates were extracted using
radioimmunoprecipitation lysis buffer (10 mM Tris pH 7.4, 20 mM
NaCl, 5 mM MgCl2, 0.5% Nonidet P-40 and 0.1 mM PMSF),
and then protein lysates were centrifuged at 12,000 × g for
5 min at 4°C. The protein concentrations were measured using a
bicinchoninic acid assay kit (Beyotime Institute of Biotechnology,
Haimen, China). Protein samples (25 µg) were separated on 10%
SDS-PAGE gels, and the gels were electropheretically transferred to
polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA,
USA). Membranes were blocked in 5% skim milk at 4°C for 1 h and
incubated with anti-AhR (1:500 dilution), anti-KGFR (1:200
dilution), anti-E2F1 (1:1,000 dilution) or anti-GAPDH (1:1,000
dilution) antibodies for 24 h at 4°C, followed by incubation for 1
h with the peroxidase-conjugated secondary antibody (1:5,000
dilution; cat no. BA1055/BA1051; Beyotime Institute of
Biotechnology). Protein bands were detected using a Kodak Gel Logic
4000R Imaging System (Kodak, Rochester, NY, USA) with an enhanced
chemiluminescence reagent (Boster Systems Inc., Pleasanton, CA,
USA). The results were quantified using Carestream Molecular
Imaging Software version 5.2.2 (Kodak).
Immunohistochemistry
Tissues were fixed in 4% paraformaldehyde at 4°C for
48 h, and embedded in paraffin. Tissue sections (5 µm thick) were
immersed in 3% hydrogen peroxide for 10 min to eliminate endogenous
peroxidase activity and then were microwaved in 0.01 mol/l citrate
buffer (pH 6.0) thrice for 5 min. PBS was used to wash the sections
for each step. Samples were incubated at 4°C overnight with
anti-PCNA antibody (1:100 dilution; cat. no. 10205-2-AP; Sanying
Biotechnology, Wuhan, China), followed by a standard staining
procedure using an LSAB kit (cat. no. SP-9000, Zhongshan
Biotechnology Co, Ltd., Beijing, China). The crypt cell
proliferation rate was calculated as the ratio of crypt cells that
were positive for PCNA to the total number of crypt cells. The
total number of proliferating cells/crypt was defined as the mean
number of proliferating cells in ten crypts.
Nuclear protein extraction
Nuclear protein extraction was performed using the
NE-PER Nuclear and Cytoplasmic Extraction Reagents kit (cat. no.
78833; Thermo Fisher Scientific, Inc.). Cells were harvested with
trypsin-EDTA then centrifuged at 500 × g at 4°C for 5 min.
For suspension, cells were harvested by centrifugation at 500 ×
g at 4°C for 5 min. Cells were washed by re-suspending the
cell pellet in PBS. A total of 5×106 cells were
transferred to a 1.5 ml microcentrifuge tube and pelleted by
centrifugation at 500 × g at 4°C for 3 min. The supernatants
were discarded, leaving the cell pellet as dry as possible.
Ice-cold CERI reagent (40 µl) was added to the cell pellet, and
then the tube was vortexed vigorously at the highest setting for 15
sec and incubated on ice for 10 min. Ice-cold CERII reagent (22 µl)
was added to the tube and then vortexed for 5 sec. Following
incubating the tube on ice for 1 min, the tube was vortexed for 5
sec on the highest setting. The tube was centrifuged at maximum
speed (16,000 × g) at 4°C for 5 min and then was immediately
transferred to a clean pre-chilled tube. NER reagent (200 µl) was
then added, and the tube was vortexed for 15 sec. The sample was
placed on ice and then continually vortexed for 15 sec every 10 min
for a total of 40 min. The tube was centrifuged at maximum speed
(16,000 × g) at 4°C for 10 min, leaving the supernatant
(nuclear extract). Nuclear proteins were stored at −80°C until
use.
Statistical analysis
Data are presented as the mean ± standard deviation.
All data were analyzed using the SPSS statistical software package
(version, 19.0; IBM SPSS, Armonk, NY, USA). The statistical
significance of differences in mean values was determined using the
Student's t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
AhR knockout mice are not sensitized
to KGF-induced intestinal epithelial cell proliferation
KGF, a classical growth factor, serves an important
role in intestinal epithelial cell proliferation (20,21).
Immunohistochemical analysis was used to detect intestinal
epithelial cell proliferation, and PCNA-positive cells were
distributed in the crypt of Lieberkuhn of the small intestine. As
presented in Fig. 1A and C, it was
identified that KGF significantly increased the number of
PCNA-positive cells in the KGF group (42.8%) when compared with the
control group (23.28%). However, the delivery of siRNA to target
AhR mRNA transcripts resulted in a reduced rate of KGF-induced
epithelial cell proliferation (24.06%). These data indicated that
the absence of AhR resulted in reduced KGF-induced intestinal
epithelial cell proliferation. Subsequently, changes in villus
height and crypt depth were assessed. AhR knockdown led to a
significant reduction in jejunal villus height (274±70.9 µm)
compared with the KGF group (411±79.3 µm; P<0.05). In addition,
crypt depth was significantly reduced in the KGF +
AhR−/− group (64.8±13.4 µm) when compared with the KGF
group (96±1.2 µm; P<0.05; Fig. 1B
and D). Furthermore, intestinal wet weight and assessments of
RNA and protein expression levels indicated that AhR deficiency
abolished the sensitivity of the intestine to KGF (Table I). These findings suggested that
AhR knockdown in intestinal epithelial cells results in reduced
sensitivity to KGF.
 | Table I.Intestinal wet weight and levels of
jejunal protein and RNA expression. |
Table I.
Intestinal wet weight and levels of
jejunal protein and RNA expression.
| Variable | Control group | KGF group | KGF + AhRKO
group | AhRKO group |
|---|
| Protein
(mg/cm) | 2.65±0.13 |
3.02±0.56a,b | 2.28±0.32 | 1.86±0.27 |
| RNA (mg/cm) | 29.9±6.1 |
38.8±0.83a,b | 25.2±1.8 | 20.9±5.1 |
| Intestinal wet
weight (mg/10 cm) | 488.7±6.9 |
553.4±8.6a,b | 449.5±10.8 | 401.8±9.5 |
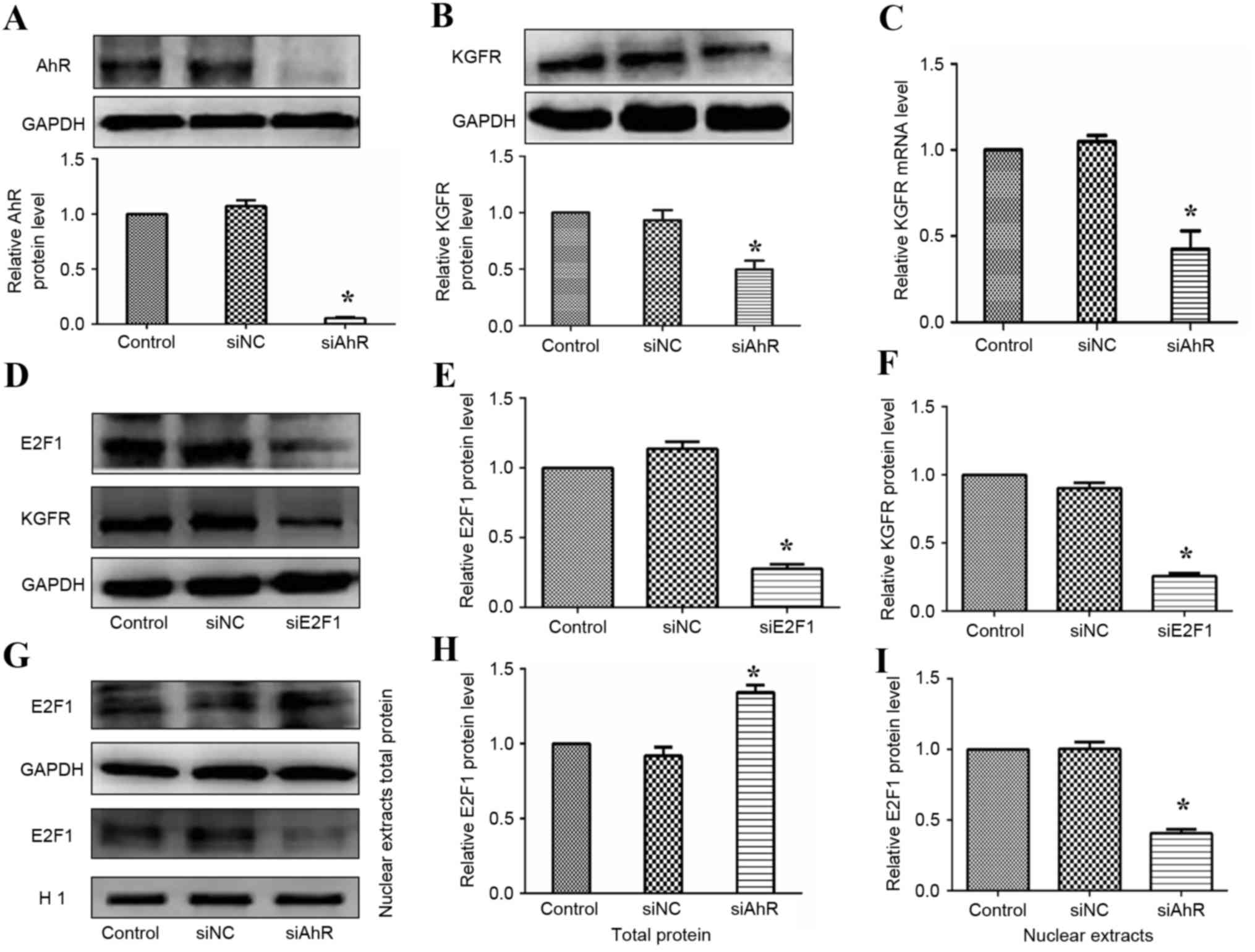
Downregulation of KGFR after AhR
knockout in vivo
KGF can modulate the proliferation of intestinal
epithelial cells that specifically express KGFR, and this effect
can be reversed by administering a blocking anti-KGFR antibody
(22). As demonstrated in Fig. 2A and B, the effect of AhR knockout
was assessed by western blotting, and it was identified that AhR
expression was significantly knocked down when compared with the
control. To investigate the mechanism whereby AhR knockout mice
were made less sensitive to KGF in the small intestine, western
blotting was performed to measure the expression of KGFR in
AhR−/− mice. In physiological conditions, AhR knockout
significantly reduced the expression of KGFR (0.65-fold, six
mice/group; P<0.05; Fig. 2C).
Additionally, reduced levels of KGFR mRNA transcripts were measured
by RT-qPCR in AhR−/− epithelial cells (0.54-fold, six
mice per group, P<0.05; Fig.
2D). These findings indicated that AhR deletion induced the
downregulation of KGFR expression, resulting in reduced intestinal
cell sensitivity to growth stimulation induced by KGF.
Regulation of KGFR expression by the
AhR-E2F1 pathway in vitro
To further investigate the effects of AhR on KGF
signaling, LoVo cells were transfected with siRNA specific for AhR
(siAhR). As presented in Fig. 3A,
the effect of siAhR was assessed by western blotting, and it was
demonstrated that AhR expression was significantly knocked down
when compared with the control. Furthermore, following transfecting
LoVo cells with siAhR for 24 h, western blotting results indicated
reduced levels (0.57) of KGFR protein expression in AhR-silenced
cells compared against the control group (Fig. 3B). In accordance with the western
blotting results, downregulated levels (0.46) of KGFR mRNA
transcripts were detected by RT-qPCR in AhR-silenced epithelial
cells (Fig. 3C). Therefore, these
findings indicated that KGFR expression may be regulated by AhR.
Following this, the authors explored the mechanism whereby AhR
regulates KGFR expression. As previous studies have reported that
FGFR expression can be regulated by E2F family members (8,9,23),
the present study focused on the nuclear transcription factor E2F.
As detailed in Fig. 3D-F, reduced
protein levels of KGFR could be observed in LoVo cells in which
E2F1 was knocked-out compared with the control group, suggesting
that this transcription factor (E2F1) may affect KGFR expression.
Following demonstrating that the transcription factor E2F1 may
meditate KGFR expression, whether AhR could influence the function
of E2F1 to regulate the expression of KGFR was assessed. Cells were
transfected with siRNA to target AhR, and there was a significant
reduction of E2F1 in the nucleus detected (Fig. 3G-I). In conclusion, these findings
demonstrated that the AhR-E2F1 pathway regulates KGFR expression in
intestinal epithelial cells.
AhR knockdown LoVo cells are less
sensitized to KGF stimulation in vitro
To test whether knockdown of AhR expression altered
KGF signaling, serum-starved LoVo cells with or without AhR
knockdown were stimulated with KGF (100 ng/ml); cell counting and
flow cytometry analysis were conducted to assess cell viability.
Following adding KGF, the proliferation rate of LoVo cells
increased significantly compared with untreated control cells, and
this effect was abolished by siAhR treatment. The present finding
of siAhR-meditated inhibition of cell number in LoVo cells was
confirmed by trypan blue/hemocytometer and flow cytometric
analyses. In the KGF group, cell numbers were significantly
increased compared with the other groups (Fig. 4A). KGF caused a significant
reduction in the relative number of G0/G1 cells and a significant
associated increase in S phase cells. At 24 h, 66.64% cells were in
G0/G1 phase for untreated cells compared with 39.18% for
KGF-treated cells. Knockdown of AhR expression alone had a slight
influence on the number of cells in G0/G1 phase. However, for cells
treated with siAhR + KGF compared to KGF alone, the % of cells in
G0/G1 phase was 59.69% compared with 39.18% (Fig. 4B and C), respectively. These data
indicated that AhR knockdown may attenuate KGF-induced cell
proliferation.
Discussion
In the current study, it was identified that
AhR−/− mice were not responsive to KGF-induced
intestinal proliferation effects or intestinal morphological
changes under physiological conditions. Moreover, KGFR expression
levels were significantly lower in AhR knockout intestinal
epithelial cells. Silencing of E2F1 may significantly reduce the
expression of KGFR, while silencing of AhR could markedly lower the
expression of E2F1. Together, these findings demonstrated that the
AhR-E2F1-KGFR signaling pathway is involved in KGF signaling in
intestinal epithelial cells.
KGF is a classical cell growth factor that has been
widely studied. For example, KGF improves epithelial function after
massive small bowel resection (11) and KGF gene therapy was demonstrated
to ameliorate ulcerative colitis in rats (16). Furthermore, in the present study,
knockdown of AhR expression may abolish KGF-induced intestinal
proliferation in AhR knockout mice. Villus height, crypt depth,
intestinal wet weight, RNA levels and protein expression analysis
also indicated that AhR knockout mice presented compromised
KGF-induced intestinal changes. Previous studies have reported that
AhR, as a conserved nuclear transcription protein, serves an
important role in cell proliferation (24–27).
Interestingly, other work in the field found that AhR regulates the
expression of receptors that meditated cell survival and growth.
For example, Lee et al (16) reported that AHR supported the
presence of NKp46+ ILCs and, partially, lymphoid tissue
inducer-like ILCs in the intestinal lamina propria through the
induction of Notch receptors. Additionally, AhR-dependent c-Kit
expression is critical for the homeostasis of invariant γδ T cells
in the murine epidermis (28).
In the present study, KGFR expression was
downregulated in AhR knockout mice and AhR knockdown LoVo cells.
These findings suggested that AhR ablation induced the
downregulation of KGFR expression, thereby reducing intestinal cell
sensitively to KGF-stimulated proliferation.
Few studies have characterized the molecular
mechanisms that regulate human KGFR expression in intestinal
epithelial cells. Koga et al (29) reported that the expression level of
KGFR significantly increased when angiotensin II (ATII) was blocked
by Losartan in a small bowel resection mouse model. Recently, it
has been reported that the transcription factor E2F1 can bind to
the KGFR promoter and activate KGFR expression in MCF-7 and HEK293
cells (30). Furthermore,
expression of the human FGFR1 and murine FGFR2 genes can be
directly regulated by E2F1 (8,23).
The present finding that KGFR expression was reduced in colon cells
with knockdown of E2F1 is consistent with these previous studies
(8,23). As a nuclear transcription factor,
once E2F1 traffics to the nucleus, it exerts its transcription
activity on the FGFR2 gene by direct binding to the KGFR promoter
(30). In the current study,
significantly reduced levels of nuclear E2F1 in the AhR knockout
group were detected when compared with the control group. This
finding suggested that KGFR expression may be regulated via the
AhR-E2F1 pathway. However, the mechanism whereby AhR regulates
trafficking of the E2F1 transcription factor from the cytoplasm to
the nucleus remains unclear. Moreover, cell counting revealed that
KGF-induced cell proliferation was reduced and flow cytometric
analyses revealed that the cell cycle profile was notably altered
when AhR was knocked-down, as there was a significant increase in
the percentage of cells in G0/G1 phase compared with the KGF-alone
treated group.
In summary, the AhR-E2F1-KGFR signaling pathway was
demonstrated to be involved in KGF-induced intestinal epithelial
cell proliferation. Further studies are required to understand the
molecular mechanisms of KGF-induced intestinal epithelial cell
growth and proliferation in greater detail.
Acknowledgements
The present study was supported by grants from the
National Natural Science Foundation of China (grant nos. NSFC
81330013 to Hua Yang, and NSFC 81300275 to Lihua Sun) and the
Program of Changjiang Scholars and Innovative Research (grant no.
IRT 13050 to H.Y.).
References
|
1
|
Boismenu R and Havran WL: Modulation of
epithelial cell growth by intraepithelial gamma delta T cells.
Science. 266:1253–1255. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cai Y, Wang W, Liang H, Sun L, Teitelbaum
DH and Yang H: Keratinocyte growth factor pretreatment prevents
radiation-induced intestinal damage in a mouse model. Scand J
Gastroenterol. 48:419–426. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen Y, Chou K, Fuchs E, Havran WL and
Boismenu R: Protection of the intestinal mucosa by intraepithelial
gamma delta T cells. Proc Natl Acad Sci USA. 99:14338–14343. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Burbach KM, Poland A and Bradfield CA:
Cloning of the Ah-receptor cDNA reveals a distinctive
ligand-activated transcription factor. Proc Natl Acad Sci USA.
89:8185–8189. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wormke M, Stoner M, Saville B, Walker K,
Abdelrahim M, Burghardt R and Safe S: The aryl hydrocarbon receptor
mediates degradation of estrogen receptor through activation of
proteasomes. Mol Cell Biol. 23:1843–1855. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cai Y, Wang W, Liang H, Sun L, Teitelbaum
DH and Yang H: Keratinocyte growth factor improves epithelial
structure and function in a mouse model of intestinal
ischemia/reperfusion. PLoS One. 7:e447722012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hankinson O: The aryl hydrocarbon receptor
complex. Annu Rev Pharmacol Toxicol. 35:307–340. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tashiro E, Minato Y, Maruki H, Asagiri M
and Imoto M: Regulation of FGF receptor-2 expression by
transcription factor E2F-1. Oncogene. 22:5630–5635. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tashiro E, Maruki H, Minato Y, Doki Y,
Weinstein IB and Imoto M: Overexpression of cyclin D1 contributes
to malignancy by up-regulation of fibroblast growth factor receptor
1 via the pRB/E2F pathway. Cancer Res. 63:424–431. 2003.PubMed/NCBI
|
|
10
|
Yang H, Wildhaber BE and Teitelbaum DH:
2003 Harry M. Vars Research Award. Keratinocyte growth factor
improves epithelial function after massive small bowel resection.
JPEN J Parenter Enteral Nutr. 27:198–207. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu CJ, Jin JD, Lv TD, Wu ZZ and Ha XQ:
Keratinocyte growth factor gene therapy ameliorates ulcerative
colitis in rats. World J Gastroenterol. 17:2632–2640. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tsuji N, Fukuda K, Nagata Y, Okada H, Haga
A, Hatakeyama S, Yoshida S, Okamoto T, Hosaka M, Sekine K, et al:
The activation mechanism of the aryl hydrocarbon receptor (AhR) by
molecular chaperone HSP90. FEBS Open Bio. 4:796–803. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hao N and Whitelaw ML: The emerging roles
of AhR in physiology and immunity. Biochem Pharmacol. 86:561–570.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Furumatsu K, Nishiumi S, Kawano Y, Ooi M,
Yoshie T, Shiomi Y, Kutsumi H, Ashida H, Fujii-Kuriyama Y, Azuma T
and Yoshida M: A role of the aryl hydrocarbon receptor in
attenuation of colitis. Dig Dis Sci. 56:2532–2544. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Monteleone I, MacDonald TT, Pallone F and
Monteleone G: The aryl hydrocarbon receptor in inflammatory bowel
disease: Linking the environment to disease pathogenesis. Curr Opin
Gastroenterol. 28:310–313. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee JS, Cella M, McDonald KG, Garlanda C,
Kennedy GD, Nukaya M, Mantovani A, Kopan R, Bradfield CA, Newberry
RD and Colonna M: AHR drives the development of gut ILC22 cells and
postnatal lymphoid tissues via pathways dependent on and
independent of Notch. Nat Immunol. 13:144–151. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Qiu J, Heller JJ, Guo X, Chen ZE, Fish K,
Fu YX and Zhou L: The aryl hydrocarbon receptor regulates gut
immunity through modulation of innate lymphoid cells. Immunity.
36:92–104. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kiss EA, Vonarbourg C, Kopfmann S, Hobeika
E, Finke D, Esser C and Diefenbach A: Natural aryl hydrocarbon
receptor ligands control organogenesis of intestinal lymphoid
follicles. Science. 334:1561–1565. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jonas CR, Gu LH, Nkabyo YS, Mannery YO,
Avissar NE, Sax HC, Jones DP and Ziegler TR: Glutamine and KGF each
regulate extracellular thiol/disulfide redox and enhance
proliferation in Caco-2 cells. Am J Physiol Regul Integr Comp
Physiol. 285:R1421–R1429. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hille A, Grüger S, Christiansen H, Wolff
HA, Volkmer B, Lehmann J, Dörr W and Rave-Fränk M: Effect of
tumour-cell-derived or recombinant keratinocyte growth factor (KGF)
on proliferation and radioresponse of human epithelial tumour cells
(HNSCC) and normal keratinocytes in vitro. Radiat Environ Biophys.
49:261–270. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mehta M, Kesinger JW, Zang XP, Lerner ML,
Brackett DJ, Brueggemeier RW, Li PK and Pento JT: Influence of
novel KGFR tyrosine kinase inhibitor on KGF-mediated proliferation
of breast cancer. Anticancer Res. 30:4883–4889. 2010.PubMed/NCBI
|
|
23
|
Kanai M, Tashiro E, Maruki H, Minato Y and
Imoto M: Transcriptional regulation of human fibroblast growth
factor receptor 1 by E2F-1. Gene. 438:49–56. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Furukawa K, Matsumoto K, Nagayasu T,
Yamamoto-Fukuda T, Tobinaga S, Abo T, Yamasaki N, Tsuchiya T,
Miyazaki T, Kamohara R, et al: Intratracheal administration of
recombinant human keratinocyte growth factor promotes alveolar
epithelial cell proliferation during compensatory lung growth in
rat. Acta Histochem Cytochem. 46:179–185. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hirobe T, Hasegawa K, Furuya R, Fujiwara R
and Sato K: Effects of fibroblast-derived factors on the
proliferation and differentiation of human melanocytes in culture.
J Dermatol Sci. 71:45–57. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li GJ, Jiang DY, Zong XL and Xu X:
Keratinocyte growth factor phage model peptides can promote human
oral mucosal epithelial cell proliferation. Oral Surg Oral Med Oral
Pathol Oral Radiol. 116:e92–e97. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Playford RJ, Marchbank T, Mandir N, Higham
A, Meeran K, Ghatei MA, Bloom SR and Goodlad RA: Effects of
keratinocyte growth factor (KGF) on gut growth and repair. J
Pathol. 184:316–322. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kadow S, Jux B, Zahner SP, Wingerath B,
Chmill S, Clausen BE, Hengstler J and Esser C: Aryl hydrocarbon
receptor is critical for homeostasis of invariant gammadelta T
cells in the murine epidermis. J Immunol. 187:3104–3110. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Koga H, Yang H, Haxhija EQ and Teitelbaum
DH: The role of angiotensin II type 1a receptor on intestinal
epithelial cells following small bowel resection in a mouse model.
Pediatr Surg Int. 24:1279–1286. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
D'Amici S, Ceccarelli S, Vescarelli E,
Romano F, Frati L, Marchese C and Angeloni A: TNFα modulates
fibroblast growth factor receptor 2 gene expression through the
pRB/E2F1 pathway: Identification of a non-canonical E2F binding
motif. PLoS One. 8:e614912013. View Article : Google Scholar : PubMed/NCBI
|