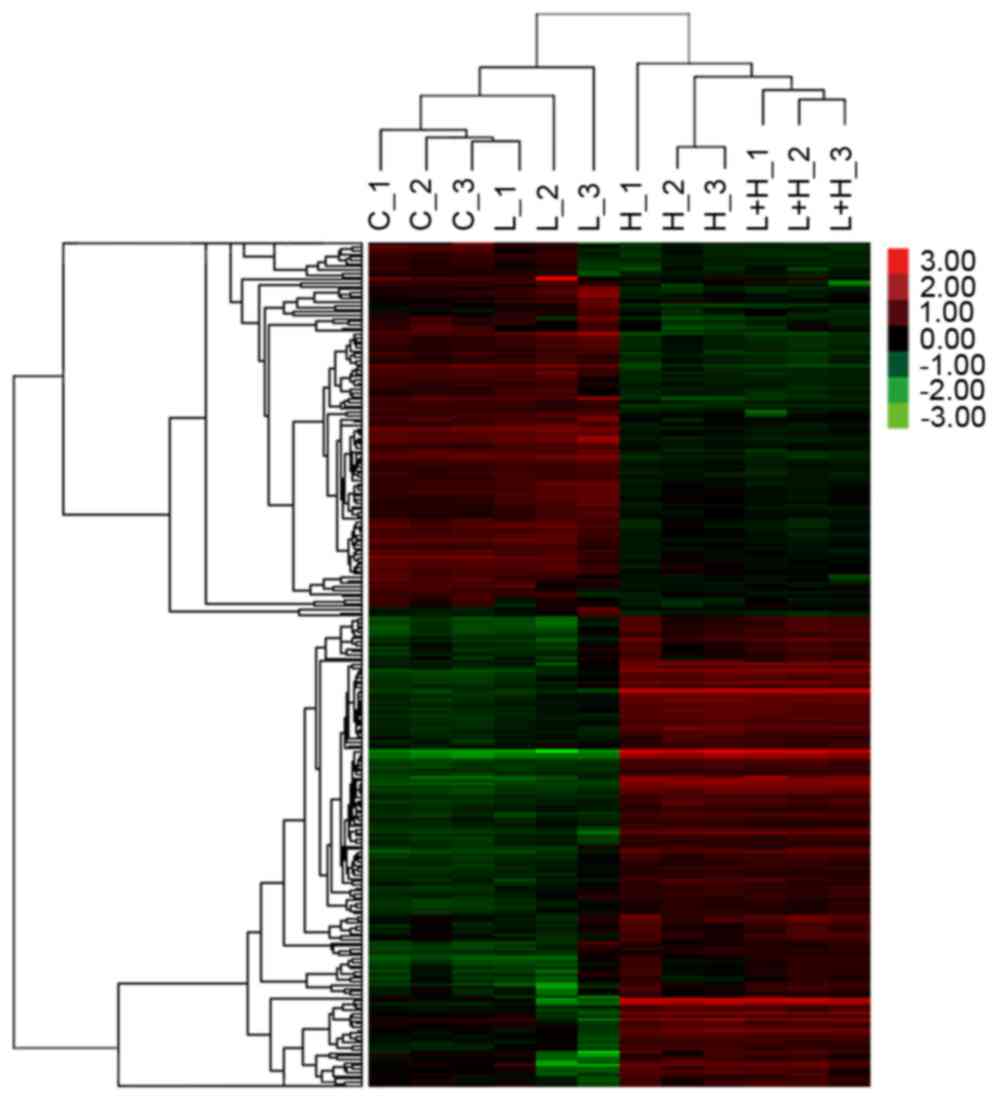
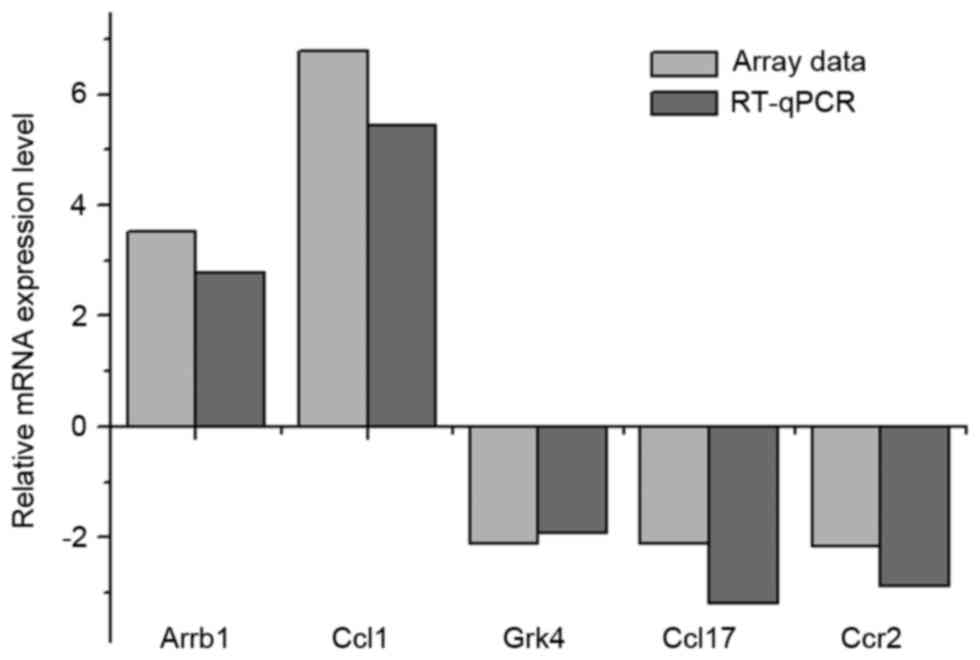
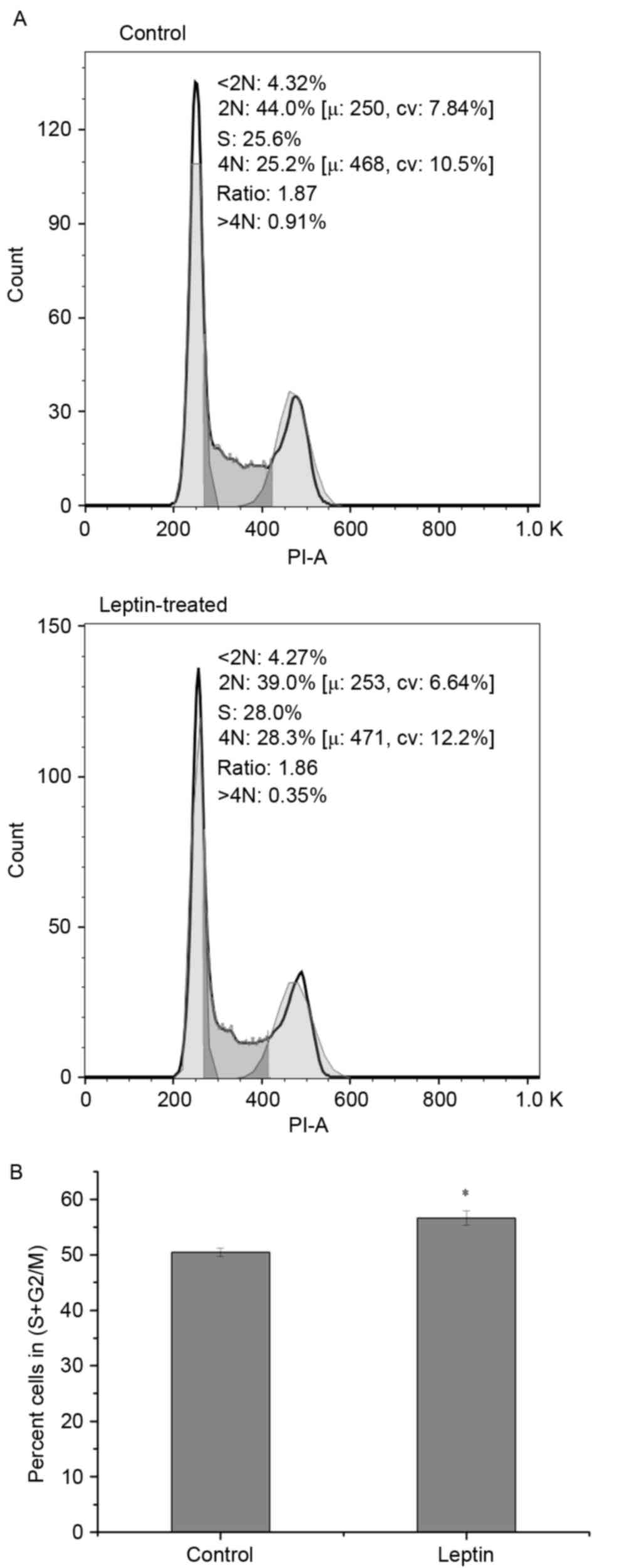
|
1
|
Sziksz E, Pap D, Lippai R, Béres NJ,
Fekete A, Szabó AJ and Vannay Á: Fibrosis related inflammatory
mediators: Role of the IL-10 cytokine family. Mediators Inflamm.
2015:7646412015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tacke F and Trautwein C: Mechanisms of
liver fibrosis resolution. J Hepatol. 63:1038–1039. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wynn TA: Cellular and molecular mechanisms
of fibrosis. J Pathol. 214:199–210. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Peters S: Cystic fibrosis: A review of
pathophysiology and current treatment recommendations. S D Med.
67:148–151, 153. 2014.PubMed/NCBI
|
|
5
|
Tao H, Shi KH, Yang JJ, Huang C, Liu LP
and Li J: Epigenetic regulation of cardiac fibrosis. Cell Signal.
25:1932–1938. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Uhal BD: Epithelial apoptosis in the
initiation of lung fibrosis. Eur Respir J Suppl. 44:S7–S9. 2003.
View Article : Google Scholar
|
|
7
|
Friedman SL: Hepatic fibrosis-overview.
Toxicology. 254:120–129. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhan L, Huang C, Meng XM, Song Y, Wu XQ,
Yang Y and Li J: Hypoxia-inducible factor-1alpha in hepatic
fibrosis: A promising therapeutic target. Biochimie. 108:1–7. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lokmic Z, Musyoka J, Hewitson TD and Darby
IA: Hypoxia and hypoxia signaling in tissue repair and fibrosis.
Int Rev Cell Mol Biol. 296:139–185. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Martínez-Martínez E, Jurado-López R,
Valero-Muñoz M, Bartolomé MV, Ballesteros S, Luaces M, Briones AM,
López-Andrés N, Miana M and Cachofeiro V: Leptin induces cardiac
fibrosis through galectin-3, mTOR and oxidative stress: Potential
role in obesity. J Hypertens. 32:1104–1114. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Handy JA, Fu PP, Kumar P, Mells JE, Sharma
S, Saxena NK and Anania FA: Adiponectin inhibits leptin signalling
via multiple mechanisms to exert protective effects against hepatic
fibrosis. Biochem J. 440:385–395. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chuang JH, Wang PW and Tai MH: An
adipocentric view of liver fibrosis and cirrhosis. Chang Gung Med
J. 27:855–868. 2004.PubMed/NCBI
|
|
13
|
Zibadi S, Cordova F, Slack EH, Watson RR
and Larson DF: Leptin's regulation of obesity-induced cardiac
extracellular matrix remodeling. Cardiovasc Toxicol. 11:325–333.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Koyama Y and Brenner DA: New therapies for
hepatic fibrosis. Clin Res Hepatol Gastroenterol. 39:(Suppl 1).
S75–S79. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xie S, Chen H, Li F, Wang S and Guo J:
Hypoxia-induced microRNA-155 promotes fibrosis in proximal tubule
cells. Mol Med Rep. 11:4555–4560. 2015.PubMed/NCBI
|
|
16
|
Tang J, Jiang X, Zhou Y and Dai Y: Effects
of A2BR on the biological behavior of mouse renal fibroblasts
during hypoxia. Mol Med Rep. 11:4397–4402. 2015.PubMed/NCBI
|
|
17
|
Fan GP, Wang W, Zhao H, Cai L, Zhang PD,
Yang ZH, Zhang J and Wang X: Pharmacological inhibition of focal
adhesion kinase attenuates cardiac fibrosis in mice cardiac
fibroblast and post-myocardial-infarction models. Cell Physiol
Biochem. 37:515–526. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Melo NC, Amorim FF and Santana AN:
Connecting the dots: Hypoxia, pulmonary fibrosis, obstructive sleep
apnea, and aging. Am J Respir Crit Care Med. 191:9662015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Koyama D, Maruoka S, Gon Y, Shintani Y,
Sekiyama T, Hiranuma H, Shikano S, Kuroda K, Takeshita I, Tsuboi E,
et al: Myeloid differentiation-2 is a potential biomarker for the
amplification process of allergic airway sensitization in mice.
Allergol Int. 64:(Suppl). S37–S45. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang WM, Zhao ZL, Zhang WF, Zhao YF, Zhang
L and Sun ZJ: Role of hypoxia-inducible factor-1α and CD146 in
epidermal growth factor receptor-mediated angiogenesis in salivary
gland adenoid cystic carcinoma. Mol Med Rep. 12:3432–3438.
2015.PubMed/NCBI
|
|
21
|
Kulsum U, Singh V, Sharma S, Srinivasan A,
Singh TP and Kaur P: RASOnD-a comprehensive resource and search
tool for RAS superfamily oncogenes from various species. BMC
Genomics. 12:3412011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bouchard L, Thibault S, Guay SP, Santure
M, Monpetit A, St-Pierre J, Perron P and Brisson D: Leptin gene
epigenetic adaptation to impaired glucose metabolism during
pregnancy. Diabetes Care. 33:2436–2441. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
da Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Baba SA, Mohiuddin T, Basu S, Swarnkar MK,
Malik AH, Wani ZA, Abbas N, Singh AK and Ashraf N: Comprehensive
transcriptome analysis of Crocus sativus for discovery and
expression of genes involved in apocarotenoid biosynthesis. BMC
Genomics. 16:6982015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Munson ME: An improved technique for
calculating relative response in cellular proliferation
experiments. Cytometry A. 77:909–910. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Okada H, Ban S, Nagao S, Takahashi H,
Suzuki H and Neilson EG: Progressive renal fibrosis in murine
polycystic kidney disease: An immunohistochemical observation.
Kidney Int. 58:587–597. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhao X, Harashima H, Dissmeyer N, Pusch S,
Weimer AK, Bramsiepe J, Bouyer D, Rademacher S, Nowack MK, Novak B,
et al: A general G1/S-phase cell-cycle control module in the
flowering plant Arabidopsis thaliana. PLoS Genet. 8:e10028472012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chang CN, Feng MJ, Chen YL, Yuan RH and
Jeng YM: p15(PAF) is an Rb/E2F-regulated S-phase protein essential
for DNA synthesis and cell cycle progression. PLoS One.
8:e611962013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Paquin MC, Cagnol S, Carrier JC, Leblanc C
and Rivard N: ERK-associated changes in E2F4 phosphorylation,
localization and transcriptional activity during mitogenic
stimulation in human intestinal epithelial crypt cells. BMC Cell
Biol. 14:332013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Garneau H, Paquin MC, Carrier JC and
Rivard N: E2F4 expression is required for cell cycle progression of
normal intestinal crypt cells and colorectal cancer cells. J Cell
Physiol. 221:350–358. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Oda A, Taniguchi T and Yokoyama M: Leptin
stimulates rat aortic smooth muscle cell proliferation and
migration. Kobe J Med Sci. 47:141–150. 2001.PubMed/NCBI
|
|
32
|
Si HF, Li J, Lü XW and Jin Y: Suppressive
effects of leflunomide on leptin-induced collagen I production
involved in hepatic stellate cell proliferation. Exp Biol Med
(Maywood). 232:427–436. 2007.PubMed/NCBI
|
|
33
|
Garofalo C, Koda M, Cascio S, Sulkowska M,
Kanczuga-Koda L, Golaszewska J, Russo A, Sulkowski S and Surmacz E:
Increased expression of leptin and the leptin receptor as a marker
of breast cancer progression: Possible role of obesity-related
stimuli. Clin Cancer Res. 12:1447–1453. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Simerly RB: Wired on hormones: Endocrine
regulation of hypothalamic development. Curr Opin Neurobiol.
15:81–85. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Harrold JA: Leptin leads hypothalamic
feeding circuits in a new direction. Bioessays. 26:1043–1045. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kitase Y, Yokozeki M, Fujihara S, Izawa T,
Kuroda S, Tanimoto K, Moriyama K and Tanaka E: Analysis of gene
expression profiles in human periodontal ligament cells under
hypoxia: The protective effect of CC chemokine ligand 2 to oxygen
shortage. Arch Oral Biol. 54:618–624. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Agostini C and Gurrieri C:
Chemokine/cytokine cocktail in idiopathic pulmonary fibrosis. Proc
Am Thorac Soc. 3:357–363. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kim JW, Tchernyshyov I, Semenza GL and
Dang CV: HIF-1-mediated expression of pyruvate dehydrogenase
kinase: A metabolic switch required for cellular adaptation to
hypoxia. Cell Metab. 3:177–185. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xu J and Kisseleva T: Bone marrow-derived
fibrocytes contribute to liver fibrosis. Exp Biol Med (Maywood).
240:691–700. 2015. View Article : Google Scholar : PubMed/NCBI
|