Introduction
Hepatitis B virus (HBV) infection remains a major
public health problem worldwide at present. Patients with chronic
HBV infection are at high risk of progressing to cirrhosis,
hepatocellular carcinoma and liver failure. It is estimated that
~800,000 people die from HBV-related diseases per year (1). Although the introduction of the
hepatitis B vaccine into national immunization programs has
dramatically reduced the incidence of HBV infection, the rate of
vertical transmission, especially for hepatitis B virus e
antigen-positive mothers, is up to 9.8% in China in 2002 (2). Nucleoside analogues (NAs), such as
entecavir (ETV), lamivudine and adefovir dipivoxil can inhibit the
reverse transcription of HBV mRNA and have been used as primary
antiviral agents for the treatment of HBV infection in clinical
practice. Nevertheless, they cannot radically eliminate HBV
covalently closed circular (cccDNA) in the nucleus of hepatocytes,
which is the template for HBV replication (3). Moreover, drug resistance may occur
following long-term use of NAs. Thus, it is urgent to find out new
efficient methods to eliminate HBV cccDNA.
The clustered regularly interspaced short
palindromic repeat (CRISPR)/CRISPR-associated (Cas) system,
originally identified in bacteria and archaea, is the third
generation of genome-editing technology (4). The type II CRISPR/Cas system from
Streptococcus pyogenes and its simplified derivative, the
Cas9/single guide RNA (sgRNA) system (5,6), has
emerged as a potent new tool for targeted gene modification in
humans and several other species (7–10).
Since 2013, CRISPR/cas9-mediated genome editing has been
successfully finished on several human viruses, such as papilloma
virus, human immunodeficiency virus and Epstein-Barr virus
(11–13).
So far, several reports have indicated that HBV
total DNA and HBV cccDNA in infected hepatocytes can be reduced by
the CRISPR/cas9 system (14–16).
However, most of these studies have used HBV-infected human
hepatoma cell lines, which are not considered as optimal cell
models for human HBV infection, to evaluate drug efficacy.
Therefore, it is essential to evaluate the inhibition efficacy of
the CRISPR/Cas9 system in the setting of primary hepatocytes with
natural HBV infection. As a model virus of HBV, duck hepatitis B
virus (DHBV) shares the similar virus structure and replication
features with HBV (17). In this
respect, primary duck hepatocytes (PDHs) naturally infected with
DHBV provide valuable model systems for studying HBV infection
(18). In light of this, the
authors hypothesized that the CRISPR/Cas9 system may suppress DHBV
DNA. In order to validate the hypothesis, firstly, DHBV-specific
sgRNA/Cas9 dual expression vector was constructed and transfected
into DHBV-infected PDHs. Secondly, the inhibition efficacy on DHBV
total DNA and cccDNA by the CRISPR/Cas9 system was evaluated.
Finally, the combined inhibition of CRISPR/cas9 system and ETV was
assessed.
Materials and methods
Design and construction of
DHBV-specific sgRNA/Cas9 plasmids
sgRNAs were designed using the CRISPR/Cas system
(Cas9/gRNA) Off-Targeter (CasOT) tool (http://casot.cbi.pku.edu.cn/; Peking University,
Beijing, China) to minimize potential off-target effects. The
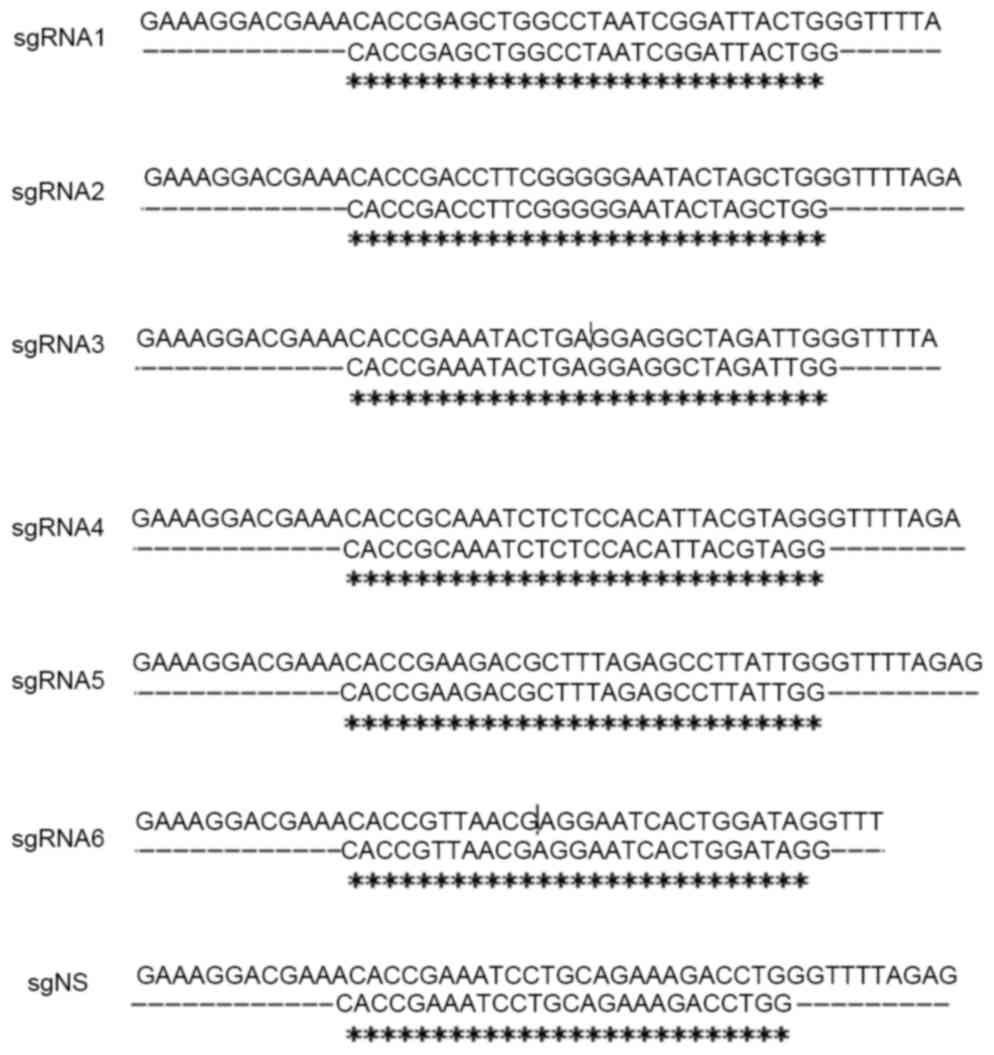
sequences of six sgRNAs targeting DHBV genome (GenBank: K01834.1)
and one nonsense sgRNA (sgNS) are presented in Table I.
 | Table I.Sequence of the protospacer and
protospacer adjacent motif targeted by DHBV-specific sgRNAs in the
DHBV genome. |
Table I.
Sequence of the protospacer and
protospacer adjacent motif targeted by DHBV-specific sgRNAs in the
DHBV genome.
| Name | Nucleotide
position | Gene region | Sequence
(GN18–20NGG) |
|---|
| sgRNA1 | 1,310–1,332 | S | F:
5′-GAGCTGGCCTAATCGGATTACTGG-3′ |
|
|
|
| R:
5′-CCAGTAATCCGATTAGGCCAGCTC-3′ |
| sgRNA2 | 1,293–1,315 | S | F:
5′-GACCTTCGGGGGAATACTAGCTGG-3′ |
|
|
|
| R:
5′-CCAGCTAGTATTCCCCCGAAGGTC-3′ |
| sgRNA3 | 1,363–1,385 | S | F:
5′-GAAATACTGAGGAGGCTAGATTGG-3′ |
|
|
|
| R:
5′-CCAATCTAGCCTCCTCAGTATTTC-3′ |
| sgRNA4 | 1,449–1,472 | S | F:
5′-GCAAATCTCTCCACATTACGTAGG-3′ |
|
|
|
| R:
5′-CCTACGTAATGTGGAGAGATTTGC-3′ |
| sgRNA5 | 2,738–2,760 | C | F:
5′-GAAGACGCTTTAGAGCCTTATTGG-3′ |
|
|
|
| R:
5′-CCAATAAGGCTCTAAAGCGTCTTC-3′ |
| sgRNA6 | 29–51 | P | F:
5′-GTTAACGAGGAATCACTGGATAGG-3′ |
|
|
|
| R:
5′-CCTATCCAGTGATTCCTCGTTAAC-3′ |
| sgNS |
|
| F:
5′-GAAATCCTGCAGAAAGACCTGG-3′ |
|
|
|
| R:
5′-CCAGGTCTTTCTGCAGGATTTC-3′ |
The PSpCas9(BB)-2A-GFP (PX458) plasmid was obtained
from Addgene, Inc. (Cambridge, MA, USA). The plasmid was extracted
using the EndoFree Mini Plasmid Kit II (Tiangen Biotech Co., Ltd.,
Beijing, China). PX458 was digested with BbsI (New England
Biolabs, Inc., Ipswich, MA, USA), and then the linearized vector
was purified using the Gel Extraction kit (Omega Bio-tek, Inc.,
Norcross, GA, USA) according to the manufacturer's instructions.
Each pair of sgRNAs was annealed to double strands, which was
ligated to the linearized vector with T4 DNA ligase (New England
Biolabs, Inc.). The co-expression plasmid sgRNA/Cas9 was identified
through sequencing (Fig. 1).
Isolation, infection and transfection
of PDHs
Ducklings (1-day-old) were purchased from Qianjin
Duck Farm (Beijing, China). All animal care and experimental
procedures were performed with the approval of the Institutional
Animal Care and Use Committee at Beijing Youan Hospital affiliated
to Capital Medical University (Beijing, China) according to the
Guide for the Care and Use of Laboratory Animals (National
Institutes of Health, Bethesda, MD, USA). DHBV-positive and
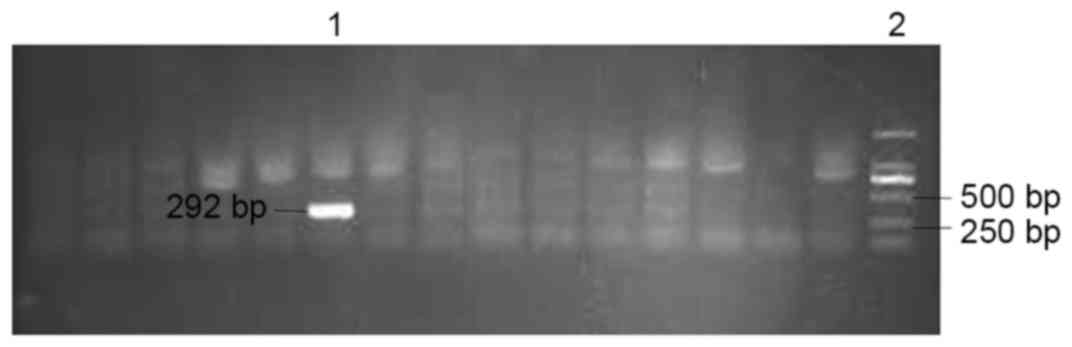
-negative ducklings were identified by polymerase chain reaction
(PCR) using Ex Taq DNA polymerase (Takara Biotechnology Co., Ltd.,
Dalian, China) using serum samples. Sample processing, the
corresponding primers (DHBV2548 and DHBV2840R), PCR reaction
mixture and amplification cycle was described previously (18). The PCR product was verified by
agarose gel electrophoresis (Fig.
2).
PDHs were isolated from 7-day-old DHBV-free Pekin
ducklings (19). Isolated PDHs
were seeded in a 24-well plate at 1.0×105/well, and were
cultured in L-15 medium (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum
(Hyclone; GE Healthcare Life Sciences, Logan, UT, USA), 10 uM
hydrocortisone (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), 10
µg/ml insulin (Cell Applications, Inc., San Diego, CA, USA), 20 mM
HEPES, and 1% penicillin/streptomycin (both from Sigma-Aldrich;
Merck KGaA) in a humidified chamber at 39°C without CO2.
The next day, PDHs were infected with DHBV-positive serum
(~4×106 copies/well). On the third day following
seeding, DHBV-infected PDHs were transfected with DHBV-specific
sgRNA/Cas9 dual expression vector using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). The ratio between
Lipofectamine 2000 (µl) and plasmid (µg) was 3:1. Some wells were
treated with ETV (China Food and Drug Administration, Beijing,
China) at 0.13 nM (20) on days 3,
5, and 7 following transfection. The culture medium and cells were
harvested on day 5 or day 9 following transfection for
analyses.
Extraction of DHBV total DNA and
cccDNA
DHBV total DNA in the culture medium was extracted
using TIANamp Virus DNA/RNA kit (Tiangen Biotech Co., Ltd.).
Following the removal of the culture medium, the cells were lysed
and total DNA was extracted using TIANamp Genomic DNA kit (Tiangen
Biotech Co., Ltd.). For the purification of DHBV cccDNA, DHBV total
DNA was further treated with Plasmid-Safe™ ATP-Dependent DNase
(Epicenter; Illumina, Inc., San Diego, CA, USA) at 37°C for 30 min
followed by 70°C for 30 min to digest linear double-stranded DNA,
and the resulting product was recycled using Cycle Pure kit (Omega
Bio-tek, Inc.) according to the instructions of manufacturer.
Quantification of DHBV DNA in the
culture medium and PDHs
DHBV total DNA and cccDNA were then quantified by
reverse transcription-quantitative PCR with specific primers and
TaqMan MGB probes (Sangon Biotech Co., Ltd, Shanghai, China). The
sequences of the primers and the corresponding TaqMan probes are
displayed in Table II. PCR was
performed using the StepOnePlus Real-time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). In a final volume of
20 µl, the following was added: 10 µl SsoAdvanced™ Universal Probes
SuperMix (Bio-Rad Laboratories, Inc., Hercules, CA, USA), 2 µl DNA
template, 2 µl forward and reverse mixed primers (5 mM), 1 µl
TaqMan probe (2.5 mM) and 5 µl double distilled water.
Amplification was performed under the following conditions: 95°C
for 30 sec, 40 cycles of 95°C for 5 sec, 60°C for 30 sec, and 72°C
for 30 sec. Duck β-globin gene was used as an internal reference.
The relative quantification of total DNA and cccDNA was
standardized to that of the sgNS group.
 | Table II.DHBV primers for polymerase chain
reaction. |
Table II.
DHBV primers for polymerase chain
reaction.
| Name | Primer | Sequence
(5′-3′) |
|---|
| cccDNA | Forward |
TGCCATAAGCGTTATCAGACGTT |
|
| Reverse |
GGCTAAGGCTCTAGAAGCATTGA |
|
| TaqMan probe |
ATATAATCCTGCTGACGGCC |
| Total DNA | Forward |
TTCGGAGCTGCTTGCCAA |
|
| Reverse |
TCATACACATTGGCTAAGGCTCT |
|
| TaqMan probe |
CGTCTACATTGCTGTTGTCGTGTGTGAC |
| β-globin | Forward |
AGCAGTTGTTGGAGCAGGAA |
|
| Reverse |
TCTTTGGCTGTTGGCATCTA |
|
| TaqMan probe |
AGAGGAGTGATGAGCAAGAGACAGTGGC |
Statistical analysis
Normally distributed data were presented as mean ±
standard error of the mean and analyzed by Student's t-test.
Non-normally distributed data were expressed as the median (range)
and was analyzed by the Mann-Whitney U test. Statistical analysis
was conducted using SPSS software (version, 19.0; IBM SPSS,
Chicago, IL, USA). A two-sided P<0.05 was considered to indicate
a statistically significant difference.
Results
sgRNA4 and sgRNA6 significantly
suppressed DHBV DNA on day 5 following transfection
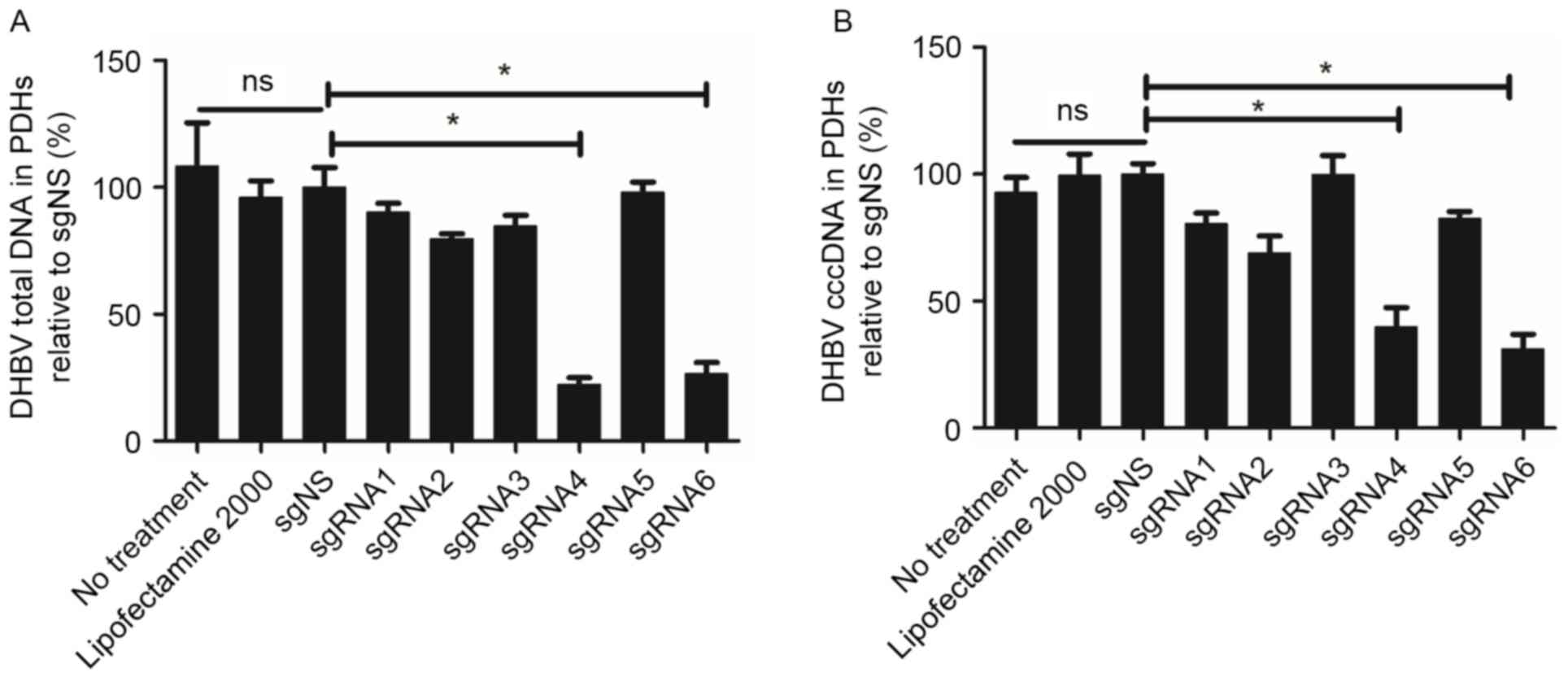
To analyze the inhibition efficacy of six sgRNAs,
the levels of DHBV DNA in PDHs were detected at various time points
following transfection. Fig. 3
indicated that there was no significant difference among the three
groups: No treatment group, Lipofectamine 2000 group and sgNS
group. Compared with the sgNS group, sgRNA4 and sgRNA6 exhibited
higher efficacy in suppressing DHBV DNA on day 5 following
transfection. For sgRNA4, DHBV total DNA in PDHs was reduced by
77.64%, (for example, from 1.30×103 copies/cell to
2.96×102 copies/cell). A similar reduction (60.19%) was
observed for sgRNA6. In addition, DHBV cccDNA was also suppressed
significantly by sgRNA4 and sgRNA6 (60.19 and 68.82%,
respectively). The inhibition efficacy of sgRNA4 and sgRNA6 on DHBV
total DNA and cccDNA presented a significant difference compared
with sgNS (P=0.002 and 0.015 respectively for sgRNA4, P=0.003 and
0.004 respectively for sgRNA6).
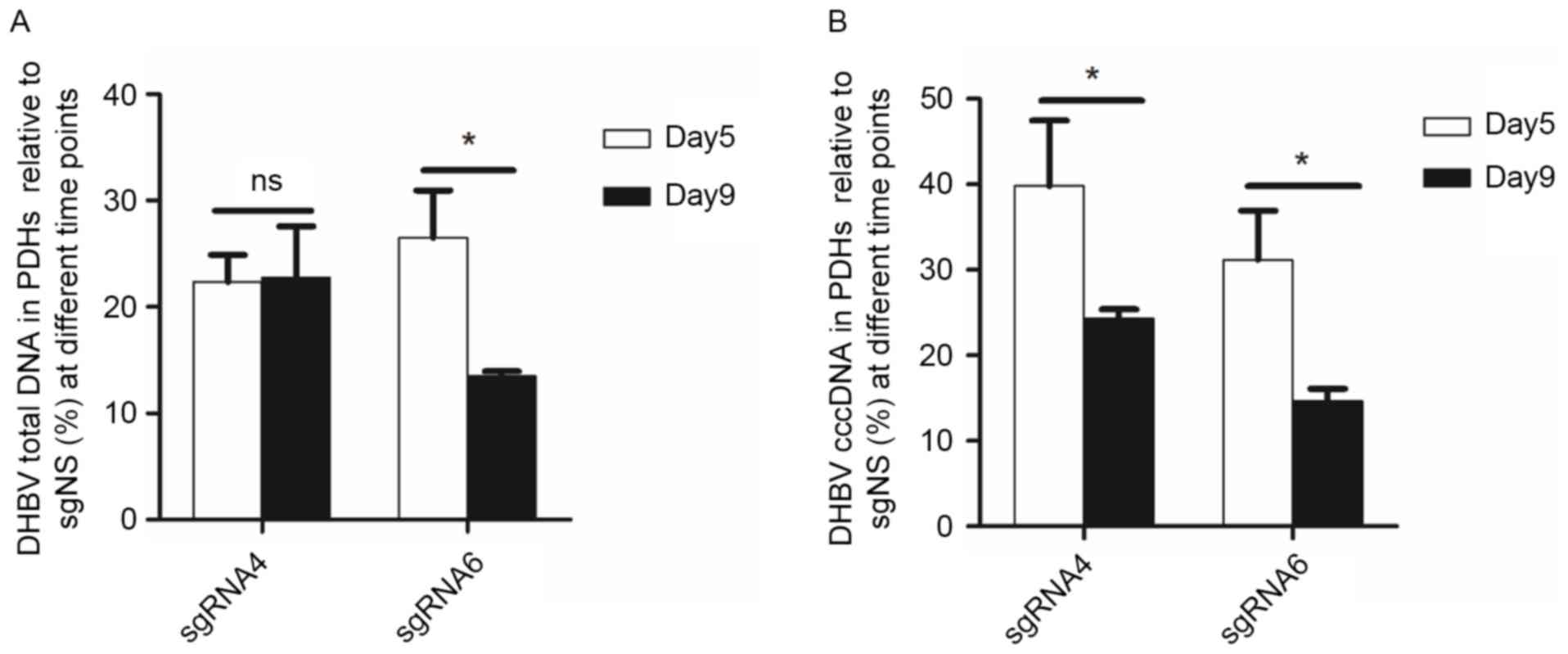
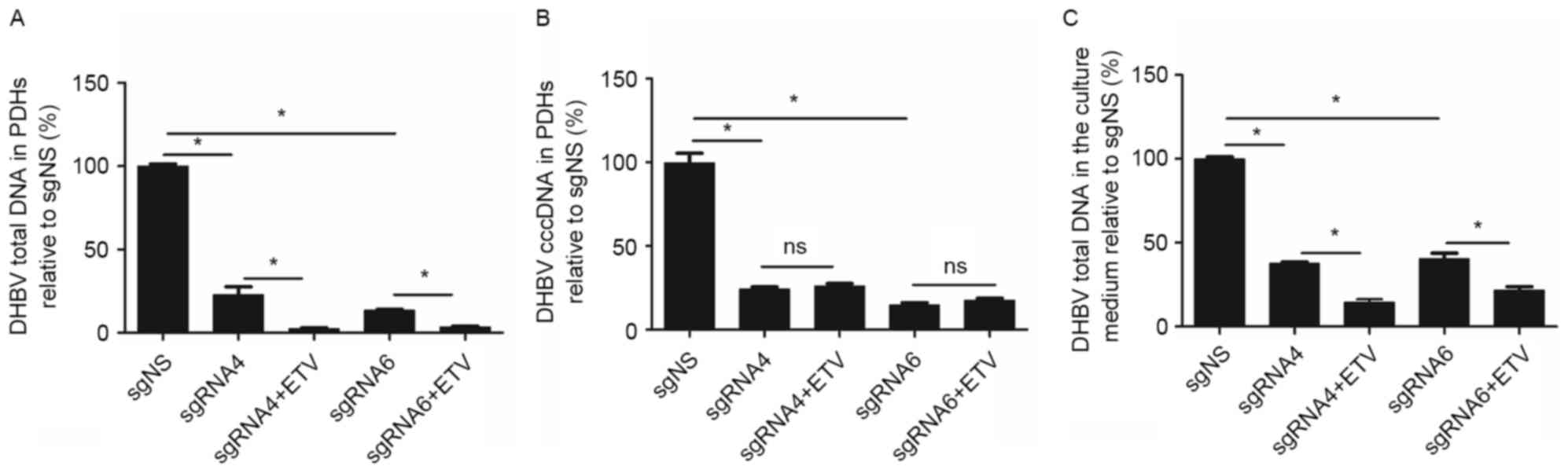
The inhibition efficacy of sgRNA4 and
sgRNA6 remained or improved on day 9 following transfection
From day 5 to day 9 following transfection, the
inhibition on DHBV total DNA remained (from 77.64 to 77.23%) by
sgRNA4, but increased (from 73.51 to 86.51%) by sgRNA6. DHBV cccDNA
in PDHs was further inhibited by sgRNA4 (from 60.19 to 75.67%) and
sgRNA6 (from 68.82 to 85.34%). Moreover, DHBV total DNA in the
culture medium was reduced by 62.17 and 59.52%, respectively for
sgRNA4 and sgRNA6 (Fig. 4).
ETV enhanced the suppression on DHBV
DNA accumulation by CRISPR/Cas9 system
Considering that the suppression on DHBV DNA by
CRISPR/Cas9 system was incomplete, it would be interesting to
further assess the combined inhibition of CRISPR/Cas9 system and
ETV, the first-line treatment option for patients with HBV
infection. As presented in Fig. 5,
on day 9 following transfection (the sixth day following ETV
treatment), the inhibition efficacy on DHBV total DNA in PDHs was
higher in sgRNA4+ETV (97.52%) and sgRNA6+ETV (96.57%) groups
compared with sgRNA4 (77.23%) and sgRNA6 (86.51%) groups (P=0.006
and 0.005 respectively). Similarly, the inhibition efficacy on DHBV
total DNA in the culture medium was higher in sgRNA4+ETV (85.45%)
and sgRNA6+ETV (78.29%) groups compared with sgRNA4 (62.17%) and
sgRNA6 (59.52%) groups (P=0.006 and 0.011, respectively). However,
ETV treatment did not enhance the inhibition on DHBV cccDNA in PDHs
by sgRNA4 (sgRNA4 vs. sgRNA4+ETV: 75.67% vs. 73.90%; P=0.144) and
sgRNA6 (sgRNA6 vs. sgRNA6+ETV: 85.34% vs. 82.60%; P=0.144). Thus,
the combination of ETV and CRISPR/Cas9 system led to a further
reduction of DHBV total DNA in PDHs and culture medium, but not
DHBV cccDNA.
Discussion
In the present study, two sgRNAs (sgRNA4 and sgRNA6)
targeting the DHBV genome were demonstrated to suppress DHBV total
DNA and cccDNA successfully. ETV enhanced the inhibition of the
CRISPR/Cas9 system on DHBV total DNA. The current findings
suggested that the CRISPR/Cas9 system targeting specific sites of
the DHBV genome may be an effective technology to inhibit DHBV
infection in PDHs. To the best of the authors' knowledge, the
present study is the first reporting the targeted inhibition of
DHBV DNA by the CRISPR/Cas9 system.
Genetic modifications have been achieved
successfully using genome-editing technologies, including zinc
finger nucleases (ZFN) (21),
transcription activator-like effector nucleases (TALEN) (22) and the CRISPR/Cas9 system (23). As the classical method for genome
engineering, ZFN is used to invalidate the HIV co-receptor C-C
chemokine receptor type 5, which is currently in clinical trials
(NCT01252641, NCT00842634 and NCT01044654). It is reported that
TALEN plasmids for 18,740 human protein-coding genes have been
assembled (24). However, both
ZFNs and TALENs utilize protein-based programmable,
sequence-specific DNA-binding modules, whose construction is
usually complex. The emergence of the CRISPR/Cas9 system in 2013
(25) makes it a facile and
efficient alternative to ZFNs and TALENs. Only sgRNAs targeting
specific genes are required for highly efficient gene modification
using the CRISPR/Cas9 system (26,27).
In the present study, the effective sgRNAs targeting
the DHBV genome were located at the S and P regions. Previous
studies also screened out effective sgRNAs located at different
regions of HBV genome, such as X, core, polymerase and surface ORFs
(14,15,28).
sgRNAs targeting the conserved HBV sequence were effective for HBV
genomes of different genotypes (29). Thus, it may be necessary to
construct the sgRNAs library targeting different regions of HBV
genome in order to screen out the most efficient sgRNAs.
Several previous reports used either Huh7 cells
(29) transfected with the
HBV-expression vector pAAV/HBV1.2 (genotype A) or HepAD38 cells
(30) stably expressing HBV DNA to
evaluate the targeted inhibition on HBV genome by the CRISPR/Cas9
system. In terms of biological characteristics and HBV infection
mode, PDHs infected with DHBV-positive serum have advantages over
immortalized cell lines, because the former may mimic the natural
process of HBV infection. In the present study, we chose
DHBV-infected PDHs as an in vitro model to assess the
inhibition efficiency of the CRISPR/Cas9 system. In this regard,
the result that the CRISPR/Cas9 system can efficiently inhibit DHBV
cccDNA is more close to the real-world HBV infection, thus
exhibiting great clinical importance. Moreover, the study
investigated whether ETV can improve the anti-viral effects of
CRISPR/Cas9 system. The results indicated that the combination of
the CRISPR/Cas9 system and ETV enhanced the suppression of DHBV
total DNA, but not cccDNA. One possible explanation is that ETV, as
a nucleoside reverse transcriptase inhibitor, could only reduce
DHBV total DNA, but has little or no effect on DHBV cccDNA.
Therefore, it was speculated that the combined application of the
CRISPR/Cas9 system and ETV may have the potential to control DHBV
infection more effectively.
There are two limitations in the present study.
Firstly, sgRNAs targeting different regions of DHBV genome need to
be designed in order to screen out the most effective ones.
Secondly, in vivo studies need to be performed to validate
the inhibitory effect of the CRISPR/Cas9 system on DHBV cccDNA.
Although great advancements have been made in the
prevention and treatment of HBV infection, the high morbidity of
HBV-associated complications is still a huge threat for human
health. The present study is thought to be the first to demonstrate
the efficient inhibition of the CRISPR/Cas9 system on DHBV cccDNA.
Even though further study is required to improve the inhibitory
efficiency, the current findings pave the way for eliminating HBV
cccDNA using the CRISPR/Cas9 system in clinical practice in the
future.
Acknowledgements
The present study was supported by the National
Science and Technology Key Project (grant nos. 2017ZX10201201,
2017ZX10203201-005, 2017ZX10202203-006-001 and
2017ZX10302201-004-002), the Beijing Municipal Administration of
Hospital's Ascent Plan (grant no. DFL20151601), the Beijing
Municipal Science and Technology Commission (grant no.
Z151100004015066), the Basic-Clinical Cooperation Project of
Capital Medical University (grant nos. 15JL67 and 17JL47) and the
YouAn Fund for Liver Diseases and AIDS (grant no.
YNKT20160012).
Glossary
Abbreviations
Abbreviations:
|
HBV
|
hepatitis B virus
|
|
cccDNA
|
covalently closed circular DNA
|
|
CRISPR
|
clustered regularly interspaced short
palindromic repeat
|
|
Cas9
|
CRISPR-associated protein 9
|
|
PDH
|
primary duck hepatocyte
|
|
DHBV
|
duck hepatitis B virus
|
|
sgRNAs
|
single-guide RNAs
|
|
ETV
|
entecavir
|
|
NAs
|
nucleoside analogues
|
|
sgNS
|
one nonsense sgRNA
|
|
ZFN
|
zinc finger nucleases
|
|
TALEN
|
transcription activator-like effector
nucleases
|
References
|
1
|
Komatsu H: Hepatitis B virus: Where do we
stand and what is the next step for eradication? World J
Gastroenterol:. 20:8998–9016. 2014.PubMed/NCBI
|
|
2
|
Xu DZ, Yan YP, Choi BC, Xu JQ, Men K,
Zhang JX, Liu ZH and Wang FS: Risk factors and mechanism of
transplacental transmission of hepatitis B virus: A case-control
study. J Med Virol. 67:20–26. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Werle-Lapostolle B, Bowden S, Locarnini S,
Wursthorn K, Petersen J, Lau G, Trepo C, Marcellin P, Goodman Z, WE
VI Delaney, et al: Persistence of cccDNA during the natural history
of chronic hepatitis B and decline during adefovir dipivoxil
therapy. Gastroenterology. 126:1750–1758. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hsu PD, Lander ES and Zhang F: Development
and applications of CRISPR-Cas9 for genome engineering. Cell.
157:1262–1278. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Barrangou R and Marraffini LA: CRISPR-Cas
systems: Prokaryotes upgrade to adaptive immunity. Mol Cell.
54:234–244. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cong L, Ran FA, Cox D, Lin S, Barretto R,
Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA and Zhang F:
Multiplex genome engineering using CRISPR/Cas systems. Science.
339:819–823. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang T, Wei JJ, Sabatini DM and Lander ES:
Genetic screens in human cells using the CRISPR-Cas9 system.
Science. 343:80–84. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang H, Wang H, Shivalila CS, Cheng AW,
Shi L and Jaenisch R: One-step generation of mice carrying reporter
and conditional alleles by CRISPR/Cas-mediated genome engineering.
Cell. 154:1370–1379. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gratz SJ, Cummings AM, Nguyen JN, Hamm DC,
Donohue LK, Harrison MM, Wildonger J and O'Connor-Giles KM: Genome
engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease.
Genetics. 194:1029–1035. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X,
Xiong JW and Xi JJ: Genome editing with RNA-guided Cas9 nuclease in
zebrafish embryos. Cell Res. 23:465–472. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hu Z, Yu L, Zhu D, Ding W, Wang X, Zhang
C, Wang L, Jiang X, Shen H, He D, et al: Disruption of HPV16-E7 by
CRISPR/Cas system induces apoptosis and growth inhibition in HPV16
positive human cervical cancer cells. Biomed Res Int.
2014:6128232014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hu W, Kaminski R, Yang F, Zhang Y,
Cosentino L, Li F, Luo B, Alvarez-Carbonell D, Garcia-Mesa Y, Karn
J, et al: RNA-directed gene editing specifically eradicates latent
and prevents new HIV-1 infection. Proc Natl Acad Sci USA.
111:11461–11466. 2014; View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang J and Quake SR: RNA-guided
endonuclease provides a therapeutic strategy to cure latent
herpesviridae infection. Proc Natl Acad Sci USA. 111:13157–13162.
2014; View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dong C, Qu L, Wang H, Wei L, Dong Y and
Xiong S: Targeting hepatitis B virus cccDNA by CRISPR/Cas9 nuclease
efficiently inhibits viral replication. Antiviral Res. 118:110–117.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Seeger C and Sohn JA: Targeting hepatitis
B virus with CRISPR/Cas9. Mol Ther Nucleic Acids. 3:e2162014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhen S, Hua L, Liu YH, Gao LC, Fu J, Wan
DY, Dong LH, Song HF and Gao X: Harnessing the clustered regularly
interspaced short palindromic repeat (CRISPR)/CRISPR-associated
Cas9 system to disrupt the hepatitis B virus. Gene Ther.
22:404–412. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jilbert AR, Botten JA, Miller DS, Bertram
EM, Hall PM, Kotlarski J and Burrell CJ: Characterization of age-
and dose-related outcomes of duck hepatitis B virus infection.
Virology. 244:273–282. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang YY, Theele DP and Summers J:
Age-related differences in amplification of covalently closed
circular DNA at early times after duck hepatitis B virus infection
of ducks. J Virol. 79:9896–9903. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tuttleman JS, Pugh JC and Summers JW: In
vitro experimental infection of primary duck hepatocyte cultures
with duck hepatitis B virus. J Virol. 58:17–25. 1986.PubMed/NCBI
|
|
20
|
Marion PL, Salazar FH, Winters MA and
Colonno RJ: Potent efficacy of entecavir (BMS-200475) in a duck
model of hepatitis B virus replication. Antimicrob Agents
Chemother. 46:82–88. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Miller JC, Holmes MC, Wang J, Guschin DY,
Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et
al: An improved zinc-finger nuclease architecture for highly
specific genome editing. Nat Biotechnol. 25:778–785. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Miller JC, Tan S, Qiao G, Barlow KA, Wang
J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al: A TALE
nuclease architecture for efficient genome editing. Nat Biotechnol.
29:143–148. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Barrangou R: RNA-mediated programmable DNA
cleavage. Nat Biotechnol. 30:836–838. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kim Y, Kweon J, Kim A, Chon JK, Yoo JY,
Kim HJ, Kim S, Lee C, Jeong E, Chung E, et al: A library of TAL
effector nucleases spanning the human genome. Nat Biotechnol.
31:251–258. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Grissa I, Vergnaud G and Pourcel C: The
CRISPRdb database and tools to display CRISPRs and to generate
dictionaries of spacers and repeats. BMC Bioinformatics. 8:1722007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ramanan V, Shlomai A, Cox DB, Schwartz RE,
Michailidis E, Bhatta A, Scott DA, Zhang F, Rice CM, Bhatia SN, et
al: CRISPR/Cas9 cleavage of viral DNA efficiently suppresses
hepatitis B virus. Sci Rep. 5:108332015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ebina H, Misawa N, Kanemura Y and Koyanagi
Y: Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1
provirus. Sci Rep. 3:25102013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kennedy EM, Kornepati AV and Cullen BR:
Targeting hepatitis B virus cccDNA using CRISPR/Cas9. Antiviral
Res. 123:188–192. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lin SR, Yang HC, Kuo YT, Liu CJ, Yang TY,
Sung KC, Lin YY, Wang HY, Wang CC, Shen YC, et al: The CRISPR/Cas9
system facilitates clearance of the intrahepatic HBV templates In
Vivo. Mol Ther Nucleic Acids. 3:e1862014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kennedy EM, Bassit LC, Mueller H,
Kornepati AV, Bogerd HP, Nie T, Chatterjee P, Javanbakht H,
Schinazi RF, Cullen BR, et al: Suppression of hepatitis B virus DNA
accumulation in chronically infected cells using a bacterial
CRISPR/Cas RNA-guided DNA endonuclease. Virology. 476:196–205.
2015. View Article : Google Scholar : PubMed/NCBI
|