Introduction
Colorectal cancer (CRC), one of the most commonly
diagnosed cancers worldwide, is the third most prevalent and fourth
most frequent cause of cancer-related mortality (1). CRC is the third most frequently
reported cancer in males and the second most frequently reported
cancer in females (2). Globally,
approximately 1.36 million new CRC cases are diagnosed and 694,000
deaths due to CRC are recorded every year (3). Patient prognosis remains poor despite
the remarkable progress in the diagnosis and treatment of CRC
(4). Recurrence following curative
surgery and metastasis are the main reasons for the unsatisfactory
prognosis of CRC patients (5).
Therefore, fully understanding the mechanisms that underlie CRC
formation and progression is essential in identifying potential
biomarkers for CRC and improving the prognosis and treatment of CRC
patients.
MicroRNAs (miRNAs) constitute a large family of
endogenous and small noncoding RNA molecules that are 18–24
nucleotides long (6). miRNAs
negatively modulate the expression of corresponding target
messenger RNAs by binding to their 3′-untranslated regions
(3′-UTRs). This process causes translational repression or mRNA
degradation (7). Bioinformatics
studies have demonstrated that miRNAs account for approximately 1%
of all human genes and could regulate approximately 60% of human
protein-coding genes (8–10). miRNAs are aberrantly expressed in
various types of human cancers, including CRC (11,12).
MiRNA dysregulation is involved in tumor occurrence and progression
by regulating numerous biological functions, such as cellular
proliferation, cycle, differentiation, apoptosis,
epithelial-mesenchymal transition (EMT), migration, invasion,
metastasis, angiogenesis and chemoresistance (9,13,14).
Increasing evidence has indicated that miRNAs act as oncogenes or
tumour suppressors in tumorigenesis and tumor development by
regulating corresponding target genes (15–17).
Therefore, investigating the expression pattern and biological
roles of miRNAs in CRC would provide novel and effective
therapeutic targets for patients with this disease.
miR-760 is abnormally expressed in breast (18) and ovarian cancers (19). Plasma miR-760 expression is lower
in CRC patients than in healthy participants (20). However, miR-760 expression, its
roles and underlying regulatory mechanism in CRC tissues remain to
be fully elucidated. Therefore, the present study aims to
investigate the expression and roles of miR-760 in CRC and
determine the underlying regulatory mechanism. The results of this
study, for the first time, demonstrated that miR-760 was
downregulated in CRC tissues and cell lines. The low expression
levels of miR-760 were associated with the tumor size, lymph node
metastasis and TNM stage of CRC. In addition, miR-760 upregulation
suppressed CRC cell proliferation and invasion in vitro.
Moreover, SP1 was confirmed to be a novel direct target gene of
miR-760 in CRC cells. miR-760 was found to be participated in the
regulation of PTEN/AKT pathway in CRC was the novelty of this
research.
Materials and methods
Tissue samples
This study was approved by the Ethics Committee of
Linyi Central Hospital. Written informed consents were also
obtained from patients prior to sampling. The CRC tissues and
matched adjacent normal tissues were surgically resected from 49
patients with CRC in the Department of General Surgery, Linyi
Central Hospital, between April 2014 to March 2016. All these CRC
patients did not undergo chemotherapy or radiotherapy before
surgery. All tissue samples were immediately snap-frozen in liquid
nitrogen and stored at −80°C until use.
Cell lines and transfection
Human CRC cell lines (HCT116, HT29, LoVo, SW480 and
SW620) and normal human colon epithelium cell line FHC were
acquired from American Type Culture Collection (Manassas, VA, USA).
All cells were cultured in Dulbecco's modified Eagle's medium
(DMEM) supplemented with 10% fetal bovine serum (FBS) (both from
Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 100 U/ml
penicillin and 100 mg/ml streptomycin in a humidified incubator
with a mixture of 5% CO2 at 37°C.
miR-760 mimics and corresponding negative control
miRNA (miR-NC) were obtained from GenePharma (Shanghai, China).
specificity protein 1 (SP1) overexpression plasmid, pcDNA3.1-SP1
and blank plasmid pcDNA3.1 were synthesized by Chinese Academy of
Sciences (Changchun, China). Cells were seeded in 6-well plates
with a density of 8×105 cells each well. After
incubation overnight, cells were transfected with these
oligonucleotides using Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.), according to the manufacturer's
instructions. Transfected cells were then incubated at 37°C with 5%
CO2. After incubation 6 h, cell culture medium was
replaced with fresh DMEM containing 10% FBS.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA of clinical tissue specimens and cells
were isolated with TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's instructions. To
determine miR-760 (accession no. MI0005567) expression level, total
RNA (1 µg) was reverse-transcribed to cDNA using TaqMan MicroRNA
Reverse Transcription kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.). Real-time PCR was carried out with TaqMan
MicroRNA PCR kit on the Applied Biosystems 7500 Sequence Detection
system (both from Applied Biosystems; Thermo Fisher Scientific,
Inc.). The reaction system contained 1 µl TaqMan® small
RNA assay (20X), 1.33 µl cDNA, 10 µl TaqMan® Universal
PCR Master Mix II (2X) and 7.67 µl nuclease-free water. The cycling
conditions were as follows: 50°C for 2 min, 95°C for 10 min; 40
cycles of denaturation at 95°C for 15 sec; and annealing/extension
at 60°C for 60 sec. To detect SP1 (accession no. NM_003109) mRNA
expression, cDNA was synthesized using M-MLV reverse transcriptase
(Fermentas; Thermo Fisher Scientific, Inc., Pittsburgh, PA, USA).
The relative expression of SP1 mRNA was detected using SYBR Premix
Ex Taq™ (Takara Biotechnology Co., Ltd., Dalian, China).
The reaction system contained 10 µl SYBR Premix Ex Taq, 2 µl cDNA
(200 ng), 0.8 µl forward primer, 0.8 µl reverse primer, 0.4 µl ROX
reference dye and 6 µl ddH2O. The amplification was
performed with cycling conditions as follows: 5 min at 95°C,
followed by 40 cycles of 95°C for 30 sec and 65°C for 45 sec. U6
small nuclear RNA (U6) and glyceraldehyde-3-phosphate dehydrogenase
(GAPDH) were used as endogenous control in the detection of miR-760
and SP1 mRNA, respectively. The primers were designed as follows:
miR-760, 5′-GTCGAGCGGCTCTGGGTCTGTG-3′ (forward) and
5′-TCCAGTGCAGGGTCCGAGGT-3′ (reverse); U6, 5′-CTCGCTTCGGCAGCACA-3′
(forward) and 5′-AACGCTTCACGAATTTGCGT-3′ (reverse); SP1,
5′-TGGTGGGCAGTATGTTGT-3′ (forward) and 5′-GCTATTGGCATTGGTGAA−3′
(reverse); and GAPDH, 5′-AGAAGGCTGGGGCTCATTTG-3′ (forward) and
5′-AGGGGCCTCCACAGTCTTC-3′ (reverse). Relative expression levels
were calculated using the 2−∆∆Ct method (21). Each assay was performed in
triplicate and repeated three times.
Cell Counting Kit (CCK)8 assay
CCK8 assay was performed to measure cell
proliferation. Transfected cells were collected at 24 h
post-transfection, and seeded into 96-well plates at a density of
3×103 cells/well. Cells were then maintained at 37°C
with 5% CO2 for 0, 24, 48 or 72 h. At these time-points,
10 µl of CCK8 solution (Dojindo Molecular Technologies, Kumamoto,
Japan) were added into each well, and the cells were incubated for
addition 2 h at 37°C. The absorbance at a wavelength of 450 nm was
determined using Victor 3 Multi-Label microplate reader
(PerkinElmer, Inc., Waltham, MA, USA). Each assay was performed in
five parallel wells and repeated three times.
Cell invasion assay
Cell invasion assays were performed in 24-well
Transwell® chambers with 8 µm pores (Costar; Corning
Incorporated, Corning, NY, USA) coated with Matrigel (BD
Biosciences, Franklin Lakes, NJ, USA). Transfected cells were
harvested 48 h following transfection. Transfected cells
(5.0×104) in FBS-free medium were added into the upper
chamber, and 600 µl DMEM medium with 10% FBS was supplemented into
the matched lower chamber. After 24 h incubation at 37°C with 5%
CO2, the non-invading cells were wiped out carefully
with cotton swabs. Cells that invaded to the bottom chamber were
fixed with 100% methanol, stained with 0.1% crystal violet, washed
in PBS and dried in air. The invasive cells were photographed and
counted in five randomly selected visual fields under an inverted
microscope (×200 magnifications; Olympus Corporation, Tokyo,
Japan). Each assay was repeated three times.
Bioinformatics analysis
The potential target genes of miR-760 was predicted
using TargetScan (www.targetscan.org/) and miRanda (www.microrna.org).
Luciferase reporter assay
The luciferase plasmids, including the
psiCHECK2-SP1-3′-UTR-wild-type (Wt) and psiCHECK2-SP1-3′-UTR-mutant
(Mut), were synthesized and obtained from GenePharma. For the
luciferase reporter assays, cells were seeded into 24-well plates
at a density of 60–70% confluence, and co-transfected with
luciferase reporter plasmid, and miR-760 mimics or miR-NC using
Lipofectamine 2000, according to the manufacturer's instructions.
After 48 h of transfection, the firefly and Renilla
luciferase activities were detected with Dual-Luciferase Reporter
Assay system (Promega, Manheim, Germany) in accordance with the
manufacturer's suggestions. Renilla luciferase activity was
normalized to firefly luciferase activity. Each assay was repeated
three times.
Western blotting
Whole protein extracts form tissues and cells were
lysed by ice-cold radioimmunoprecipitation assay (RIPA) buffer
(Beyotime Institute of Biotechnology, Haimen, China), according to
the manufacturer's protocol. The concentration of total protein was
measured using a BCA protein assay kit (Beyotime Institute of
Biotechnology, Haimen, China). Equal amounts of protein were
separated through a 10% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) gel and transferred onto nitrocellulose
membranes (Millipore, Billerica, MA, USA).
The membranes were then blocked in 5% nonfat milk in
TBST and incubated with primary antibodies overnight at 37°C: Mouse
anti-human monoclonal SP1 antibody (1:1,000 dilution; sc-420),
mouse anti-human monoclonal PTEN antibody (1:1,000 dilution;
sc-7974), mouse anti-human monoclonal p-AKT antibody (1:1,000
dilution; sc-271966), mouse anti-human monoclonal AKT antibody
(1:1,000 dilution; sc-56878), and mouse anti-human monoclonal GAPDH
antibody (1:1,000 dilution; sc-32233) (all from Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA). After washing three
times with TBST, the membranes were probed with a goat-anti-mouse
horseradish peroxidase (HRP)-conjugated secondary antibody (1:5,000
dilution; sc-2005; Santa Cruz Biotechnology, Inc.). Protein bands
were visualized by incubating the membranes with ECL detection kit
(GE Healthcare Life Sciences, Chalfont, UK). Protein expression
levels were normalized to GAPDH. Each assay was repeated three
times.
Statistical analysis
Data are presented as the mean ± standard deviation
and compared using SPSS software (version 13.0; SPSS, Inc.,
Chicago, IL, USA). The differences between two groups were analyzed
using Students t-test, or assessed by one-way ANOVA when there were
more than two groups. Student-Newman-Keuls test was used as a post
hoc test following ANOVA. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-760 is frequently downregulated in
CRC tissues and cell lines
RT-qPCR was used to detect miR-760 expression levels
in 49 CRC tissue samples and matching adjacent normal tissue
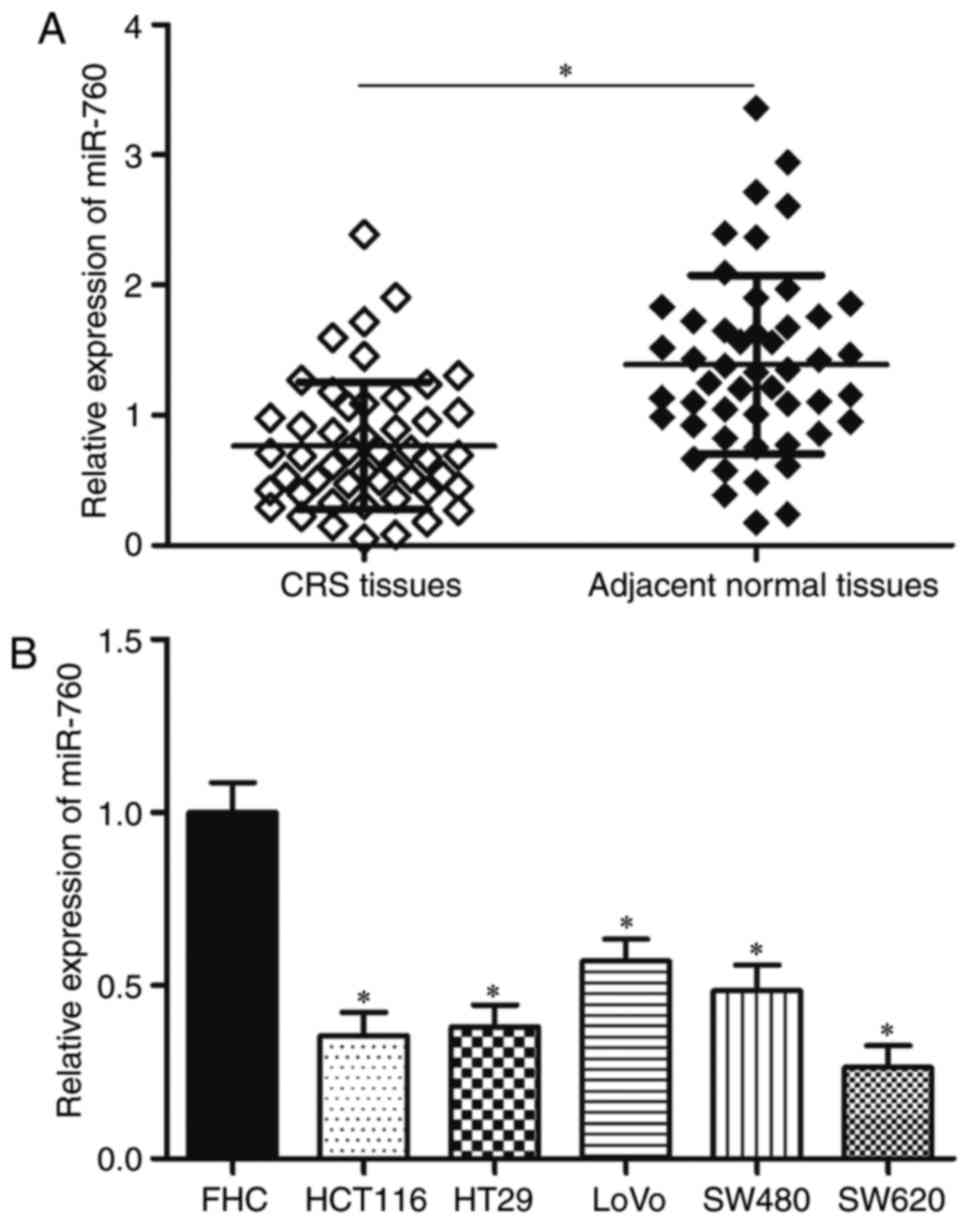
samples. The results showed that miR-760 expression was lower in
CRC tissues than in matching adjacent normal tissues (Fig. 1A, P<0.05). Then, miR-760
expression levels in the CRC cell lines HCT116, HT29, LoVo, SW480
and SW620 and in the normal human colon epithelial cell line FHC
were determined. RT-qPCR results revealed that miR-760 was
significantly lower in CRC cell lines compared with in FHC
(Fig. 1B, P<0.05). These
results suggested that miR-760 might play important roles in CRC
progression.
miR-760 underexpression is correlated
with the adverse clinicopathological parameters of CRC
patients
To investigate the correlation of miR-760 expression
with the clinicopathological factors of CRC, patients were divided
into miR-760 low- and miR-760 high-expression groups based on the
median expression of miR-760. As shown in Table I, low miR-760 expression was
associated with the tumor size (P=0.015), lymph node metastasis
(P=0.027) and TNM stage (P=0.006) of CRC. However, miR-760
expression was not correlated with sex (P=0.483), age (P=0.282),
tumor location (P=0.680) or differentiation (P=0.470). These
results suggested that miR-760 might be a prognostic biomarker for
CRC patients.
 | Table I.Correlation between microRNA-760
expression and clinicopathological factors of colorectal
cancer. |
Table I.
Correlation between microRNA-760
expression and clinicopathological factors of colorectal
cancer.
| Clinicopathological
factors | No. of cases | miR-760 low
group | miR-760 high
group | P-value |
|---|
| Sex |
|
|
| 0.483 |
|
Male | 31 | 17 | 14 |
|
|
Female | 18 | 8 | 10 |
|
| Age (years) |
|
|
| 0.282 |
|
<55 | 18 | 11 | 7 |
|
|
≥55 | 31 | 14 | 17 |
|
| Tumor location |
|
|
| 0.680 |
|
Colon | 28 | 15 | 13 |
|
|
Rectum | 21 | 10 | 11 |
|
| Tumor
differentiation |
|
|
| 0.470 |
| Well
and Moderate | 35 | 19 | 16 |
|
|
Poor | 14 | 6 | 8 |
|
| Tumor size
(cm) |
|
|
| 0.015 |
|
<5 | 26 | 9 | 17 |
|
| ≥5 | 23 | 16 | 7 |
|
| Lymph node
metastasis |
|
|
| 0.027 |
|
Absence | 29 | 11 | 18 |
|
|
Presence | 20 | 14 | 6 |
|
| TNM stage |
|
|
| 0.006 |
|
I–II | 21 | 6 | 15 |
|
|
III–IV | 28 | 19 | 9 |
|
miR-760 upregulation represses cell
proliferation and invasion in CRC
Given that miR-760 is significantly downregulated in
CRC, the tumor-suppressing roles of miR-760 in CRC were examined.
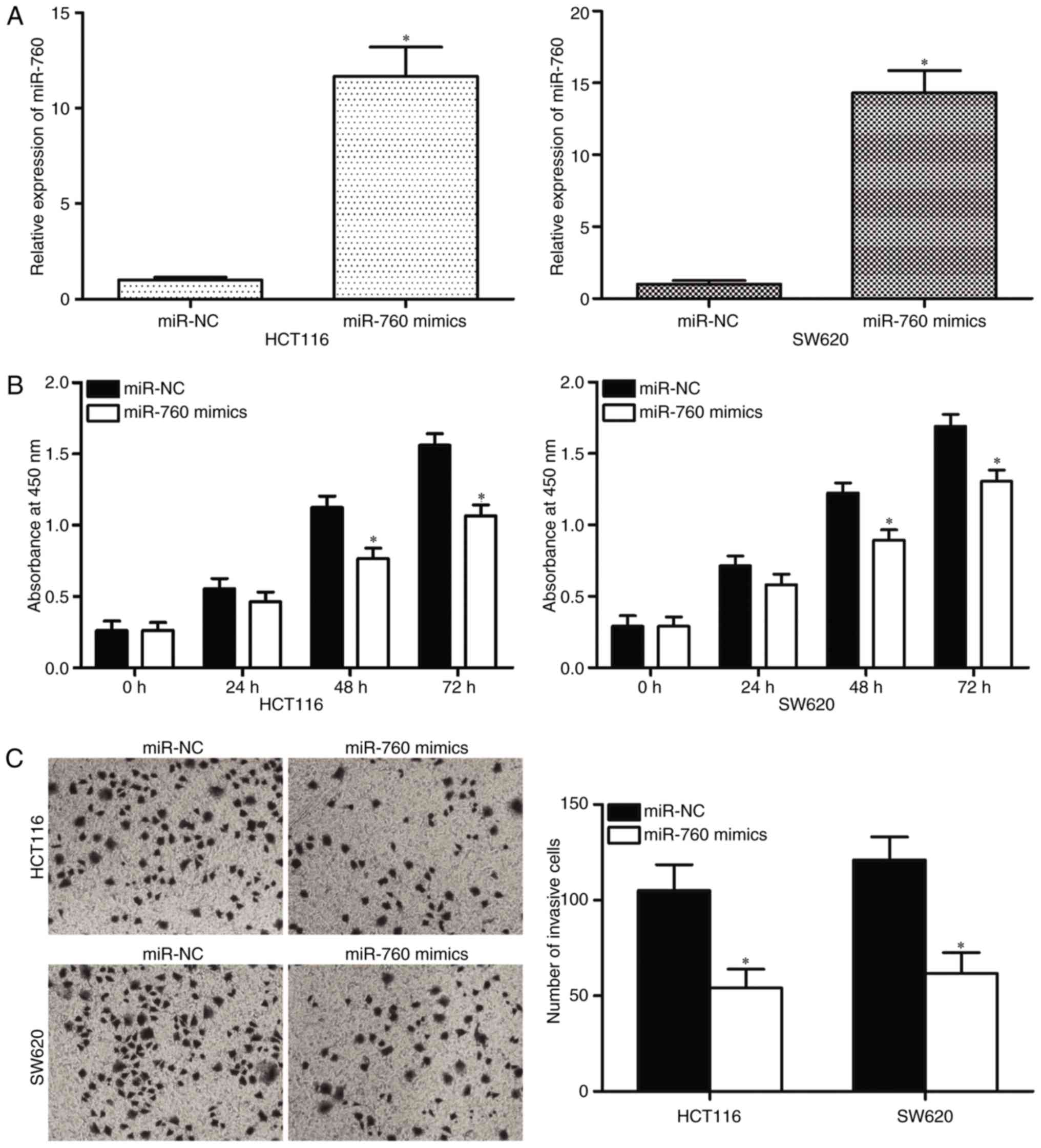
HCT116 and SW620 cells, which both express low levels of endogenous
miR-760, were selected for the transfection of miR-760 mimics.
RT-qPCR was performed to determine transfection efficiency, and the
results indicated that miR-760 was markedly upregulated in HCT116
and SW620 cells transfected with miR-760 mimics (Fig. 2A, P<0.05). The effects of
miR-760 overexpression on the cell proliferation and invasion
capacity of CRC were investigated. CCK8 assay results revealed that
the ectopic expression of miR-760 attenuated HCT116 and SW620 cell
proliferation compared with transfection with miR-NC (Fig. 2B, P<0.05). Cell invasion assays
indicated that the restored expression of miR-760 in HCT116 and
SW620 cells significantly inhibited cell invasion capacities
compared with that in miR-NC groups (Fig. 2C, P<0.05). These results
demonstrate that miR-760 may act as a tumor suppressor in CRC
progression.
SP1 is a direct target of miR-760 in
CRC
To determine the molecular mechanisms of miR-760 in
the regulation of CRC cell proliferation and invasion,
bioinformatics analysis was used to predict the potential target
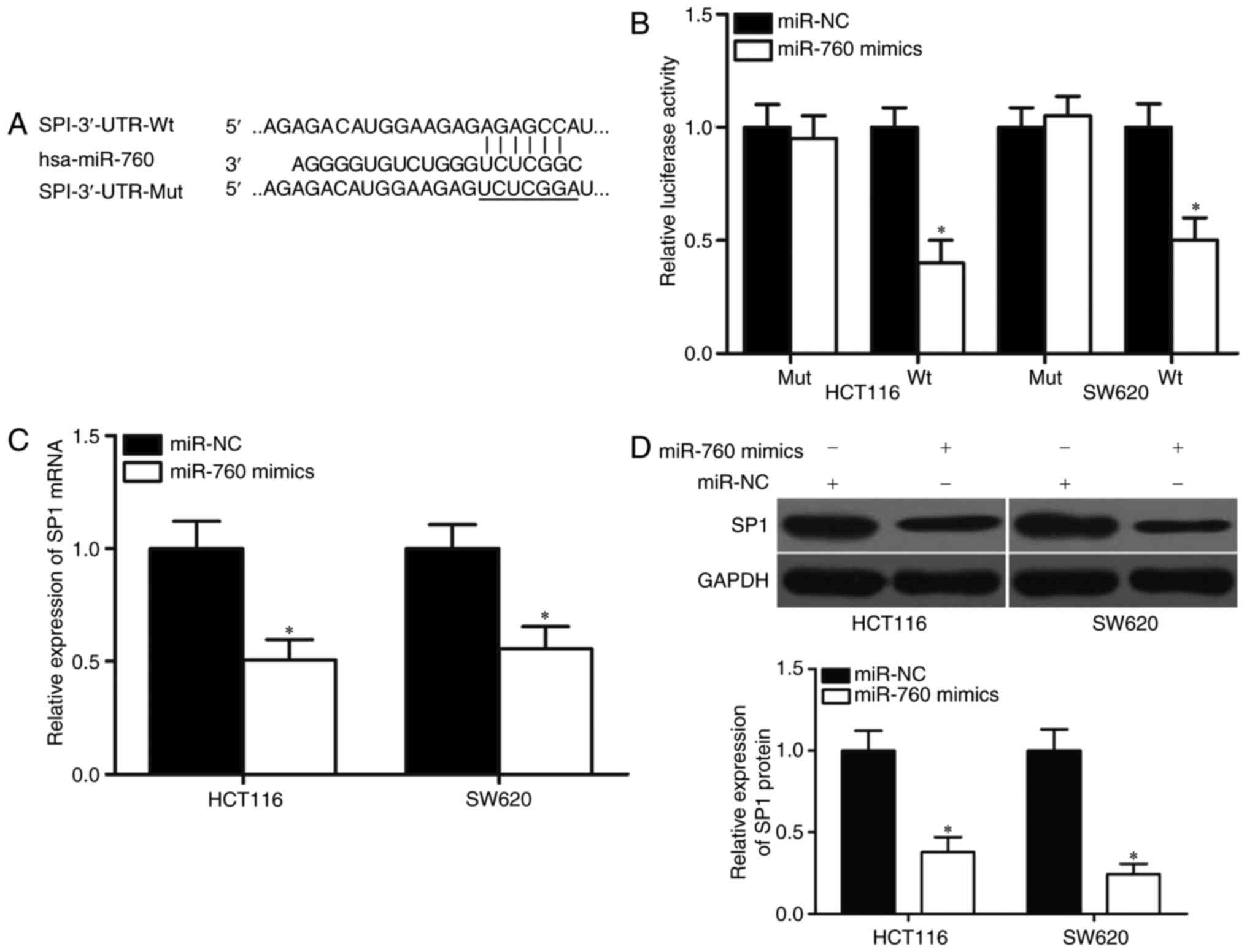
genes of miR-760. Among candidate genes, the SP1 gene (Fig. 3A), which is upregulated in CRC and
is associated with CRC progression (22–27),
was identified as a major target of miR-760 and selected for
further analysis. Luciferase reporter assays were performed on
HCT116 and SW620 cells transfected with luciferase plasmids that
contained the wild-type or mutant miR-760-binding site in the SP1
3′-UTR, together with miR-760 mimics or miR-NC. As shown in
Fig. 3B, luciferase activities in
the reporter that contained the wild-type SP1 3′-UTR markedly
decreased upon cotransfection with miR-760 mimics (P<0.05),
whereas those in the reporter that contained the mutant binding
site were unaffected.
Furthermore, the mRNA and protein levels of SP1 in
HCT116 and SW620 cells that were transfected with miR-760 mimics or
miR-NC were detected using RT-qPCR and Western blot. The results
showed that miR-760 overexpression decreased SP1 expression in
HCT116 and SW620 cells at the mRNA and protein levels (Fig. 3C and D, P<0.05). These findings
suggested that SP1 is a direct target of miR-760 in CRC.
SP1 is upregulated in CRC tissues and
its expression is inversely correlated with miR-760 expression
Given that SP1 is a direct target gene of miR-760 in
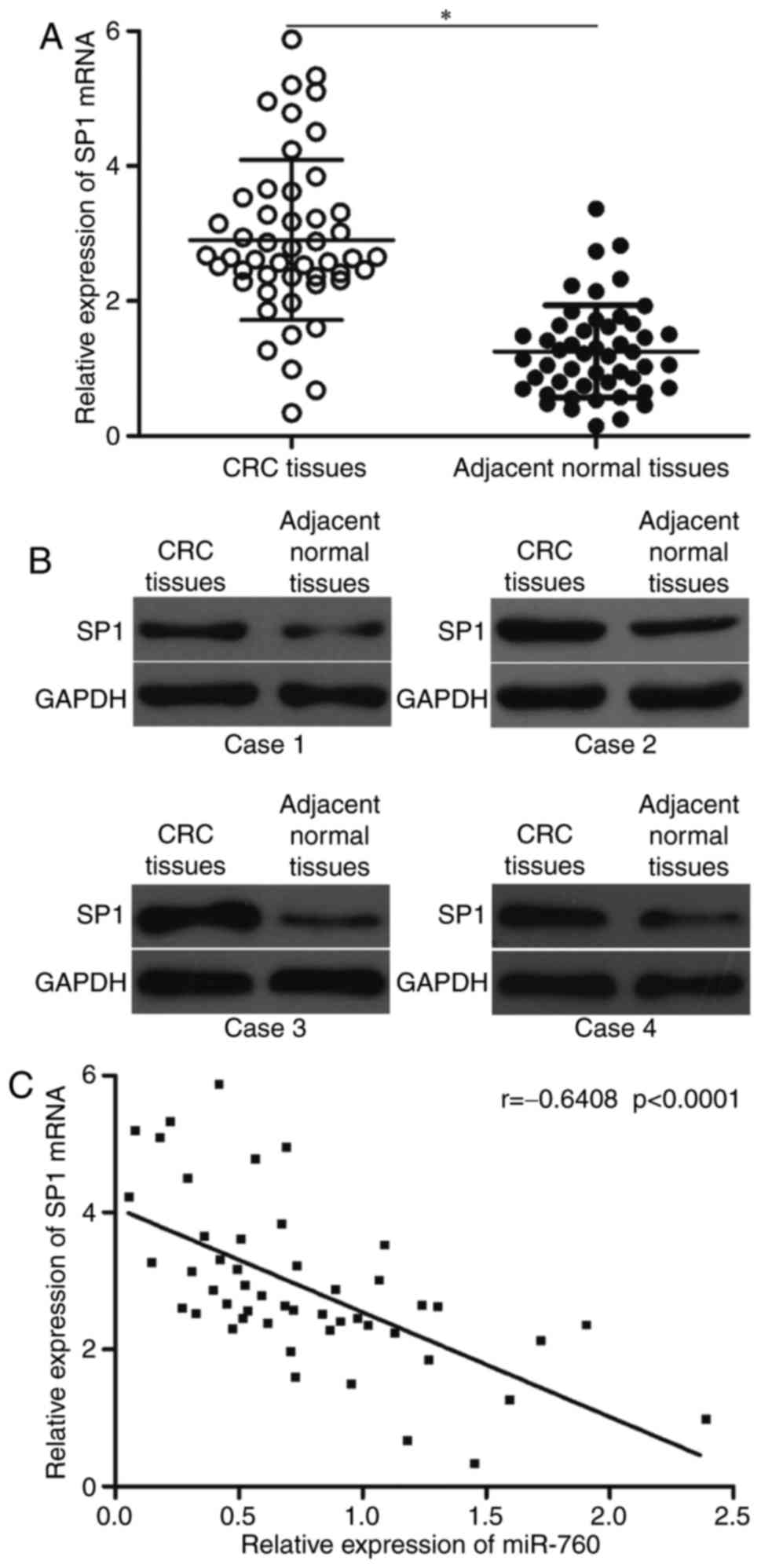
CRC, its expression in CRC tissues was measured and its association
with miR-760 expression levels was investigated. SP1 expression at
the mRNA and protein levels significantly increased in CRC tissues
compared with that in the matching adjacent normal tissues
(Fig. 4A and B). In addition,
Spearman's correlation analysis revealed an inverse association
between miR-760 and SP1 mRNA levels in CRC tissues (Fig. 4C; r=-0.6408, P<0.0001).
SP1 reverses the tumor-suppressing
effects of miR-760 on CRC cell proliferation and invasion
Given that SP1 is a direct target of miR-760, rescue
experiments were performed to determine whether SP1 restoration
could abolish the tumor-suppressing roles of miR-760 in CRC cells.
HCT116 and SW620 cells were transfected with miR-760 mimics with or
without SP1 overexpression (pcDNA3.1-SP1). Western blot analysis
indicated that SP1 was downregulated in HCT116 and SW620 cells
after transfection with miR-760 mimics; meanwhile, pcDNA3.1-SP1
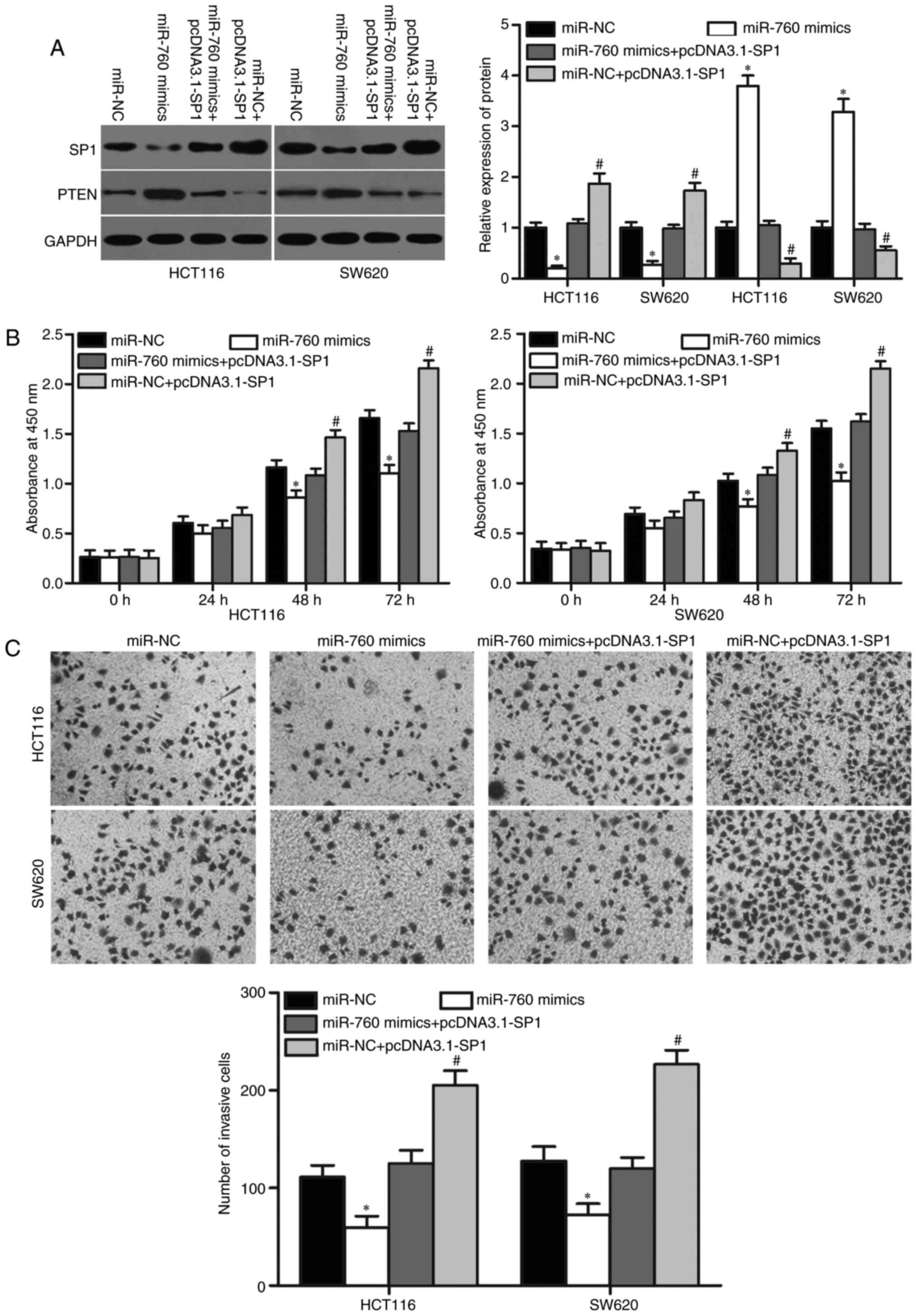
cotransfection could recover SP1 expression (Fig. 5A, P<0.05) SP1 was identified as
an important regulator of PTEN in cancer (28,29).
Hence, we detected PTEN expression in above cells. As shown in
Fig. 5A, PTEN expression was
downregulated in HCT116 and SW620 cells transfected with miR-760
mimics, and pcDNA3.1-SP1 cotransfection could recover PTEN
expression. Moreover, cotansfection of miR-NC and pcDNA3.1-SP1
could decrease PTEN expression (Fig.
5A, P<0.05).
Subsequently, CCK8 and cell invasion assays revealed
that SP1 upregulation markedly reversed the inhibitory effects of
miR-760 overexpression on cell proliferation (Fig. 5B, P<0.05) and invasion (Fig. 5C, P<0.05) in HCT116 and SW620
cells. Collectively, these results suggested that miR-760 partly
inhibits CRC cell proliferation and invasion by regulating SP1.
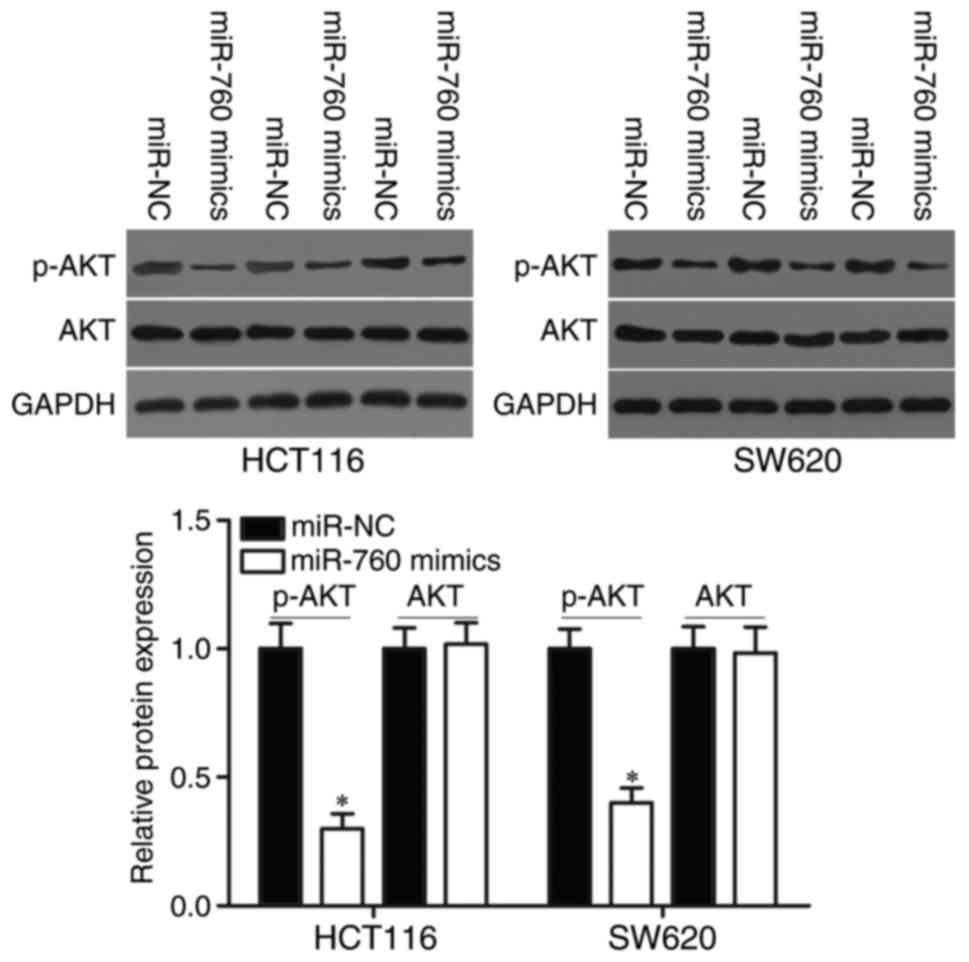
MiR-760 regulates the PTEN/AKT pathway
in CRC
We demonstrated that miR-760 participated in the
regulation of PTEN expression in CRC cells. Therefore, AKT and
p-AKT protein expression were measured in HCT116 and SW620 cells
transfected with miR-760 mimics or miR-NC. As shown in Fig. 6, miR-760 overexpression decreased
p-AKT expression without changing total AKT expression in HCT116
and SW620 cells (P<0.05). These results suggested that miR-760
directly targets SP1 and indirectly regulates the PTEN/AKT
signalling pathways, thus playing tumor-suppressing roles in
CRC.
Discussion
Dysregulated miRNAs have been recently implicated in
the development and progression of different cancers (30–32).
Further investigating the miRNAs involved in CRC formation and
progression may lead to the development of more effective
therapeutic strategies for CRC. Previous studies have reported that
plasma miR-760 expression is lower in CRC patients than in healthy
participants. Plasma miR-760 expression has a significant
diagnostic value for advanced neoplasia (20). However, the biological roles and
molecular mechanism of miR-760 expression in CRC tissues remain to
be fully elucidated.
The present study revealed that miR-760 is
significantly downregulated in CRC tissues and cell lines compared
with in matching adjacent normal tissues and the normal human colon
epithelial cell line FHC. Further correlation analysis showed that
downregulated miR-760 expression is associated with the tumor size,
lymph node metastasis and TNM stage of CRC. Cell function
investigation showed that miR-760 upregulation inhibits CRC cell
proliferation and invasion in vitro. SP1 was confirmed as a
novel direct target of miR-760 in CRC. Moreover, miR-760 was found
to regulate PTEN/AKT signalling pathway in CRC. These results
suggested that miR-760 might serve as a novel biomarker and
therapeutic target for CRC.
miR-760 is involved in the development and progress
of certain cancers. For example, miR-760 expression is
downregulated in doxorubicin (DOX)-resistant MCF-7/DOX cells and in
chemoresistant breast cancer tissues. MiR-760 overexpression
increases the chemosensitivity of breast cancer cells to anticancer
agents. Additionally, the restored expression of miR-760 decreases
the expression level of Nanog, a transcriptional factor involved in
chemoresistance, thus reversing EMT in breast cancer cells
(33). Han et al reported
that miR-760 upregulation represses the subpopulation of cancer
stem cells and the proliferation and migration of breast cancer
cells (18). miR-760 is
upregulated in the tumor tissues and cell lines of ovarian cancer.
The high level of miR-760 expression is associated with an
aggressive phenotype and poor prognosis in ovarian cancer. miR-760
acts as an oncogene in ovarian cancer by promoting cellular
proliferation (19). These
findings suggest that miR-760 is a potential target for the
treatment of these cancers.
miRNAs negatively regulate their target genes by
binding to the 3′UTR. Therefore, identifying the direct target
genes of miR-760 is important in understanding the roles of miR-760
in tumorigenesis and tumor development. Several miR-760 targets,
including RHOB (34), ANGOTL4
(34), ABCA1 (34) and NANOG (18) in breast cancer and PHLPP2 (19) in ovarian cancer, have been
identified. In this study, SP1 was identified as a novel and
functional target of miR-760 in CRC. SP1, a sequence-specific
DNA-binding protein, is located at 12q13.1 and encodes a protein of
785 amino acids (35). SP1 is
highly expressed in multiple types of human cancers, such as
gastric cancer (36),
hepatocellular carcinomas (37),
prostate cancer (38), thyroid
cancer (39), breast cancer
(40), pancreatic cancer (41) and lung cancer (23). SP1 plays important roles in
numerous pathophysiological processes, such as cell growth,
differentiation, apoptosis, survival, metastasis and invasion
(42). SP1 expression is increased
in CRC tumor tissues (27).
Subsequent functional assays have demonstrated that SP1 acts as an
oncogene in CRC progression by regulating cell proliferation,
apoptosis and metastasis (22–26).
Therefore, SP1 may be developed as a therapeutic target for the
suppression of tumorigenesis and tumor development in CRC.
In conclusion, this research demonstrated that
miR-760 is downregulated in CRC and is markedly associated with
cancer development. The restored expression of miR-760 in CRC cells
inhibits cell proliferation and invasion through the regulation of
SP1/PTEN/AKT signalling pathways, thus indicating that miR-760 has
a therapeutic value in CRC. However, future studies are needed to
investigate the feasibility of exploiting the miR-760/SP1 pathway
in a therapeutic approach for CRC. The absence of the normal colon
epithelium cell line (FHC) as a control group may have been a
weakness of the study, and that this is something that will be
included in future studies.
References
|
1
|
Center MM, Jemal A, Smith RA and Ward E:
Worldwide variations in colorectal cancer. CA Cancer J Clin.
59:366–378. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Haggar FA and Boushey RP: Colorectal
cancer epidemiology: Incidence, mortality, survival, and risk
factors. Clin Colon Rectal Surg. 22:191–197. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Manfredi S, Lepage C, Hatem C, Coatmeur O,
Faivre J and Bouvier AM: Epidemiology and management of liver
metastases from colorectal cancer. Ann Surg. 244:254–259. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010.PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lim LP, Glasner ME, Yekta S, Burge CB and
Bartel DP: Vertebrate microRNA genes. Science. 299:15402003.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Oliveto S, Mancino M, Manfrini N and Biffo
S: Role of microRNAs in translation regulation and cancer. World J
Biol Chem. 8:45–56. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Manikandan J, Aarthi JJ, Kumar SD and
Pushparaj PN: Oncomirs: The potential role of non-coding microRNAs
in understanding cancer. Bioinformation. 2:330–334. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li W, Chen A, Xiong L, Chen T, Tao F, Lu
Y, He Q, Zhao L, Ou R and Xu Y: miR-133a acts as a tumor suppressor
in colorectal cancer by targeting eIF4A1. Tumour Biol.
39:10104283176983892017.PubMed/NCBI
|
|
16
|
Ding L, Yu LL, Han N and Zhang BT: miR-141
promotes colon cancer cell proliferation by inhibiting MAP2K4.
Oncol Lett. 13:1665–1671. 2017.PubMed/NCBI
|
|
17
|
Quan Y, Song Q, Wang J, Zhao L, Lv J and
Gong S: MiR-1202 functions as a tumor suppressor in glioma cells by
targeting Rab1A. Tumour Biol. 39:10104283176975652017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Han ML, Wang F, Gu YT, Pei XH, Ge X, Guo
GC, Li L, Duan X, Zhu MZ and Wang YM: MicroR-760 suppresses cancer
stem cell subpopulation and breast cancer cell proliferation and
metastasis: By down-regulating NANOG. Biomed Pharmacother.
80:304–310. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liao Y, Deng Y, Liu J, Ye Z, You Z, Yao S
and He S: MiR-760 overexpression promotes proliferation in ovarian
cancer by downregulation of PHLPP2 expression. Gynecol Oncol.
143:655–663. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Q, Huang Z, Ni S, Xiao X, Xu Q, Wang
L, Huang D, Tan C, Sheng W and Du X: Plasma miR-601 and miR-760 are
novel biomarkers for the early detection of colorectal cancer. PLoS
One. 7:e443982012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bajpai R and Nagaraju GP: Specificity
protein 1: Its role in colorectal cancer progression and
metastasis. Crit Rev Oncol Hematol. 113:1–7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang X, Wang J, Lin S, Geng Y, Wang J and
Jiang B: Sp1 is involved in H2O2-induced PUMA gene expression and
apoptosis in colorectal cancer cells. J Exp Clin Cancer Res.
27:442008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dong W, Shen R, Wang Q, Gao Y, Qi X, Jiang
H, Yao J, Lin X, Wu Y and Wang L: Sp1 upregulates expression of
TRF2 and TRF2 inhibition reduces tumorigenesis in human colorectal
carcinoma cells. Cancer Biol Ther. 8:2166–2174. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao Y, Zhang W, Guo Z, Ma F, Wu Y, Bai Y,
Gong W, Chen Y, Cheng T, Zhi F, et al: Inhibition of the
transcription factor Sp1 suppresses colon cancer stem cell growth
and induces apoptosis in vitro and in nude mouse xenografts. Oncol
Rep. 30:1782–1792. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yu MH and Zhang W: TEAD1 enhances
proliferation via activating SP1 in colorectal cancer. Biomed
Pharmacother. 83:496–501. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hosoi Y, Watanabe T, Nakagawa K, Matsumoto
Y, Enomoto A, Morita A, Nagawa H and Suzuki N: Up-regulation of
DNA-dependent protein kinase activity and Sp1 in colorectal cancer.
Int J Oncol. 25:461–468. 2004.PubMed/NCBI
|
|
28
|
Kou XX, Hao T, Meng Z, Zhou YH and Gan YH:
Acetylated Sp1 inhibits PTEN expression through binding to PTEN
core promoter and recruitment of HDAC1 and promotes cancer cell
migration and invasion. Carcinogenesis. 34:58–67. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xia SS, Zhang GJ, Liu ZL, Tian HP, He Y,
Meng CY, Li LF, Wang ZW and Zhou T: MicroRNA-22 suppresses the
growth, migration and invasion of colorectal cancer cells through a
Sp1 negative feedback loop. Oncotarget. 8:36266–36278.
2017.PubMed/NCBI
|
|
30
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death, and tumorigenesis. Br J Cancer. 96
Suppl:R40–R44. 2007.PubMed/NCBI
|
|
32
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:pp. 2257–2261.
2006; View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hu SH, Wang CH, Huang ZJ, Liu F, Xu CW, Li
XL and Chen GQ: miR-760 mediates chemoresistance through inhibition
of epithelial mesenchymal transition in breast cancer cells. Eur
Rev Med Pharmacol Sci. 20:5002–5008. 2016.PubMed/NCBI
|
|
34
|
Lv J, Fu Z, Shi M, Xia K, Ji C, Xu P, Lv
M, Pan B, Dai L and Xie H: Systematic analysis of gene expression
pattern in has-miR-760 overexpressed resistance of the MCF-7 human
breast cancer cell to doxorubicin. Biomed Pharmacother. 69:162–169.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chang WC and Hung JJ: Functional role of
post-translational modifications of Sp1 in tumorigenesis. J Biomed
Sci. 19:942012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu Y, Zhao F, Wang Z, Song Y, Luo Y, Zhang
X, Jiang L, Sun Z, Miao Z and Xu H: MicroRNA-335 acts as a
metastasis suppressor in gastric cancer by targeting Bcl-w and
specificity protein 1. Oncogene. 31:1398–1407. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yin P, Zhao C, Li Z, Mei C, Yao W, Liu Y,
Li N, Qi J, Wang L, Shi Y, et al: Sp1 is involved in regulation of
cystathionine γ-lyase gene expression and biological function by
PI3K/Akt pathway in human hepatocellular carcinoma cell lines. Cell
Signal. 24:1229–1240. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sankpal UT, Goodison S, Abdelrahim M and
Basha R: Targeting Sp1 transcription factors in prostate cancer
therapy. Med Chem. 7:518–525. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bonofiglio D, Qi H, Gabriele S, Catalano
S, Aquila S, Belmonte M and Ando S: Peroxisome
proliferator-activated receptor gamma inhibits follicular and
anaplastic thyroid carcinoma cells growth by upregulating
p21Cip1/WAF1 gene in a Sp1-dependent manner. Endocr Relat Cancer.
15:545–557. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yue L, Li L, Liu F, Hu N, Zhang W, Bai X,
Li Y, Zhang Y, Fu L, Zhang X and Ye L: The oncoprotein HBXIP
activates transcriptional coregulatory protein LMO4 via Sp1 to
promote proliferation of breast cancer cells. Carcinogenesis.
34:927–935. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Huang C and Xie K: Crosstalk of Sp1 and
Stat3 signaling in pancreatic cancer pathogenesis. Cytokine Growth
Factor Rev. 23:25–35. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hsu TI, Wang MC, Chen SY, Yeh YM, Su WC,
Chang WC and Hung JJ: Sp1 expression regulates lung tumor
progression. Oncogene. 31:3973–3988. 2012. View Article : Google Scholar : PubMed/NCBI
|