Introduction
Back pain is a common and costly neurological
ailment (1). Approximately 85% of
people worldwide will experience back pain at some point in their
lives, and lifetime, annual and point prevalence rates are 38.9,
38.0 and 18.3%, respectively. In the United States of America, $100
billion is spent annually on treatment for individuals that suffer
with back pain (2). Chronic back
pain is closely associated with degenerative alterations in
intervertebral discs. Degenerative intervertebral disc changes are
associated with reduced proteoglycan content, loss of bound water
molecules, decreased tissue osmotic pressure and a resulting
decline in the ability of the tissues to absorb biomechanical
forces (3). Disc degeneration is
the result of various factors, including age, mechanical stress,
genetic determinants and lifestyle, which are generally considered
to be its major causes. Current treatments for intervertebral disc
degeneration and back pain range from conservative methods that are
based on alleviating the symptoms, including back exercises to
strengthen back muscles, painkillers and physiotherapy, to surgical
procedures such as spinal fusion (4). However, none of these methods are
completely successful. Although a number of studies have focused on
the etiology of intervertebral disc degeneration (IVD), including
genetics, cytokines and mechanical load, the underlying pathology
remains to be fully elucidated (5,6).
Previous studies have suggested a close association
between cellular senescence and IVD; with increasing age and
advancing disc degeneration, senescent nucleus pulposus (NP) cells
accumulate in the NP (7,8). Cellular senescence occurs when cells
stop dividing; senescent cells remain viable but exhibit
alterations in phenotype and gene expression patterns. For example,
senescent cells may have altered responsiveness to external stimuli
and may produce increased levels of matrix metalloproteinases
(MMPs), which are associated with IVD. Although senescent cells may
be observed in the aging/degenerating human disc, the mechanisms
and signaling pathways involved in senescence and how these
contribute to the process of IVD remain unknown.
The aim of the present study was to identify the
differentially expressed genes (DEGs) associated with senescence in
human annulus cells from degenerative intervertebral discs. DEGs
were subsequently subjected to functional and pathway annotations,
transcription factor (TF) enrichment analysis, construction of
protein-protein interaction (PPI) networks and module analysis, to
investigate the critical DEGs in the progression of senescence. The
findings of the present study may provide the basis for further
studies to identify novel potential hub genes associated with
senescence in the development and progression of IVD.
Materials and methods
Microarray data extraction and DEG
analysis
The gene expression profile dataset GSE17077, based
on the platform of GPL1352 (Affymetrix Human X3P Array), was
downloaded from the Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo/) database. The data
were obtained by Gruber et al (9), who collected disc tissue samples from
patients with degenerative disc disease undergoing surgical disc
procedures. Standard laser capture microdissection techniques were
used to collect senescent cells. In total, eight non-senescent cell
samples and eleven senescent cell samples were harvested for
mircoarray analysis.
Original CEL expression profiling data were
processed into expression estimates following background
correction, and quartile data normalization was performed using the
robust multi-array average algorithm (10) with the default parameters in the R
affy package (bioconductor.org/packages/release/bioc/html/affy.html)
(11). Subsequently, t-tests were
performed in the Linear Models for Microarray Data package
(bioconductor.org/packages/release/bioc/html/limma.html)
(12) to identify DEGs. Genes with
P<0.05 and |log2 fold change (FC)|>2 were
considered DEGs.
Hierarchical clustering analysis
To identify gene expression differences,
hierarchical clustering analysis of DEGs was performed using
MultiExperiment Viewer software version 4.9 (mev.tm4.org/#/welcome) (13).
Functional and pathway
annotations
To further investigate the functions and pathways of
DEGs, the Database for Annotation, Visualization and Integrated
Discovery (DAVID; version 6.7; david.ncifcrf.gov/) was used to obtain the enrichment
in Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes
(KEGG) terms. To do this, DEGs were entered into the DAVID online
tool to cluster the genes according to the GO (14) categories of cellular component
(CC), biological process (BP) and molecular function (MF) (15) and KEGG pathway though the module of
functional annotation with the count value >2 and P<0.05.
TF enrichment analysis
To identify TFs that regulate the expression of
senescence-associated DEGs, Web-based Gene Set Analysis Toolkit
(WebGestalt; www.webgestalt.org/option.php) and Enrichr (amp.pharm.mssm.edu/lib/chea.jsp) were
used. The adjusted enrichment P<0.001 was selected as the
threshold. Following analysis of DEGs associated with senescence by
these two independent methods, the TFs identified by the two
methods were selected as the key TFs.
Protein-protein interaction network
construction
As proteins seldom function in isolation, it is
important to understand the interaction of proteins by studying
larger functional groups. The Search Tool for the Retrieval of
Interacting Genes (STRING) database (string-db.org/) (16), which contains experimental and
predicted interaction information, was used to annotate functional
interactions between DEGs with the cut-off criterion of combined
score >0.7. Subsequently, the interaction network was
constructed using Cytoscape software version 3.3.0 (www.cytoscape.org/) (17). The total connectivity degree of
each node in the network was calculated by connectivity degree
analysis.
Screening for hub genes
Hub genes were identified as genes that: i) Were
involved in one of the enriched pathways; ii) had a high
connectivity degree (>5); and iii) were target genes of key
TFs.
Results
Identification of DEGs
According to the cut-off criteria of
|log2 FC|>2.0 and P<0.05, 667 DEGs, including 368
up- and 299 down-regulated genes, were obtained between senescent
cells and non-senescent cells (Tables
I and II). Of these, 41 and
18 were upregulated and downregulated by ≥5-fold, respectively.
 | Table I.Top 15 upregulated differentially
expressed genes with |log2(FC)|>2 and P<0.05. |
Table I.
Top 15 upregulated differentially
expressed genes with |log2(FC)|>2 and P<0.05.
| Gene | Gene ID | FC | P-value |
|---|
| NCK1 | 4690 | 9.56 | 0.0001 |
| HOXB7 | 3217 | 9.28 | 0.0035 |
| HNRPDL | 9987 | 9.15 | 0.0214 |
| CNTROB | 116840 | 8.03 | 0.0001 |
| SPATA6L | 55064 | 7.98 | 0.0078 |
| EFCAB7 | 84455 | 7.94 | 0.0081 |
| HOXB9 | 3219 | 7.80 | 0.0011 |
| CYP2B6 | 1555 | 7.70 | 0.0460 |
| AHSA2 | 130872 | 7.26 | 0.0002 |
| ST3GAL5 | 8869 | 7.03 | 0.0152 |
| PPME1 | 51400 | 6.95 | 0.0019 |
| NME6 | 10201 | 6.76 | 0.0006 |
| IDS | 3423 | 6.60 | 0.0001 |
| MAN2A2 | 4122 | 6.58 | 0.0005 |
| FILIP1L | 11259 | 6.57 | 0.0033 |
 | Table II.Top 15 downregulated differentially
expressed genes with |log2(FC)|>2 and P<0.05. |
Table II.
Top 15 downregulated differentially
expressed genes with |log2(FC)|>2 and P<0.05.
| Gene | Gene ID | FC | P-value |
|---|
| GTPBP8 | 29083 | 0.11 | 0.0083 |
| GPATCH2 | 55105 | 0.11 | 0.0145 |
| LOC441461 | 441461 | 0.11 | 0.0126 |
| IQCC | 55721 | 0.15 | 0.0015 |
| ZNF347 | 84671 | 0.15 | 0.0017 |
| TMEM74B | 55321 | 0.15 | 0.0062 |
| F2RL2 | 2151 | 0.17 | 0.0108 |
| ATG4B | 23192 | 0.17 | 0.0045 |
| RBM26-AS1 | 100505538 | 0.17 | 0.0152 |
| UBALD1 | 124402 | 0.17 | 0.0002 |
| CSTF3-AS1 | 338739 | 0.18 | 0.0071 |
| OBSL1 | 23363 | 0.18 | 0.0224 |
| ALPK1 | 80216 | 0.18 | 0.0147 |
| LOC155060 | 155060 | 0.18 | 0.0300 |
| C1orf116 | 79098 | 0.19 | 0.0005 |
Hierarchical clustering analysis
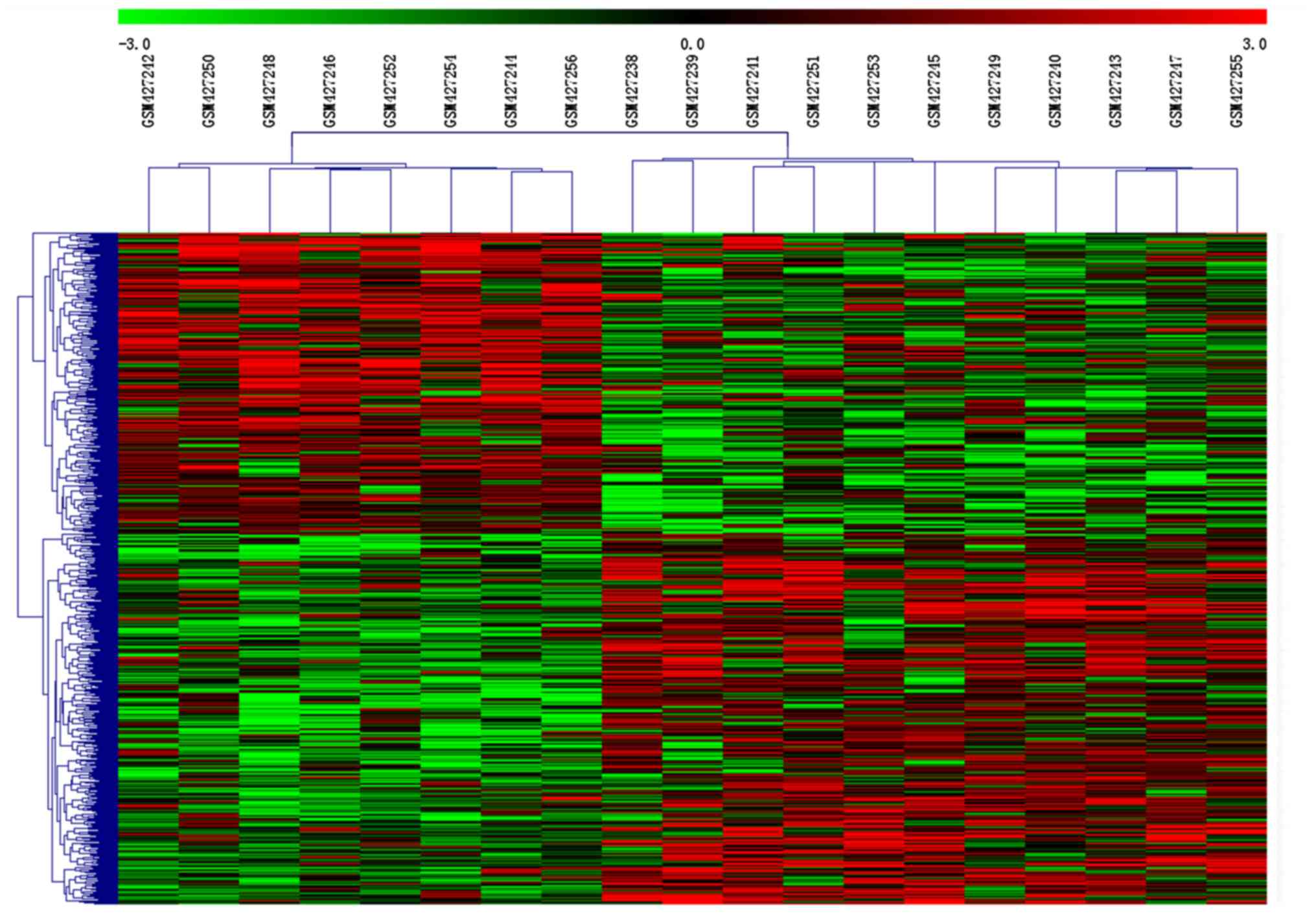
To assess whether the 667 DEGs identified could
separate senescent from non-senescent cells, hierarchical
clustering analysis was performed. Clustering of conditions divided
samples into two groups (Fig. 1).
The eight gene expression profiles of non-senescent controls were
clustered together, separated from the gene expression profiles of
senescent cells.
GO functional and pathway enrichment
analysis
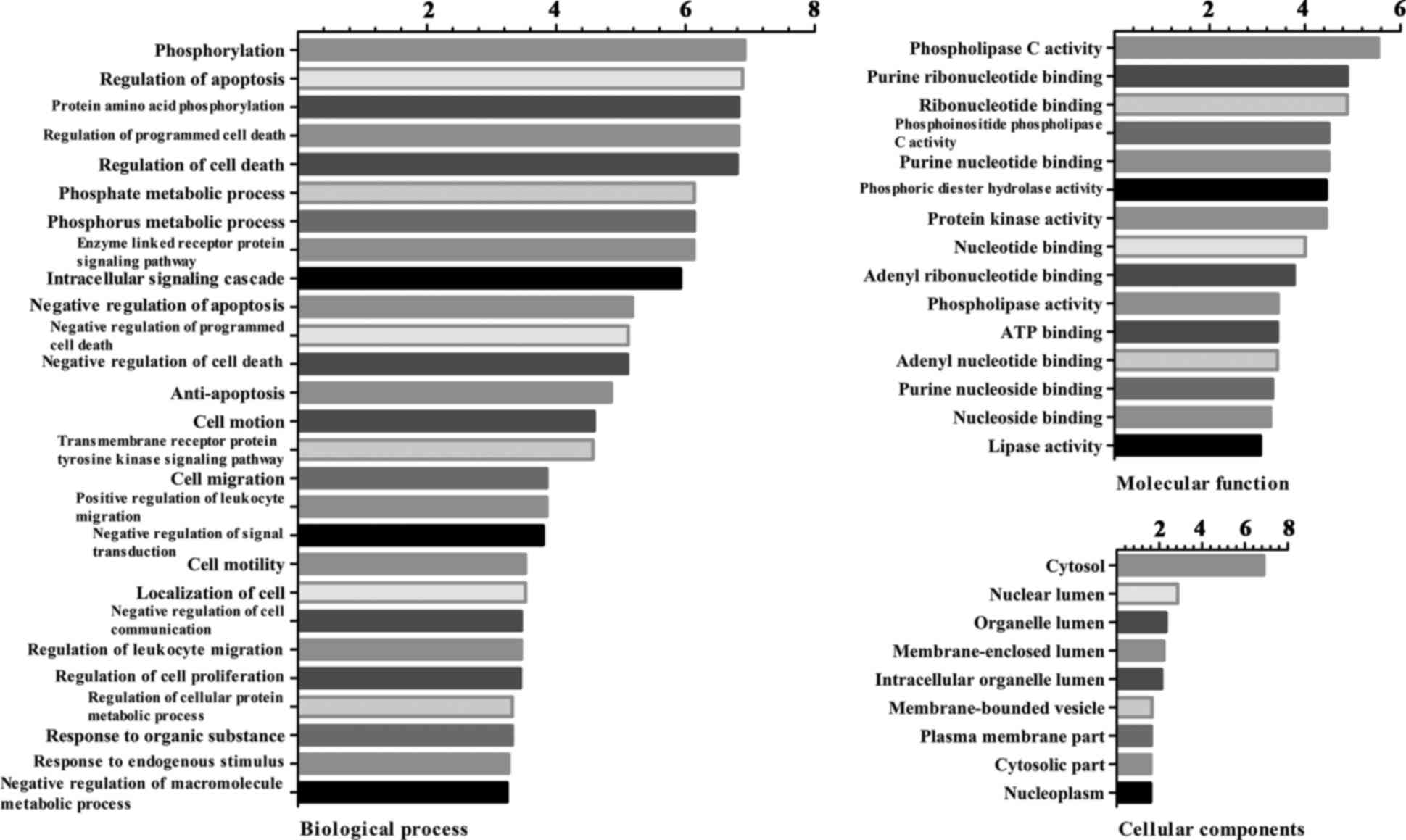
The results of the GO enrichment analysis of DEGs
based on CC, BP and MF are presented in Fig. 2. According to the BP analysis, DEGs
were primarily involved in phosphorylation, regulation of
apoptosis, protein amino acid phosphorylation and regulation of
programmed cell death. Based on the MF analysis, DEGs were
associated with phospholipase C activity, purine ribonucleotide
binding, ribonucleotide binding and phosphoinositide phospholipase
C activity. In addition, DEGs were primarily located in the
cytosol, nuclear lumen, organelle lumen and membrane-enclosed
lumen.
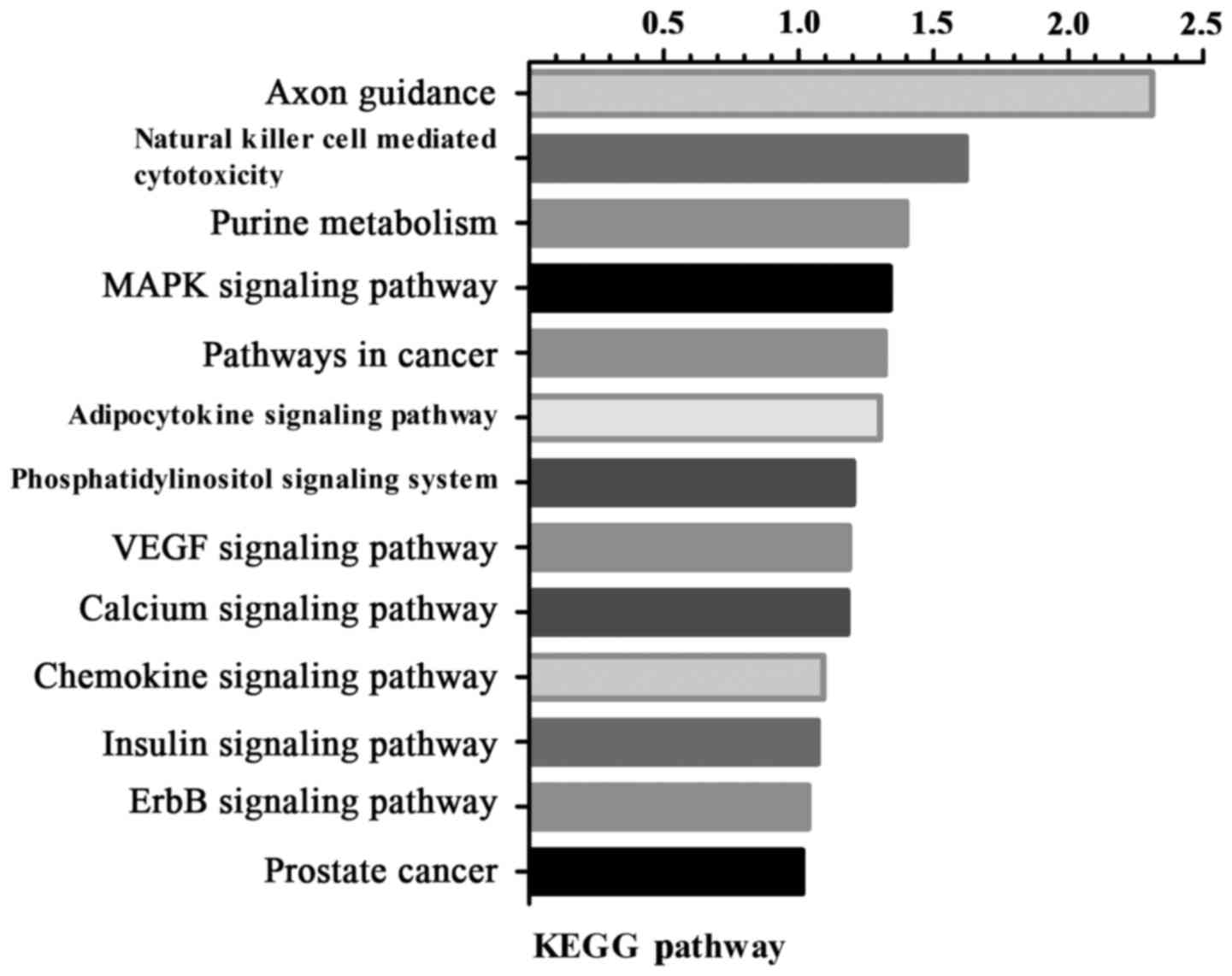
Pathway enrichment analysis revealed that the DEGs
were involved in axon guidance, natural killer cell mediated
cytotoxicity, purine metabolism and the mitogen-activated protein
kinase (MAPK) signaling pathway (Fig.
3).
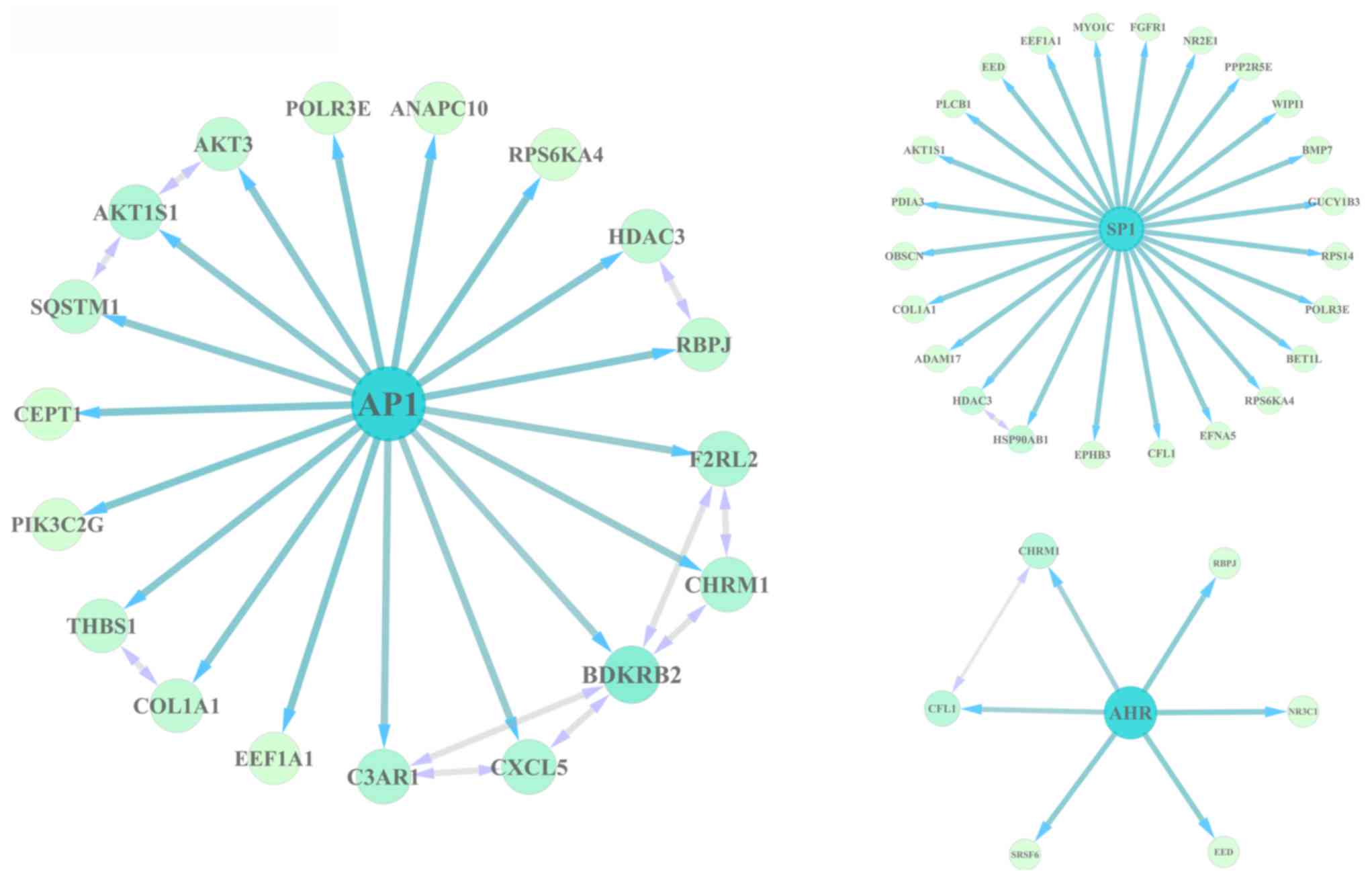
TF enrichment analysis and
transcriptional regulatory network construction
The DEGs were subjected to WebGestalt and ChEA to
identify the TFs involved in their regulation. The results revealed
that activator protein 1 (AP1), specificity protein 1 (SP1) and
aryl hydrocarbon receptor (AHR) may serve important roles in
regulating DEGs, as the AP1, SP1, AHR binding motifs were enriched
in the DEGs, and Chip-seq experiments verified that AP1-, SP1-,
AHR-targeted genes were enriched in the DEGs. Of these, AP1 and SP1
regulated 18 and 24 DEGs, respectively, in senescent cells
(Fig. 4).
PPI network construction and module
analysis
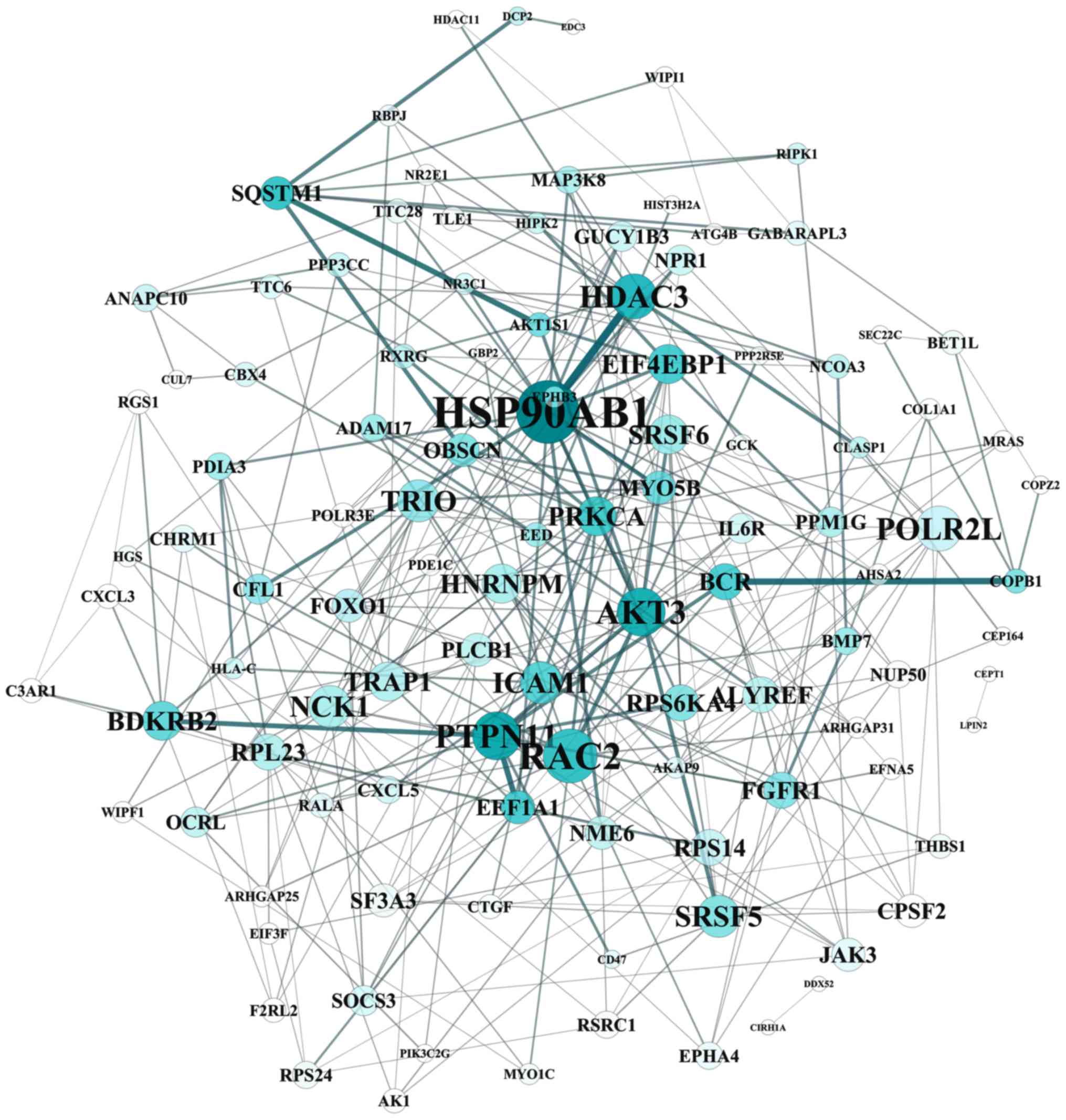
The STRING tool was used to obtain PPI associations
of DEGs. The PPI network contained 248 nodes (proteins) and 399
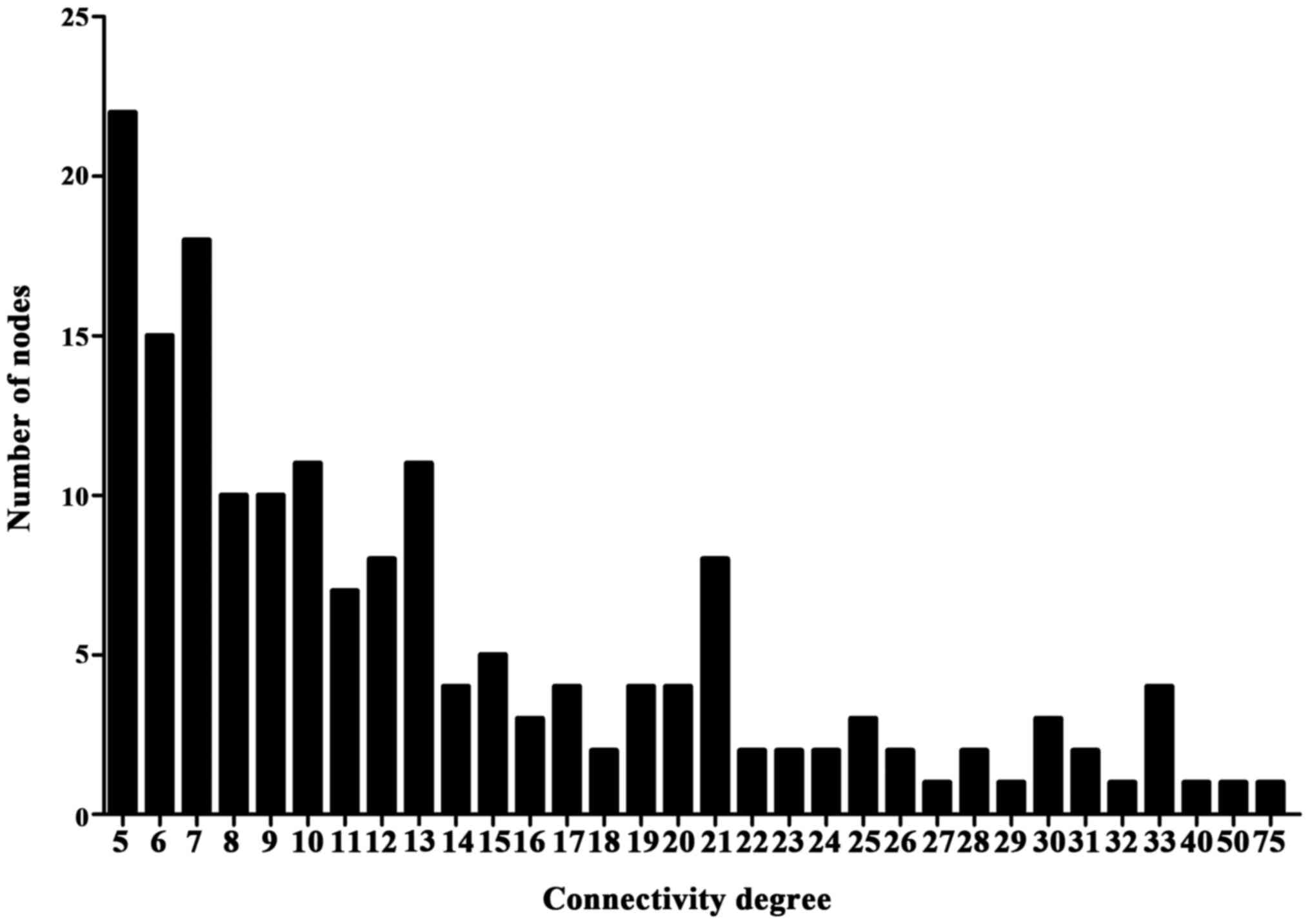
lines (interactions) with the combined score >0.7 (Fig. 5). The connectivity degree of each
node of the PPI network was calculated and the results of certain
of these nodes are presented in Fig.
6.
Hub gene identification
Two hub genes with high connectivity degree (>5)
were selected: HSP90AB1 (degree=57), encoding heat shock protein
(HSP) 90, was involved in pathways in cancer and C-X-C motif
chemokine (CXCL) 5 (degree=8) was involved in chemokine signaling
pathway. HSP90AB1 and CXCL5 were target genes of the TFs SP1 and
AP1, respectively.
Discussion
Aging is the most important single risk factor for
IVD (18). Intervertebral discs
undergo age-associated degenerative alterations that contribute to
lower back pain; an important socio-economic problem. The link
between aged degenerative discs and cellular senescence has been
previously observed in human discs (19); however, the senescence-associated
gene expression alterations and underlying molecular mechanisms
remain unknown. The present study performed an in-depth and
extensive analysis based on gene expression profiling combined with
bioinformatics. Gene expression profile data from two groups
(senescent and non-senescent cells) were downloaded from the GEO
database and subsequently analyzed by bioinformatics methods. In
total, 667 DEGs, including 368 up- and 299 down-regulated genes
were screened. Hierarchical clustering analysis revealed that these
667 DEGs could separate senescent cells from non-senescent
controls. The DEGs were primarily involved in axon guidance,
natural killer cell mediated cytotoxicity, purine metabolism and
the MAPK signaling pathway.
Transcription factors are central regulators of gene
expression. Following analysis of the DEGs with ChEA and
WebGestalt, the TFs AP1, SP1 and AHR were identified as potential
regulators of senescence in disc degeneration. Of these, AP1, which
is involved in numerous cellular processes, including
proliferation, transformation and death, regulated 18 DEGs and SP1,
which is involved in cell differentiation, growth and apoptosis,
regulated 24 DEGs in the senescent cells. AP1 and SP1 have been
demonstrated to promote the activation of beta1,3-glucuronosyl
transferase 1, a key enzyme required for chondroitin sulfate
biosynthesis, in NP cells treated with growth factors, and this
process was mediated through the MAPK signaling pathway (20). Importantly, in various cell types,
AP1 was highly activated in senescence (21,22).
Notably, one of the enriched pathways of the DEGs identified by the
present study is the MAPK signaling pathway, thus suggesting that
this pathway may regulate the activation of AP1, therefore serving
an important role in cell senescence and contributing to the
process of disc degeneration. However, this hypothesis requires
further investigation.
The present study identified two hub genes, HSP90AB1
and CXCL5. HSP90AB1 belongs to the HSP family, which has a number
of members now recognized to be associated with cell senescence,
including HSP70, which has been revealed to be downregulated in
senescent annulus cells (9).
Additionally, HSP90 inhibition has been demonstrated to induce
cellular senescence in certain cancer cells (23). However, in contrast to previous
studies, the results of the present study demonstrated that
HSP90AB1, with the highest connectivity degree of 57, was
>2-fold greater expressed in senescent compared with
non-senescent annulus cells, indicating that the function of HSP90
is cell-specific.
Chemokine expression within the IVD has been
demonstrated by a number of previous reports (24,25).
In the present study, chemokine signal pathway was enriched among
the DEGs, and chemokines including CXCL5 and CXCL3 were increased
>3-fold in senescent compared with non-senescent cells with a
high degree. Notably, CXCL5 was additionally predicted as a target
gene of the TF AP1. Acosta et al (26) demonstrated that knockdown of the
chemokine receptor CXCR2 inhibits senescence and that ectopic
expression of CXCR2 results in premature senescence in human
fibroblasts, which suggests that senescent cells activate a
self-amplifying secretory network. Senescent cells may express
elevated levels of extracellular matrix-degrading proteases,
collagenases and MMPs, and decreased levels of the MMP inhibitor
tissue inhibitor of metalloproteinases 1. A similar shift has been
identified in aging and degenerated IVDs (27). Therefore, senescent disc cells may
disturb extracellular matrix homeostasis via increased production
of chemokines, resulting in reduced matrix synthesis and increased
matrix degradation compared with non-senescent cells, eventually
leading to degeneration. Additionally, the MAPK signaling pathway
and the TF AP1 may be involved in this regulation.
One of the limitations of the present study is that
the screened genes identified by statistically significant
differences were not confirmed experimentally. That these genes are
increased or decreased does not automatically imply causation, nor
does it provide much insight regarding the mechanisms underlying
these alterations. However, the results of the present study may
provide the basis for further experimental studies.
In conclusion, the present study used microarray
analysis combined with bioinformatics techniques to reveal the
involvement of CXCL5 and HSP90 in the progression of senescence. In
addition, the MAPK-regulated AP1 pathway may contribute to
senescence-associated IVD. These findings suggested potential
targets for use in the diagnosis and therapy of disc degenerative
diseases. However, further studies are required to confirm this
data.
Acknowledgements
The authors thank Dr Xinyi Liu (School of Medicine,
Shanghai Jiaotong University) for technical assistance, Dr Jia Cao
(Xinhua Hospital Affiliated to Shanghai Jiao Tong University School
of Medicine) for data collection and Mr. Chengcai Jin (School of
Medicine, Shanghai Jiaotong University) for image processing. The
present study was supported by the National Natural Science
Foundation of China (No. 81601920) and the Program for Tackling Key
Problems in Science and Technology in Songjiang District.
References
|
1
|
Katz JN: Lumbar disc disorders and
low-back pain: Socioeconomic factors and consequences. J Bone Joint
Surg Am. 88 Suppl 2:S21–S24. 2006. View Article : Google Scholar
|
|
2
|
Hoy D, Bain C, Williams G, March L, Brooks
P, Blyth F, Woolf A, Vos T and Buchbinder R: A systematic review of
the global prevalence of low back pain. Arthritis Rheum.
64:2028–2037. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Luoma K, Riihimäki H, Luukkonen R,
Raininko R, Viikari-Juntura E and Lamminen A: Low back pain in
relation to lumbar disc degeneration. Spine (Phila Pa 1976).
25:487–492. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Saltychev M, Eskola M and Laimi K: Lumbar
fusion compared with conservative treatment in patients with
chronic low back pain: A meta-analysis. Int J Rehabil Res. 37:2–8.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Walter BA, Korecki CL, Purmessur D,
Roughley PJ, Michalek AJ and Iatridis JC: Complex loading affects
intervertebral disc mechanics and biology. Osteoarthritis
Cartilage. 19:1011–1018. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Risbud MV and Shapiro IM: Role of
cytokines in intervertebral disc degeneration: Pain and disc
content. Nat Rev Rheumatol. 10:44–56. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gruber HE, Ingram JA, Norton HJ and Hanley
EN Jr: Senescence in cells of the aging and degenerating
intervertebral disc: Immunolocalization of senescence-associated
beta-galactosidase in human and sand rat discs. Spine (Phila Pa
1976). 32:321–327. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim KW, Chung HN, Ha KY, Lee JS and Kim
YY: Senescence mechanisms of nucleus pulposus chondrocytes in human
intervertebral discs. Spine J. 9:658–666. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gruber HE, Hoelscher GL, Ingram JA,
Zinchenko N and Hanley EN Jr: Senescent vs. non-senescent cells in
the human annulus in vivo: Cell harvest with laser capture
microdissection and gene expression studies with microarray
analysis. BMC Biotechnol. 10:52010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fujita A, Sato JR, Rodrigues Lde O,
Ferreira CE and Sogayar MC: Evaluating different methods of
microarray data normalization. BMC Bioinformatics. 7:4692006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ritchie ME, Silver J, Oshlack A, Holmes M,
Diyagama D, Holloway A and Smyth GK: A comparison of background
correction methods for two-colour microarrays. Bioinformatics.
23:2700–2707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Eisen MB, Spellman PT, Brown PO and
Botstein D: Cluster analysis and display of genome-wide expression
patterns. Proc Natl Acad Sci USA. 95:pp. 14863–14868. 1998;
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
von Mering C, Huynen M, Jaeggi D, Schmidt
S, Bork P and Snel B: STRING: A database of predicted functional
associations between proteins. Nucleic Acids Res. 31:258–261. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nasto LA, Robinson AR, Ngo K, Clauson CL,
Dong Q, St Croix C, Sowa G, Pola E, Robbins PD, Kang J, et al:
Mitochondrial-derived reactive oxygen species (ROS) play a causal
role in aging-related intervertebral disc degeneration. J Orthop
Res. 31:1150–1157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Le Maitre CL, Freemont AJ and Hoyland JA:
Accelerated cellular senescence in degenerate intervertebral discs:
A possible role in the pathogenesis of intervertebral disc
degeneration. Arthritis Res Ther. 9:R452007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hiyama A, Gogate SS, Gajghate S, Mochida
J, Shapiro IM and Risbud MV: BMP-2 and TGF-beta stimulate
expression of beta1,3-glucuronosyl transferase 1 (GlcAT-1) in
nucleus pulposus cells through AP1, TonEBP and Sp1: role of MAPKs.
J Bone Miner Res. 25:1179–1190. 2010.PubMed/NCBI
|
|
21
|
Sepulveda JC, Tomé M, Fernández ME,
Delgado M, Campisi J, Bernad A and González MA: Cell senescence
abrogates the therapeutic potential of human mesenchymal stem cells
in the lethal endotoxemia model. Stem cells. 32:1865–1877. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim YJ, Hwang SH, Lee SY, Shin KK, Cho HH,
Bae YC and Jung JS: miR-486-5p induces replicative senescence of
human adipose tissue-derived mesenchymal stem cells and its
expression is controlled by high glucose. Stem Cells Dev.
21:1749–1760. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chan KC, Ting CM, Chan PS, Lo MC, Lo KW,
Curry JE, Smyth T, Lee AW, Ng WT, Tsao GS, et al: A novel Hsp90
inhibitor AT13387 induces senescence in EBV-positive nasopharyngeal
carcinoma cells and suppresses tumor formation. Mol Cancer.
12:1282013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang J, Tian Y, Phillips KL, Chiverton N,
Haddock G, Bunning RA, Cross AK, Shapiro IM, Le Maitre CL and
Risbud MV: Tumor necrosis factor α- and interleukin-1β-dependent
induction of CCL3 expression by nucleus pulposus cells promotes
macrophage migration through CCR1. Arthritis Rheum. 65:832–842.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kawaguchi S, Yamashita T, Katahira G,
Yokozawa H, Torigoe T and Sato N: Chemokine profile of herniated
intervertebral discs infiltrated with monocytes and macrophages.
Spine (Phila Pa 1976). 27:1511–1516. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Acosta JC, O'Loghlen A, Banito A, Guijarro
MV, Augert A, Raguz S, Fumagalli M, Da Costa M, Brown C, Popov N,
et al: Chemokine signaling via the CXCR2 receptor reinforces
senescence. Cell. 133:1006–1018. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Roughley PJ: Biology of intervertebral
disc aging and degeneration: Involvement of the extracellular
matrix. Spine (Phila Pa 1976). 29:2691–2699. 2004. View Article : Google Scholar : PubMed/NCBI
|