Introduction
Gastric cancer (GC) is the second leading cause of
cancer-associated mortality and the fourth highest incidence of
human malignancy worldwide. It has been previously estimated that
over 70% of novel cases and deaths occur in developing countries,
including China (1). The 5-year
survival rate of GC patients with successful surgery, with or
without chemotherapy, is <40% (2).
MicroRNAs (miRNAs) are a group of endogenous, small,
non-coding single-stranded RNAs (~18–25 nucleotides in length) that
post-transcriptionally modulate target gene expression by binding
to the 3′-untranslated region (UTR) of mRNAs (3). Previous studies have revealed that
miRNA-regulated tumor initiation and progression occurs as they
function as tumor suppressors or oncogenes (4–6). The
deregulated miRNAs were closely associated with carcinogenesis and
may also function as diagnostic or prognostic markers in human
malignancies (7–9). Therefore, in order to explore the
potential target and molecular mechanism responsible for the
gastric carcinogenesis is critical for cancer treatment and
prognosis prediction.
Previous studies have reported that miR-320a may
significantly inhibit cell proliferation and invasion in colorectal
cancer (CRC), nasopharyngeal carcinoma and breast cancer (10–14).
Additionally, upregulated miR-320a was confirmed in
paclitaxel-resistant formalin-fixed paraffin-embedded tissues from
ovarian tumor samples (15).
Restoration of miR-320a expression was suggested as a potential
therapeutic approach for tamoxifen-resistant breast cancer
(16). Previous studies determined
that plasma miR-320a was significantly increased in osteosarcoma
and hepatitis B-positive hepatocellular carcinoma patients
(17,18), whereas its levels were reduced in
patients with CRC (19). However,
the function and clinical significance of miR-320a have been
investigated in various types of cancer, the importance of miR-320a
in the development of GC remains to be elucidated.
ADAM metallopeptidase domain 10 (ADAM10) belongs to
the a disintegrin and metalloproteinase (ADAM) family, which has
been widely reported to be overexpressed in human malignancies and
promote cancer development and metastasis (20,21).
ADAM10 noticeably contributed to cell motility in pancreatic
carcinoma and oral squamous cell carcinoma (22,23)
and in pituitary adenoma and lung cancer via regulation of CD44
cleavage and Notch1 signaling (24,25).
Knockdown of ADAM10 in hepatocellular carcinoma cells enhanced the
level of doxorubicin-induced apoptosis (26). Several miRNAs, including miR-140-5p
and miR-122-5p, mediated cell migration and the mRNA expression of
ADAM10 by targeting its 3′-UTR in hypopharyngeal squamous cell
carcinoma and breast cancer, respectively (27,28).
Therefore, the present study examined the association of miR-320a
and ADAM10 in gastric carcinogenesis.
The present study determined that miR-320a was
downregulated in GC cells and tissues and overexpression of
miR-320a inhibited tumor growth in vitro and in vivo.
It is of note that miR-320a increased the sensitivity of GC cells
to cisplatin. ADAM10 was a direct and functional target of
miR-320a. ADAM10 was inversely associated with miR-320a in primary
GC tissues. The findings of the present study suggested that
miR-320a had an important role in regulating cancer cell growth and
chemoresistance of GC through regulating ADAM10 expression.
Materials and methods
Cell lines and tissues
A total of 4 GC cell lines, including SGC-7901, AGS,
BGC-823 and MGC-803 were obtained from the Type Culture Collection
of the Chinese Academy of Sciences (Shanghai, China), maintained in
our laboratory and used in the present study. A normal gastric
mucosal epithelial cell line GES-1 (Type Culture Collection of the
Chinese Academy of Sciences, Shanghai, China) was also used. All
cells were cultured in RPMI-1640 (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc.), 100
µg/ml streptomycin and 100 U/ml penicillin, in a culture incubator
under the atmosphere of 5% CO2 and 37°C.
Tissue samples of GC and adjacent normal tissues
were obtained from 40 patients (male, n=24; female, n=16) who were
diagnosed at the 401 Hospital of People's Liberation Army between
January 2009 and December 2012. Participants were between 42 and 76
years of age and had no history of tumors. Written informed consent
was obtained from patients and the protocols were approved by the
ethics committee of the hospital. Non-cancerous tissues from the
macroscopic tumor margin were isolated simultaneously and used as
control. All tissues were snap frozen and stored in −80°C until
use.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
miRNAs were extracted from cells and tissues and
purified using a miRNA isolation system (Omega Bio-Tek, Inc.,
Norcross, GA, USA) and processed with the miScript II RT kit
(Qiagen GmbH, Hilden, Germany) to generate cDNA. The template cDNA
was used for the qPCR assay with the miScript SYBR-Green PCR kit
(Qiagen GmbH) following the manufacturer's protocol. The PCR
thermal parameters were: 15 min at 95°C, 40 cycles of 15 sec at
94°C followed by 30 sec at 55°C, and 30 sec at 72°C. The expression
threshold cycle values of miRNAs were calculated by normalizing
with internal reference U6 and the relative quantity of miRNAs were
calculated using the 2−ΔΔCq method (29). For mRNA analysis, total RNA was
isolated using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) and followed by RT reaction using the
Prime-script RT reagent kit (Takara Biotechnology Co., Ltd.,
Dalian, China) and the RT reaction was 37°C for 60 min, followed by
85°C for 5 sec. RT-qPCR was conducted using SYBR Premix Ex Taq
(Qiagen GmbH) and β-actin was used as internal control. The PCR
reaction was 95°C for 5 min, followed by 45 cycles of 95°C for 15
sec and 58°C for 30 sec. The sequences of the PCR primers were as
follows: miR-320a forward, 5′-AAAAGCTGGGTTGAGAGGGCG-3′; U6 forward,
5′-CTCGCTTCGGCAGCACA-3′ and U6 reverse, 5′-AACGCTTCACGAATTTGCGT-3′;
ADAM10 forward, 5′-CTGCCCAGCATCTGACCCTAA-3′ and reverse,
5′-TTGCCATCAGAACTGGCACAC-3′; GAPDH forward,
5′-GACCCAGATCATGTTTGAGACC-3′, and reverse
5′-ATCTCCTTCTGCATCCTGTCG-3′. Relative quantification of ADAM10
expression was calculated using the 2−ΔΔCq method
(29). Each sample was analyzed in
triplicate.
Transfection
miR-320a mimics, mimic control, inhibitor and
inhibitor control were synthesized by GenePharma (Shanghai, China)
and were transfected into cells (1×105) with a final
concentration of 20 nmol/l using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.). miR-320a mimics,
5′-AAAAGCUGGGUUGAGAGGGCGA-3′; mimics control,
5′-UUCUCCGAACGUGUCACGUTT-3′; inhibitor,
5′-UCGCCCUCUCAACCCAGCUUUU3′; inhibitor control,
5′-CAGUACUUUUGUGUAGUACAA-3′. The specific anti-ADAM10 small
interfering (si)RNA (siADAM10) and its non-specific scramble siRNA
(control) were purchased from Santa Cruz Biotechnology, Inc.
(Dallas, TX, USA), and then transfected into cells using
Lipofectamine® 2000 according to the manufacturer's
protocol. Briefly, cells were cultured in RPMI-1640 medium without
antibiotics. Transfection procedure was performed once the cultured
cells are at a confluency of 80%. SiRNA (5 µl) and 4.5 µl RNAi-MAX
complexes (Thermo Fisher Scientific, Inc.) were diluted in 250 µl
Opti-MEM medium and incubated for 20 min. The mixture was then
added to the cells and incubated at 37°C. The efficiency of
siADAM10 was confirmed by western blot analysis. ADAM10 cDNA with
3′-UTR were inserted into pcDNA3.1(+) vector (Invitrogen; Thermo
Fisher Scientific, Inc.) to build the plasmid pcDNA3.1(+)-ADAM10
(pcADAM10), and the empty vector (Invitrogen; Thermo Fisher
Scientific, Inc.) was used as control. The samples were collected
for gene and protein detection 48 h post-transfection.
Colony formation assay
Transfected cells were seeded into 6-well culture
dishes (400 cells/well) and cultured for 14 days. The colonies were
fixed with 75% ethanol and stained with 0.2% crystal violet (Sangon
Biotech Co., Ltd., Shanghai, China) for 20 min at room temperature
and counted under a microscope (SZ61-ILST; Olympus Corporation,
Tokyo, Japan). The number of formed colonies was counted in 6
different fields (magnification, ×100).
Viability assay
MMT was used for the viability assay identify the
effect of miR-320a. The transfected GC cells were seeded in 96-well
plates at 2,000 cells/well and maintained at 37°C and 5%
CO2 for 4 days. Then, 150 µl DMSO was added to block the
reaction and the cell proliferation was determined by measuring the
optical density at 570 nm using a spectrophotometric reader (Thermo
Fisher Scientific, Inc.). All tests were performed in
triplicate.
Luciferase reporter activity
assay
The wild-type 3′-UTR and the mutated 3′-UTR
sequences of ADAM10 were inserted into the SpeI and
HindIII sites of the pMiR-REPORT (Ambion; Thermo Fisher
Scientific, Inc.) to produce constructs of wild-type 3′-UTR segment
of ADAM10 (ADAM10-WT) and mutated 3′-UTR segment of ADAM10
(ADAM10-Mut), respectively. The following primers were used:
forward 5′-AGTCCTTATGTGGCATGCCCCCTATG-3′ and reverse
5′-ATTGCGATGATACAAGTCCATCGGATTATTTCA-3′. SGC-7901 cells were seeded
in 24-well plates (3×105 cells/well). After 1 day,
ADAM10-WT, ADAM10-Mut, and pMiR-REPORT control vectors were
co-transfected with miR-320a and β-galactosidase into SGC-7901
cells. Following incubation at 37°C for 48 h, the luciferase
activity was quantified using Dual-Light Combined Reporter Gene
Assay system (Thermo Fisher Scientific, Inc.) 48 h after
transfection.
MTS assay
The sensitivity of cells to cisplatin was determined
using the Cell Titer 96 AQueous One Solution Cell Proliferation
Assay kit (Promega Corporation, Madison, WI, USA). Cells were
cultured in 96-well plates seeded at 3,000 cells/well and different
concentrations of cisplatin were added. The RPMI-1640 medium was
replaced with fresh medium (containing cisplatin) every 24 h. After
3 days, MTS (0.02 ml/well) was added. The absorbance was recorded
at 490 nm for each well on the BioTek Synergy 2 (BioTek, Winooski,
VT, USA).
Western blot analysis
Cells were collected and treated with cell lysis
buffer (Beyotime Institute of Biotechnology, Haimen, China),
followed by centrifugation for 30 min at 4°C and 13,400 × g, and
the protein samples were collected. Protein quantity was determined
using BCA Protein Assay kit (Beyotime Institute of Biotechnology)
and then separated on a 10% SDS-PAGE gel and transferred onto PVDF
membranes. The membranes were subsequently incubated at 4°C
overnight with primary antibody ADAM10 (sc-28358; 1:500; Santa Cruz
Biotechnology, Inc.) or β-actin (sc-8432; 1:2,000; Santa Cruz
Biotechnology, Inc.), followed by incubated with horseradish
peroxidase-conjugated secondary goat anti-mouse antibodies
(STAR117P, 1:1,000; Bio-Rad Laboratories, Hercules, CA, USA) at
room temperature for 30 min. The western blots were visualized
using an enhanced chemiluminescence system (Thermo Fisher
Scientific, Inc.).
In vivo xenograft models
The miR-320a precursor sequences were cloned into
pCDH-CMV-MCS-EF1-Puro vector (System Biosciences, Palo Alto, CA,
USA) to construct stably miR-320a-expressing cells used in
xenograft models. A total of 5 male BALB/c nu/nu mice (5-weeks old;
weight, 18–20 g) were provided by the Animal Center of the Chinese
Academy of Science (Shanghai, China) and used for the in
vivo xenograft tumor model. Animals were housed in a specific
pathogen-free room with a 12-h light/dark cycle and 40–70% humidity
at 26–28°C. SGC-7901 cells (3×106) with miR-320a stable
expression vector or empty vector (control) were subcutaneously
injected into the left and right flank. Tumor volumes were
calculated using the formula: Tumor volume=(length ×
width2)/2. After 35 days, all mice were sacrificed
following the standard procedure and harvested tumors were
weighed.
Statistical analysis
Each experiment was repeated at least three times.
Data are reported as mean ± standard deviation and statistical
tests were performed using SPSS version 14.0 (SPSS, Inc., Chicago,
IL, USA) and Prism version 5.0 (GraphPad, La Jolla, CA, USA).
Statistical significance was determined using a two-sided Student's
t-test. Paired-sample t-test was used to compare the expression
levels of miR-320a and ADAM10 in clinical samples, and Pearson
correlation was used to determine if there was a relationship
between. Multiple group comparisons were analyzed using one-way
analysis of variance. Tukey post hoc tests were used when comparing
multiple parameters. P<0.05 was considered to indicate a
statistically significant difference.
Results
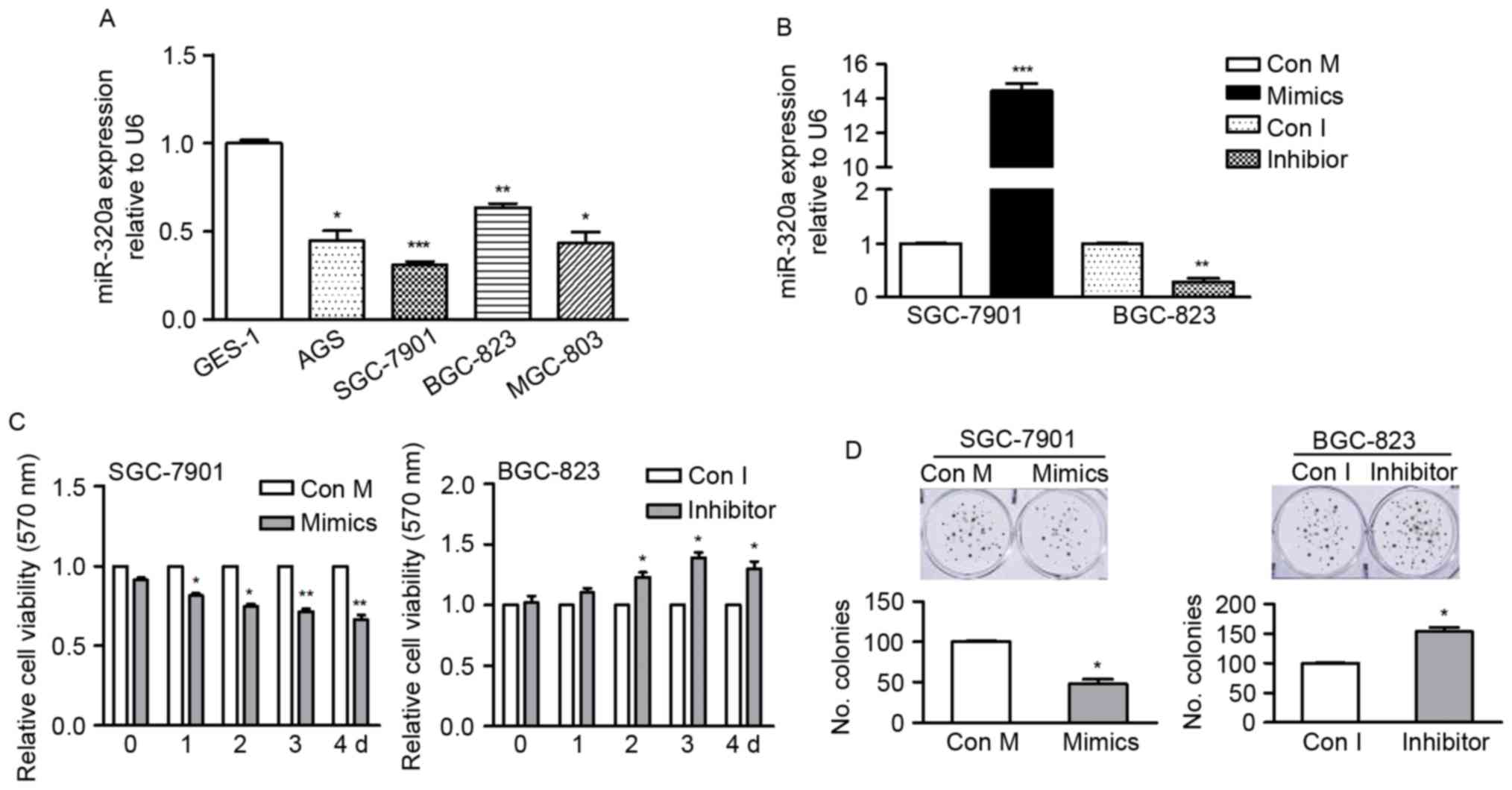
miR-320a is downregulated in GC cell
lines
In order to determine the endogenous miR-320a level
in GC cells, the present study used RT-qPCR to detect miR-320a
expression in four GC cell lines (AGS, SGC-7901, BGC-823 and
MGC-803 cells) and compared them with miR-320a expression in a
normal gastric mucosal epithelial cell line GES-1 cells. The data
revealed that compared with GES-1, all GC cells had significantly
reduced expression levels of miR-320a (Fig. 1A), suggesting miR-320a may
contribute to GC development.
miR-320a inhibits cell growth and
enhances sensitivity of GC cells to cisplatin
The present study identified the role of miR-320a in
GC cells by gain- or loss-of-function analysis (Fig. 1A). SGC-7901 cells were selected as
they have relatively low endogenous miR-320a expression and BGC-823
cells were selected due to their relatively high endogenous
miR-320a and were transfected with mimics or inhibitor. The
expression of miR-320a in transfected cells was confirmed by
RT-qPCR (Fig. 1B).
Colony formation and MTT assays were used to
identify the effect of miR-320a on GC cell growth. As presented in
Fig. 1C, the proliferation rate of
SGC-7901 cells was significantly inhibited by miR-320a mimics,
whereas the proliferation of BGC-823 cells was promoted by miR-320a
inhibitor, in comparison with their corresponding control cells.
For colony formation, the number of formed colonies was markedly
reduced in miR-320a overexpressing cells, whereas it was increased
in cells where miR-320a was inhibited (Fig. 1D). These findings indicated that
miR-320a suppressed cell growth in vitro.
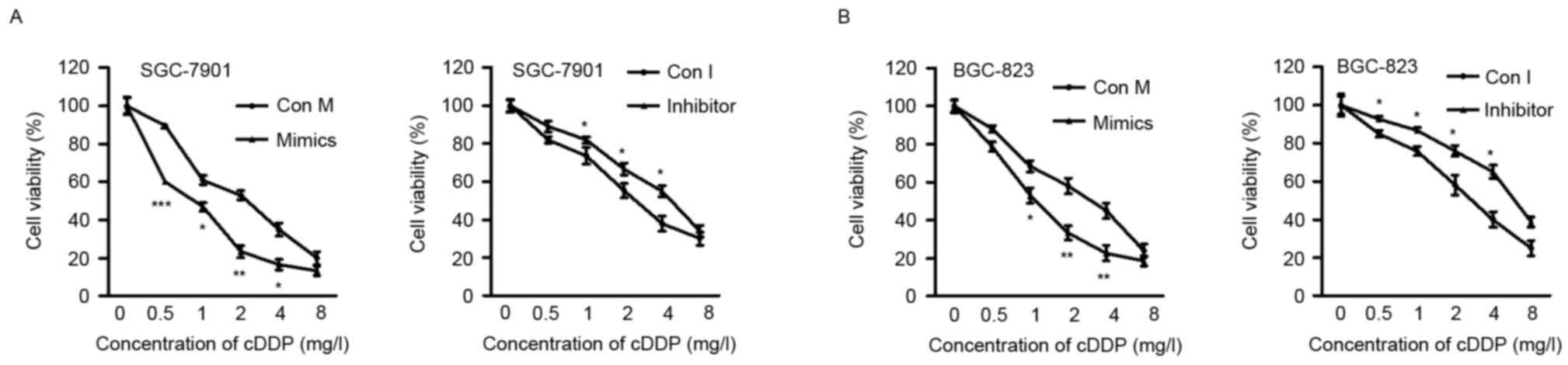
The present study determined the effects of ectopic
miR-320a expression on cell chemosensitivity. It was revealed that
miR-320a mimics effectively sensitized SGC-7901 and BGC-823 cells
to cisplatin, compared with control-transfected cells. By contrast,
the sensitivity of SGC-7901 and BGC-823 cells to cisplatin was
reduced by the miR-320a inhibitor, when compared with the control
cells (Fig. 2A and B). These
findings collectively indicated that miR-320a sensitized GC cells
to cisplatin.
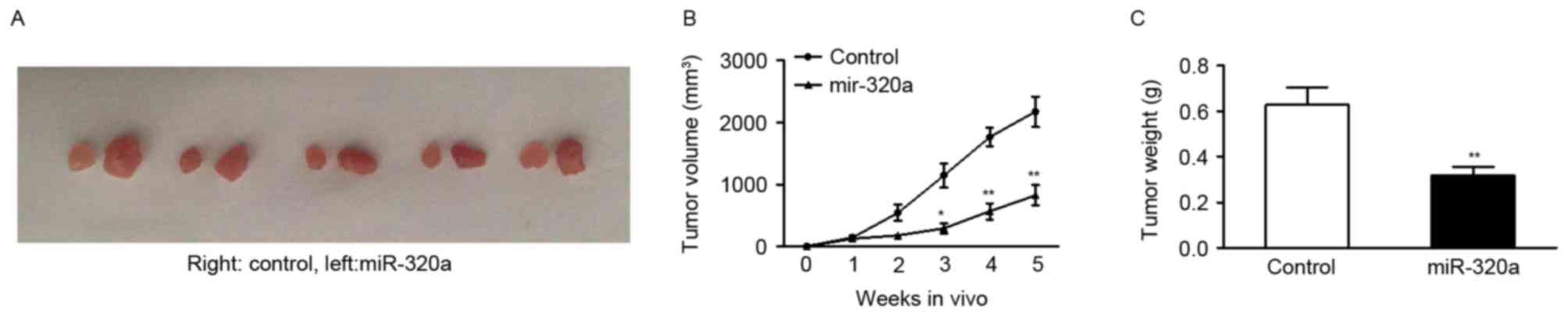
miR-320a suppresses tumor growth in
vivo
As it was evident that up or downregulating miR-320a
expression had a functional role in regulating GC cell growth in
vitro, the present study investigated whether overexpressing
miR-320a may have a similar antitumor role in inhibition of tumor
growth in vivo. SGC-7901 cells were transfected with stably
expressing miR-320a vector or control vector. Subsequently, cells
were subcutaneously injected into five null mice. The tumors were
harvested 5 weeks after injection. The findings revealed that
tumors were significantly smaller when miR-320a expression was
upregulated (Fig. 3A and B).
Quantification of tumor weight confirmed that miR-320a markedly
suppressed the ability of SGC-7901 cells to form tumors in
vivo (Fig. 3C).
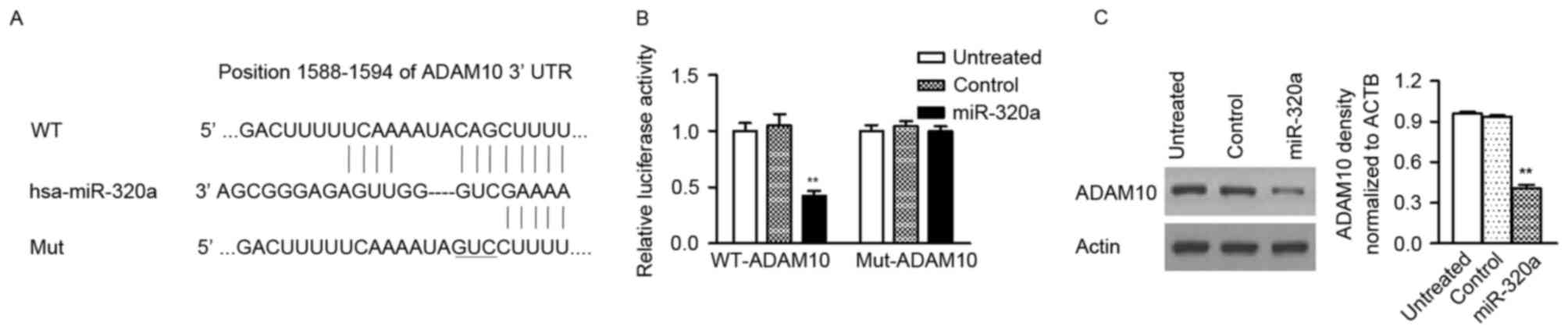
miR-320a directly targets ADAM10 in GC
cells
Three publically available databases (miRDB,
microRNA, TargetScan) were used to predict the potential targets of
miR-320a in GC cells and ADAM10 with conserved binding site in its
3′-UTR was selected for further analysis (Fig. 4A). To assess whether ADAM10 is a
direct target of miR-320a, the luciferase reporter vectors with the
putative ADAM10 3′-UTR target site for miR-320a (ADAM10-WT) and
mutant type (ADAM10-Mut) were constructed. The present study
confirmed that miR-320a significantly decreased luciferase activity
of the ADAM10-WT plasmid, but not the ADAM10-Mut plasmid (Fig. 4B). Additionally, western blot assay
also revealed that the protein level of ADAM10 was reduced in GC
cells with miR-320a mimic, compared with control-transfected cells
(Fig. 4C). These findings
indicated that miR-320a directly targets to the 3′-UTR of ADAM10
and then suppresses its protein expression.
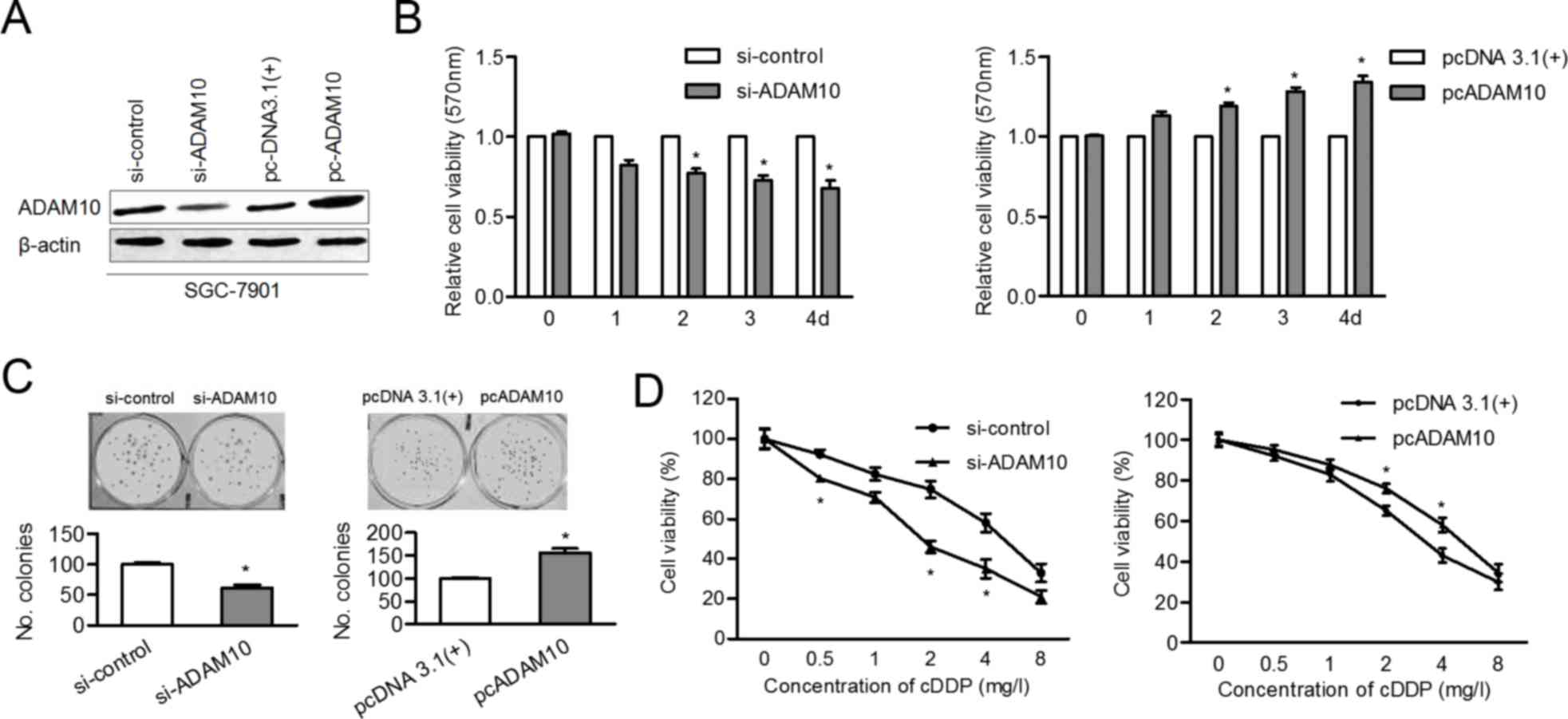
ADAM10 contributes to proliferation
and desensitization of GC cells to cisplatin
To clarify whether miR-320a regulates cell growth
and drug sensitivity by targeting ADAM10, the current study induced
knockdown or overexpression of ADAM10 in SGC-7901 cells. As
presented in Fig. 5A, the protein
level of ADAM10 was inhibited after siADAM10 transfection and ADAM
10 level was upregulated following cDNA transfection. The
proliferation ability of SGC-7901 cells was impaired by knockdown
of ADAM10 and promoted by overexpression of ADAM10 (Fig. 5B and C). Finally, the
cisplatin-sensitivity of SGC-7901 cells was enhanced by siADAM10,
whereas it was attenuated by overexpression of ADAM10 (Fig. 5D). These findings suggested that
ADAM10 is a functional target of miR-320a in GC development and
chemotherapy.
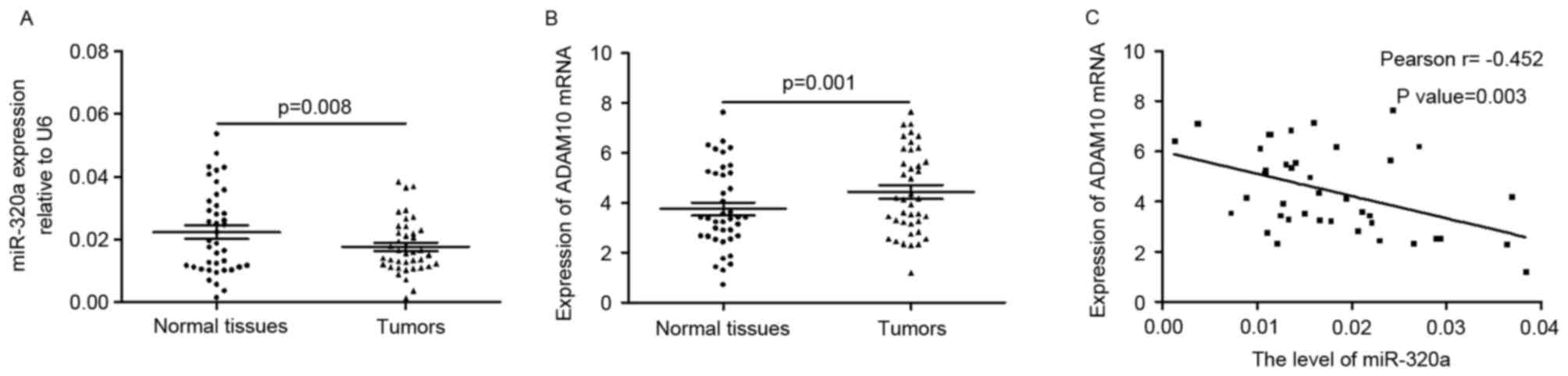
miR-320a is negatively correlated with
ADAM10 in tumors
The present study determined the expression level of
miR-320a in primary GC tissues and corresponding normal tissues. As
presented in Fig. 6A, the
expression of miR-320a was significantly downregulated in tumors
when compared with adjacent normal tissues (0.1766±0.0085 vs.
0.2230±0.0134; P=0.008). The mRNA level of ADAM10 was higher in
tumors compared with normal tissues (4.4278±1.6846 vs.
3.5373±1.6943; P=0.00; Fig. 6B).
Pearson correlation coefficient analysis revealed that the
expression of miR-320a was negatively correlated with the mRNA
levels of ADAM10 (r=-0.452; P=0.003; Fig. 6C) in tumor tissues. These findings
verified the negative correlation between miR-320a and ADAM10 in
tumors.
Discussion
Previous studies reported that miR-320a has a
functional role in proliferation, invasion and drug resistance of
various tumors (10–13,16).
However, to the best of our knowledge no studies examining the role
of miR-320a in GC development have been previously conducted. The
findings of the current study indicated that miR-320a inhibited GC
cell growth in vitro and in vivo. The sensitivity of
GC cells to cisplatin was increased by miR-320a overexpression,
whereas it was decreased in cells where miR-320a was downregulated.
ADAM10 was a direct target of miR-320a and involved in
miR-320a-regulated cell proliferation and
cisplatin-sensitivity.
RT-qPCR analysis was used in order to determine the
endogenous miR320a levels in GC cell lines and the data revealed
that miR-320a was reduced in all GC cells, suggesting the potential
functional role of miR-320a in gastric tumorigenesis. Subsequently,
by transfecting cells with mimics or inhibitor, the expression of
miR-320a was effectively upregulated or downregulated. The effect
of downregulated miR-320a on proliferative ability of GC cells was
assessed by colony formation and MTT assays in vitro, as
well as xenograft models in vivo. The presents study
determined the role of miR-320a in GC and suggested that it may
contribute to growth inhibition in gastric tumorigenesis. Similar
results were found in CRC, glioma, and breast cancer (12,14,30).
These findings indicated that miR-320a has an important role in the
inhibition of tumor growth in various types of tumor.
Considering miR-320a was also reported as a
chemotherapy-associated gene in tumors, the present study tested
whether miR-320a modulated cisplatin-sensitivity of GC cells. It
was determined that ectopic miR-320a expression significantly
enhanced cisplatin-sensitivity of GC cells. miR-320a was identified
to be significantly correlated with sensitivity to preoperative
chemoradiotherapy (31). Low
expression of miR-320a was correlated with shortened time to
imatinib resistance (32). The
function of miR-320a in chemoresistance revealed that restoration
of miR-320a may a provide novel therapeutic strategy for GC
treatment.
ADAM10, is a typical member of the ADAMs family,
which has been previously reported to be upregulated in various
types of cancers and contribute to cancer progression (33). The present study identified ADAM10
as a target of miR-320a. Knockdown of ADAM10 markedly inhibited
cell proliferation and colony formation. On the contrary,
overexpression of ADAM10 accelerated cell growth rate, which was in
consistent with a previous study (20). A previous clinical study revealed
that upregulated ADAM10 is associated with GC progression (34). The present study determined that
ADMA10 was significantly upregulated in tumors, suggesting ADAM10
involvement in gastric tumorigenesis. Direct evidence was provided
by the current study indicating that ADAM10 has an oncogene role in
GC by stimulating cell growth. It is of note that silencing of
ADAM10 impaired the cisplatin-sensitivity of GC cells, suggesting
ADAM10 may be a promising target for the improvement
chemotherapeutic efficacy in GC. The negative correlation between
miR-320a and ADMA10 in tumors also suggested the miR-320a/ADAM10
axis may have an important role in GC development.
In conclusion, to the best of our knowledge, the
present study demonstrated for the first time that downregulation
of miR-320a contributed to GC progression and chemoresistance by
targeting ADAM10. These findings collectively identified the
miR-320a/ADAM10 axis as a promising therapeutic tool for further GC
therapy.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Siegel R, Ward E, Murray T, Xu J
and Thun MJ: Cancer statistics, 2007. CA Cancer J Clin. 57:43–66.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shkumatava A, Stark A, Sive H and Bartel
DP: Coherent but overlapping expression of microRNAs and their
targets during vertebrate development. Genes Dev. 23:466–481. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ma F, Song H, Guo B, Zhang Y, Zheng Y, Lin
C, Wu Y, Guan G, Sha R, Zhou Q, et al: miR-361-5p inhibits
colorectal and gastric cancer growth and metastasis by targeting
staphylococcal nuclease domain containing-1. Oncotarget.
6:17404–17416. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ozen M, Karatas OF, Gulluoglu S, Bayrak
OF, Sevli S, Guzel E, Ekici ID, Caskurlu T, Solak M, Creighton CJ
and Ittmann M: Overexpression of miR-145-5p inhibits proliferation
of prostate cancer cells and reduces SOX2 expression. Cancer
Invest. 33:251–258. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ni F, Zhao H, Cui H, Wu Z, Chen L, Hu Z,
Guo C, Liu Y, Chen Z, Wang X, et al: MicroRNA-362-5p promotes tumor
growth and metastasis by targeting CYLD in hepatocellular
carcinoma. Cancer Lett. 356:809–818. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Organista-Nava J, Gómez-Gómez Y,
Illades-Aguiar B, Del Carmen Alarcón-Romero L, Saavedra-Herrera MV,
Rivera-Ramírez AB, Garzón-Barrientos VH and Leyva-Vázquez MA: High
miR-24 expression is associated with risk of relapse and poor
survival in acute leukemia. Oncol Rep. 33:1639–1649. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shen H, Shen J, Wang L, Shi Z, Wang M,
Jiang BH and Shu Y: Low miR-145 expression level is associated with
poor pathological differentiation and poor prognosis in non-small
cell lung cancer. Biomed Pharmacother. 69:301–305. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vilquin P, Donini CF, Villedieu M, Grisard
E, Corbo L, Bachelot T, Vendrell JA and Cohen PA: MicroRNA-125b
upregulation confers aromatase inhibitor resistance and is a novel
marker of poor prognosis in breast cancer. Breast Cancer Res.
17:132015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhao H, Dong T, Zhou H, Wang L, Huang A,
Feng B, Quan Y, Jin R, Zhang W, Sun J, et al: miR-320a suppresses
colorectal cancer progression by targeting Rac1. Carcinogenesis.
35:886–895. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang Y, He X, Liu Y, Ye Y, Zhang H, He P,
Zhang Q, Dong L, Liu Y and Dong J: microRNA-320a inhibits tumor
invasion by targeting neuropilin 1 and is associated with liver
metastasis in colorectal cancer. Oncol Rep. 27:685–694.
2012.PubMed/NCBI
|
|
12
|
Sun JY, Huang Y, Li JP, Zhang X, Wang L,
Meng YL, Yan B, Bian YQ, Zhao J, Wang WZ, et al: MicroRNA-320a
suppresses human colon cancer cell proliferation by directly
targeting β-catenin. Biochem Biophys Res Commun. 420:787–792. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qi X, Li J, Zhou C, Lv C and Tian M:
MicroRNA-320a inhibits cell proliferation, migration and invasion
by targeting BMI-1 in nasopharyngeal carcinoma. FEBS Lett.
588:3732–3738. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang B, Yang Z, Wang H, Cao Z, Zhao Y,
Gong C, Ma L, Wang X, Hu X and Chen S: MicroRNA-320a inhibits
proliferation and invasion of breast cancer cells by targeting
RAB11A. Am J Cancer Res. 5:2719–2729. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li X, Lu Y, Chen Y, Lu W and Xie X:
MicroRNA profile of paclitaxel-resistant serous ovarian carcinoma
based on formalin-fixed paraffin-embedded samples. BMC Cancer.
13:2162013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lü M, Ding K, Zhang G, Yin M, Yao G, Tian
H, Lian J, Liu L, Liang M, Zhu T and Sun F: MicroRNA-320a
sensitizes tamoxifen-resistant breast cancer cells to tamoxifen by
targeting ARPP-19 and ERRγ. Sci Rep. 5:87352015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lian F, Cui Y, Zhou C, Gao K and Wu L:
Identification of a plasma four-microRNA panel as potential
noninvasive biomarker for osteosarcoma. PLoS One. 10:e01214992015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wen Y, Han J, Chen J, Dong J, Xia Y, Liu
J, Jiang Y, Dai J, Lu J, Jin G, et al: Plasma miRNAs as early
biomarkers for detecting hepatocellular carcinoma. Int J Cancer.
137:1679–1690. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fang Z, Tang J, Bai Y, Lin H, You H, Jin
H, Lin L, You P, Li J, Dai Z, et al: Plasma levels of microRNA-24,
microRNA-320a, and microRNA-423-5p are potential biomarkers for
colorectal carcinoma. J Exp Clin Cancer Res. 34:862015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
You B, Shan Y, Shi S, Li X and You Y:
Effects of ADAM10 upregulation on progression, migration, and
prognosis of nasopharyngeal carcinoma. Cancer Sci. 106:1506–1514.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Arima T, Enokida H, Kubo H, Kagara I,
Matsuda R, Toki K, Nishimura H, Chiyomaru T, Tatarano S, Idesako T,
et al: Nuclear translocation of ADAM-10 contributes to the
pathogenesis and progression of human prostate cancer. Cancer Sci.
98:1720–1726. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gaida MM, Haag N, Günther F, Tschaharganeh
DF, Schirmacher P, Friess H, Giese NA, Schmidt J and Wente MN:
Expression of A disintegrin and metalloprotease 10 in pancreatic
carcinoma. Int J Mol Med. 26:281–288. 2010.PubMed/NCBI
|
|
23
|
Jones AV, Lambert DW, Speight PM and
Whawell SA: ADAM 10 is over expressed in oral squamous cell
carcinoma and contributes to invasive behaviour through a
functional association with αvβ6 integrin. FEBS Lett.
587:3529–3534. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pan Y, Han C, Wang C, Hu G, Luo C, Gan X,
Zhang F, Lu Y and Ding X: ADAM10 promotes pituitary adenoma cell
migration by regulating cleavage of CD44 and L1. J Mol Endocrinol.
49:21–33. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Guo J, He L, Yuan P, Wang P, Lu Y, Tong F,
Wang Y, Yin Y, Tian J and Sun J: ADAM10 overexpression in human
non-small cell lung cancer correlates with cell migration and
invasion through the activation of the Notch1 signaling pathway.
Oncol Rep. 28:1709–1718. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang CL, Jiang FQ, Xu F and Jiang GX:
ADAM10 overexpression confers resistance to doxorubicin-induced
apoptosis in hepatocellular carcinoma. Tumour Biol. 33:1535–1541.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jing P, Sa N, Liu X, Liu X and Xu W:
MicroR-140-5p suppresses tumor cell migration and invasion by
targeting ADAM10-mediated Notch1 signaling pathway in
hypopharyngeal squamous cell carcinoma. Exp Mol Pathol.
100:132–138. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ergün S, Ulasli M, Igci YZ, Igci M,
Kırkbes S, Borazan E, Balik A, Yumrutaş Ö, Camci C, Cakmak EA, et
al: The association of the expression of miR-122-5p and its target
ADAM10 with human breast cancer. Mol Biol Rep. 42:497–505. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Guo T, Feng Y, Liu Q, Yang X, Jiang T,
Chen Y and Zhang Q: MicroRNA-320a suppresses in GBM patients and
modulates glioma cell functions by targeting IGF-1R. Tumour Biol.
35:11269–11275. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Salendo J, Spitzner M, Kramer F, Zhang X,
Jo P, Wolff HA, Kitz J, Kaulfuß S, Beißbarth T, Dobbelstein M, et
al: Identification of a microRNA expression signature for
chemoradiosensitivity of colorectal cancer cells, involving
miRNAs-320a, −224, −132 and let7g. Radiother Oncol. 108:451–457.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gao X, Shen K, Wang C, Ling J, Wang H,
Fang Y, Shi Y, Hou Y, Qin J, Sun Y and Qin X: MiR-320a
downregulation is associated with imatinib resistance in
gastrointestinal stromal tumors. Acta Biochim Biophys Sin
(Shanghai). 46:72–75. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Przemyslaw L, Boguslaw HA, Elzbieta S and
Malgorzata SM: ADAM and ADAMTS family proteins and their role in
the colorectal cancer etiopathogenesis. BMB Rep. 46:139–150. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang YY, Ye ZY, Li L, Zhao ZS, Shao QS and
Tao HQ: ADAM 10 is associated with gastric cancer progression and
prognosis of patients. J Surg Oncol. 103:116–123. 2011. View Article : Google Scholar : PubMed/NCBI
|