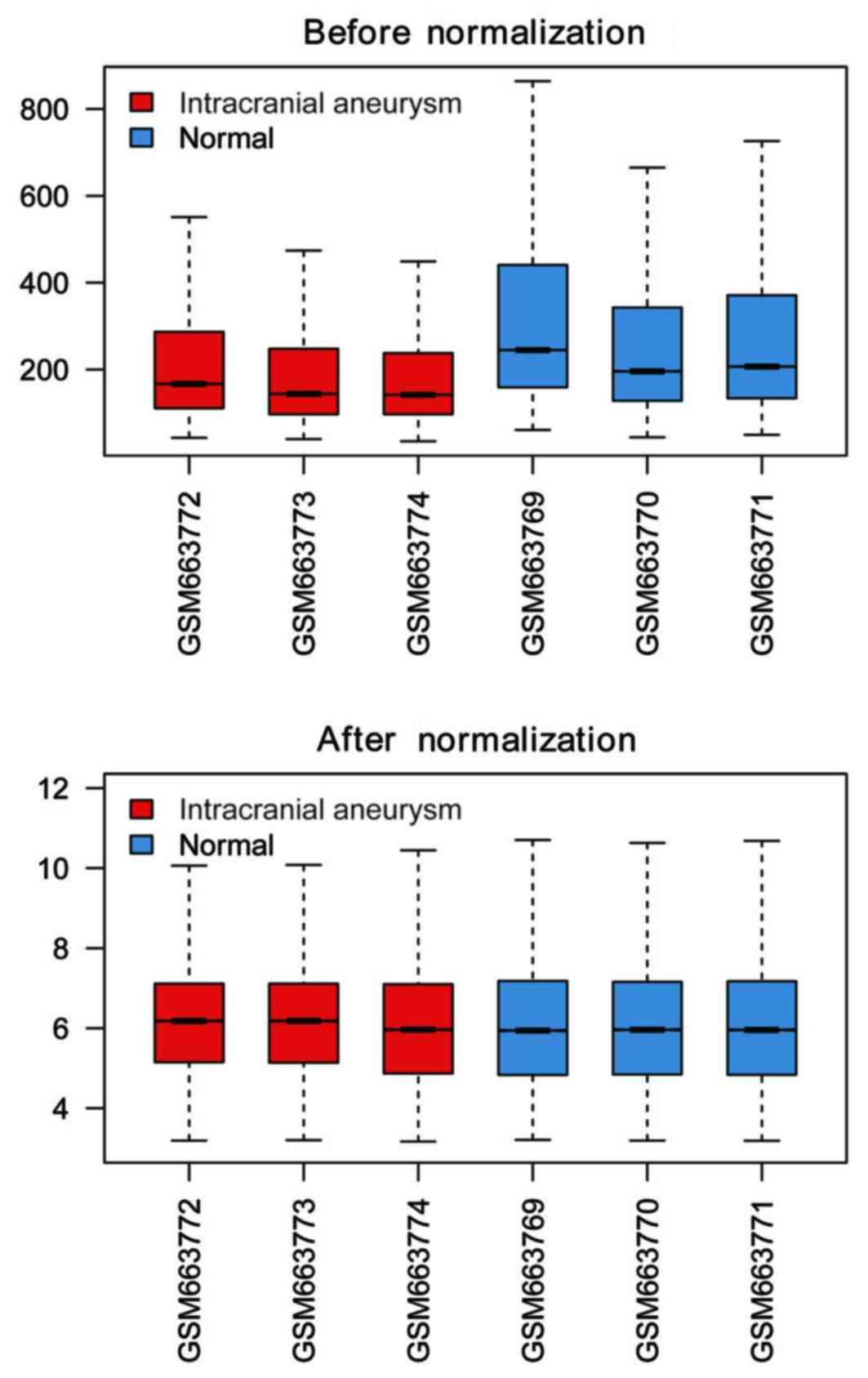
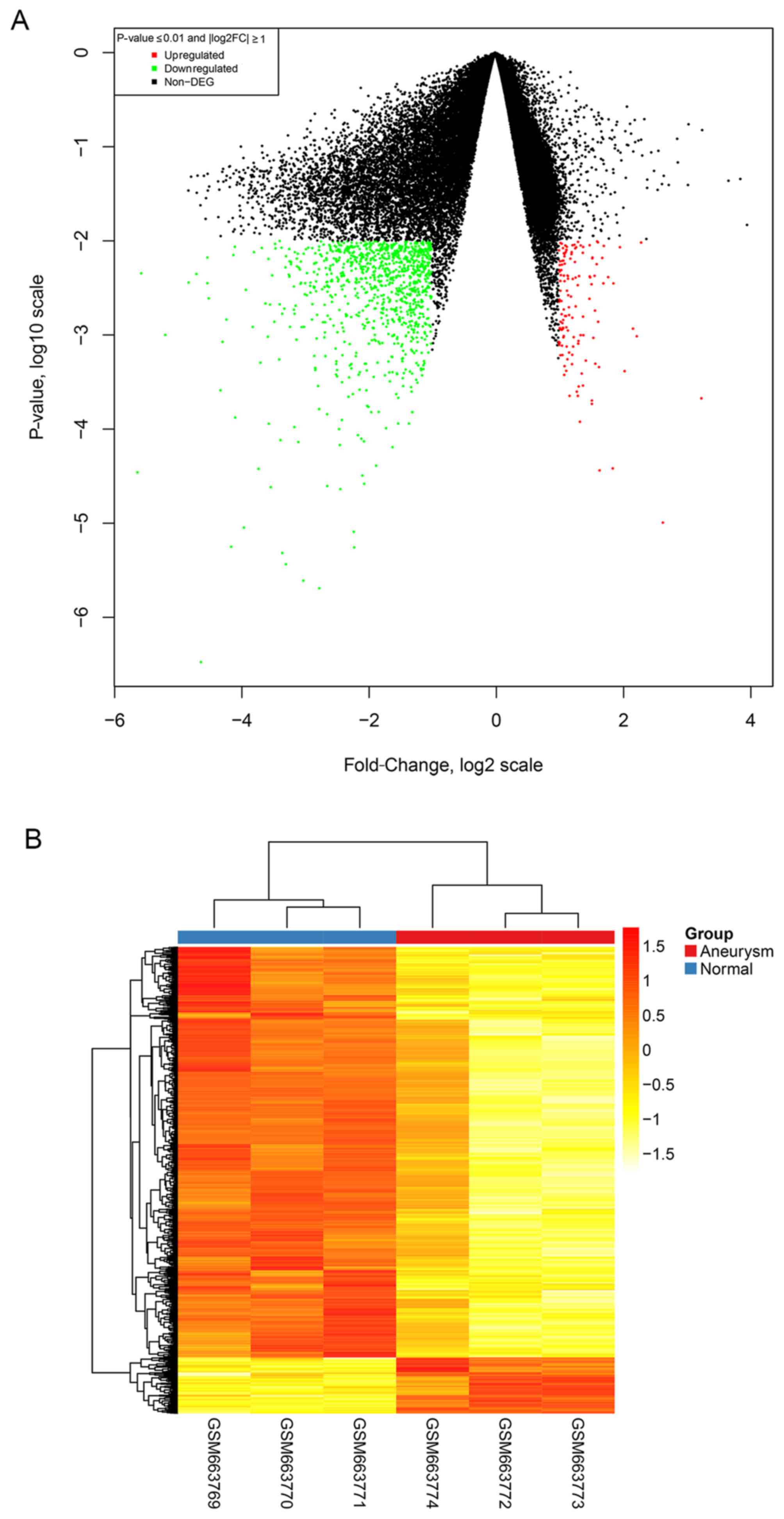
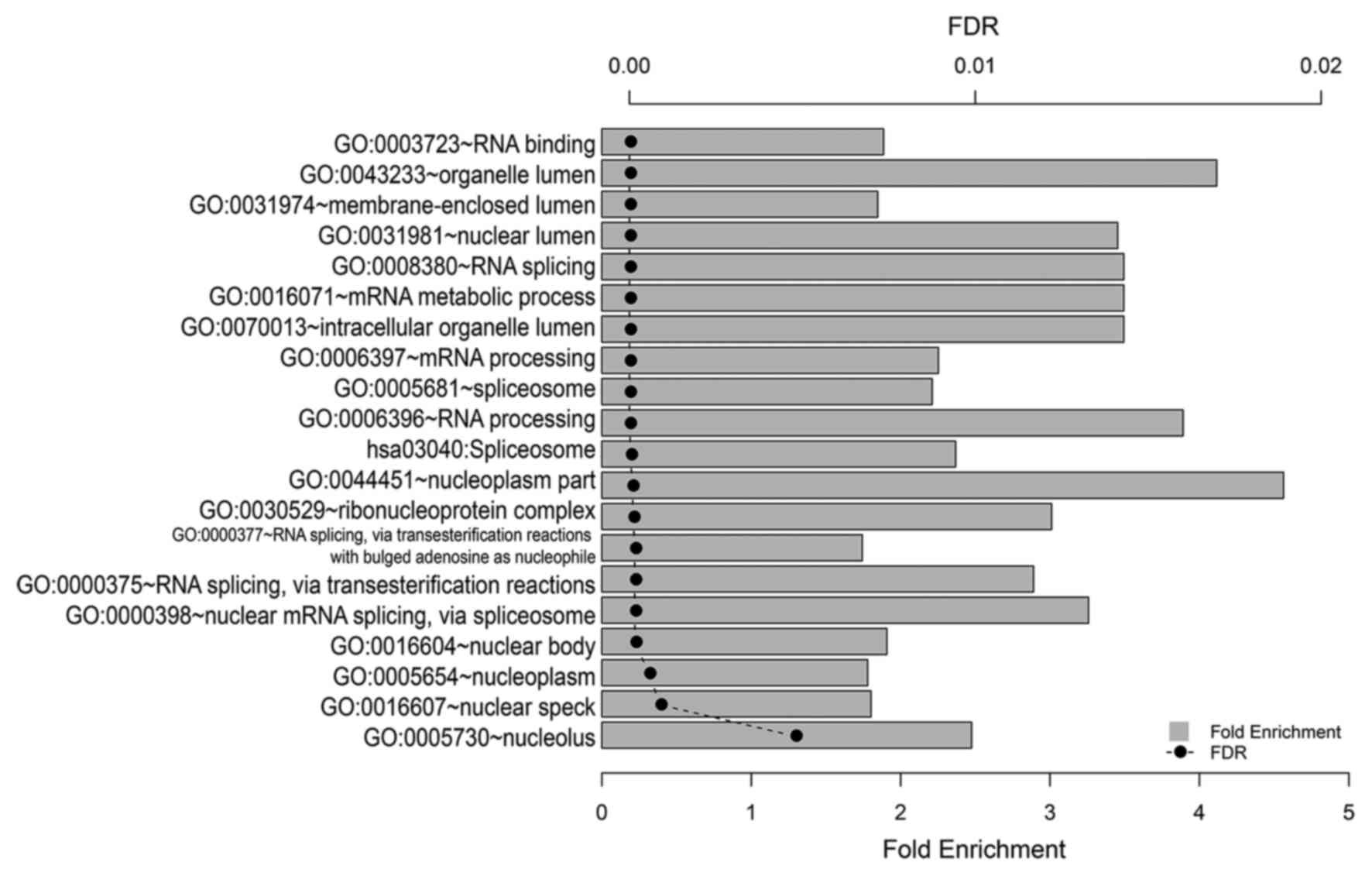
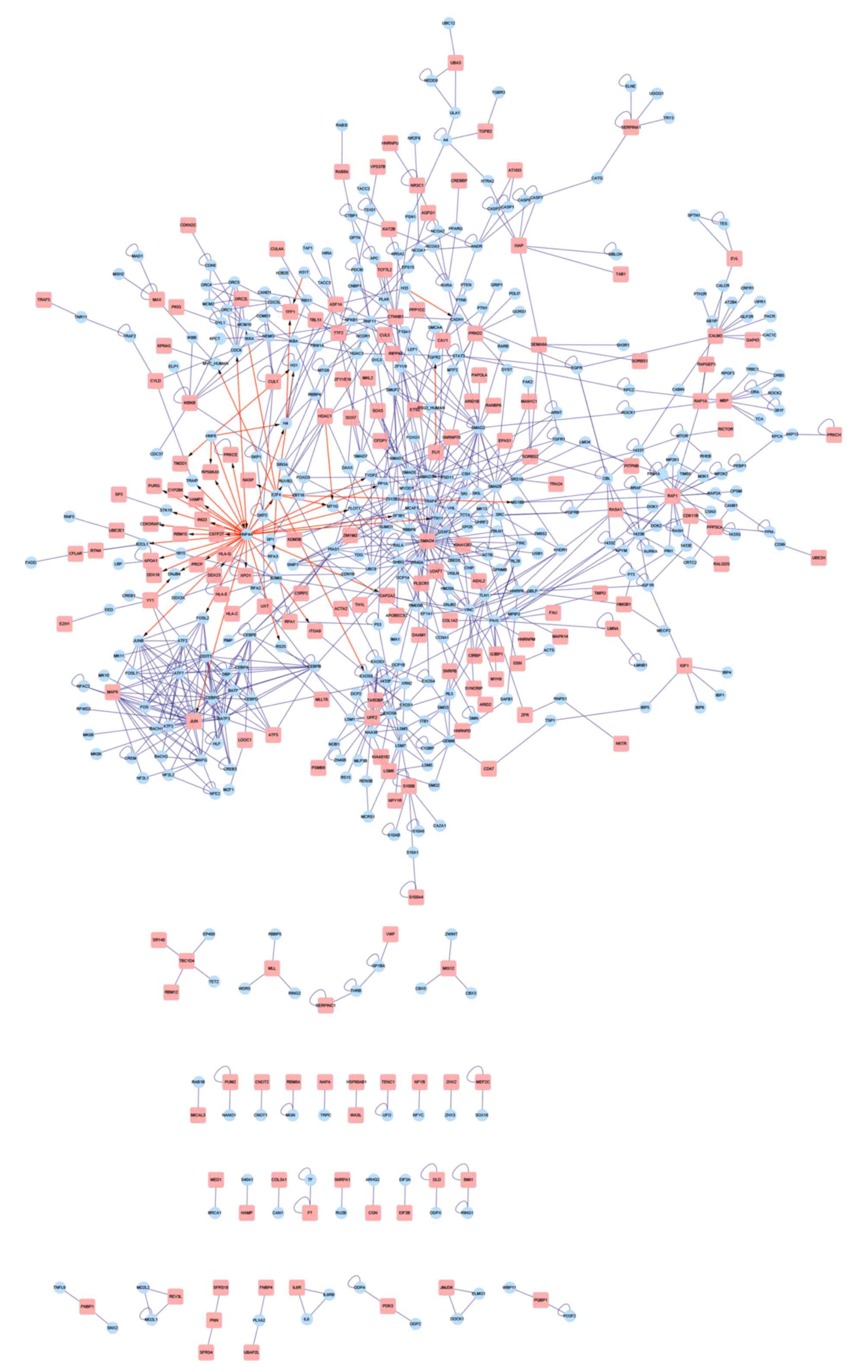
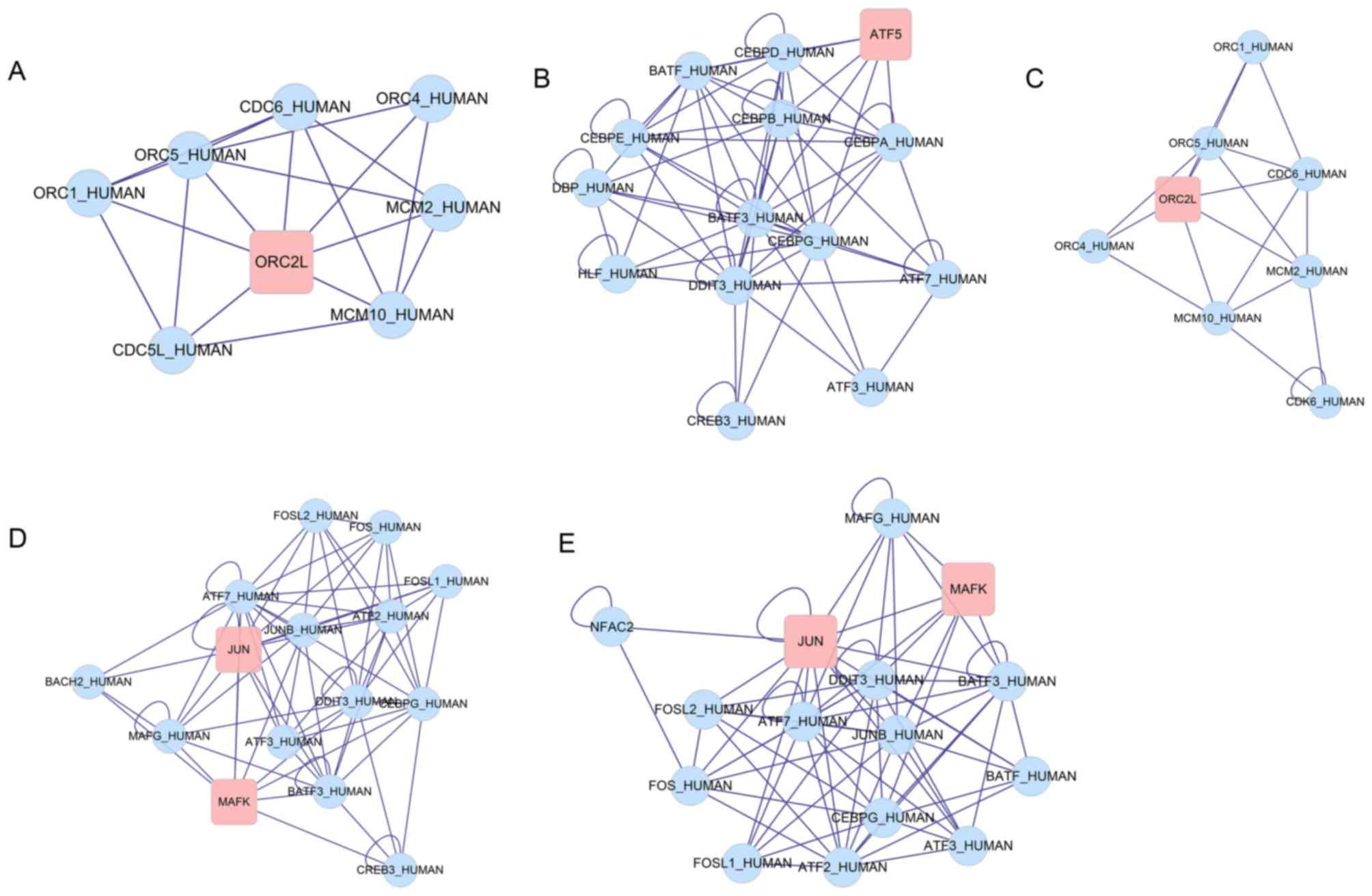
|
1
|
Alg VS, Sofat R, Houlden H and Werring DJ:
Genetic risk factors for intracranial aneurysms: A meta-analysis in
more than 116,000 individuals. Neurology. 80:2154–2165. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bakke SJ and Lindegaard KF: Subarachnoid
haemorrhage-diagnosis and management. Tidsskr Nor Laegeforen.
127:1074–1078. 2007.(In Norwegian). PubMed/NCBI
|
|
3
|
Li L, Yang X, Jiang F, Dusting GJ and Wu
Z: Transcriptome-wide characterization of gene expression
associated with unruptured intracranial aneurysms. Eur Neurol.
62:330–337. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pera J, Korostynski M, Krzyszkowski T,
Czopek J, Slowik A, Dziedzic T, Piechota M, Stachura K, Moskala M,
Przewlocki R and Szczudlik A: Gene expression profiles in human
ruptured and unruptured intracranial aneurysms: What is the role of
inflammation? Stroke. 41:224–231. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kurki MI, Hakkinen SK, Frosen J, Tulamo R,
von und zu Fraunberg M, Wong G, Tromp G, Niemelä M, Hernesniemi J,
Jääskeläinen JE and Ylä-Herttuala S: Upregulated signaling pathways
in ruptured human saccular intracranial aneurysm wall: An emerging
regulative role of Toll-like receptor signaling and nuclear
factor-κB, hypoxia-inducible factor-1A and ETS transcription
factors. Neurosurgery. 68:1666–1675. 2011. View Article : Google Scholar
|
|
6
|
Yagi K, Tada Y, Kitazato KT, Tamura T,
Satomi J and Nagahiro S: Ibudilast inhibits cerebral aneurysms by
down-regulating inflammation-related molecules in the vascular wall
of rats. Neurosurgery. 66:551–559. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Aoki T, Kataoka H, Nishimura M, Ishibashi
R, Morishita R and Miyamoto S: Ets-1 promotes the progression of
cerebral aneurysm by inducing the expression of MCP-1 in vascular
smooth muscle cells. Gene Ther. 17:1117–1123. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gentleman RC, Carey VJ, Bates DM, Bolstad
B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al:
Bioconductor: Open software development for computational biology
and bioinformatics. Genome Biol. 5:R802004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Reiner A, Yekutieli D and Benjamini Y:
Identifying differentially expressed genes using false discovery
rate controlling procedures. Bioinformatics. 19:368–375. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kolde R: Pheatmap: Pretty heatmaps. R
package version 0.6. 1:2012.
|
|
12
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kanehisa M and Goto S: KEGG: kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bader GD, Betel D and Hogue CW: BIND: The
biomolecular interaction network database. Nucleic Acids Res.
31:248–250. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nepusz T, Yu H and Paccanaro A: Detecting
overlapping protein complexes in protein-protein interaction
networks. Nat Methods. 9:471–472. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Greene LA, Lee HY and Angelastro JM: The
transcription factor ATF5: Role in neurodevelopment and neural
tumors. J Neurochem. 108:11–22. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schoenen H, Huber A, Sonda N, Zimmermann
S, Jantsch J, Lepenies B, Bronte V and Lang R: Differential control
of mincle-dependent cord factor recognition and macrophage
responses by the transcription factors C/EBPβ and HIF1α. J Immunol.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chalouhi N, Ali MS, Jabbour PM,
Tjoumakaris SI, Gonzalez LF, Rosenwasser RH, Koch WJ and Dumont AS:
Biology of intracranial aneurysms: Role of inflammation. J Cereb
Blood Flow Metab. 32:1659–1676. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wisdom R, Johnson RS and Moore C: c-Jun
regulates cell cycle progression and apoptosis by distinct
mechanisms. EMBO J. 18:188–197. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Brenner DA, O'Hara M, Angel P, Chojkier M
and Karin M: Prolonged activation of jun and collagenase genes by
tumour necrosis factor-alpha. Nature. 337:661–663. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Polavarapu R, Gongora MC, Winkles JA and
Yepes M: Tumor necrosis factor-like weak inducer of apoptosis
increases the permeability of the neurovascular unit through
nuclear factor-kappa B pathway activation. J Neurosci.
25:10094–10100. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Thomson EM, Williams A, Yauk CL and
Vincent R: Overexpression of tumor necrosis factor-α in the lungs
alters immune response, matrix remodeling and repair and
maintenance pathways. Am J Pathol. 180:1413–1430. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Moriwaki T, Takagi Y, Sadamasa N, Aoki T,
Nozaki K and Hashimoto N: Impaired progression of cerebral
aneurysms in interleukin-1beta-deficient mice. Stroke. 37:900–905.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang K and Holterman AX: Pathophysiologic
role of hepatocyte nuclear factor 6. Cell Signal. 24:9–16. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jayaraman T, Paget A, Shin YS, Li X, Mayer
J, Chaudhry H, Niimi Y, Silane M and Berenstein A:
TNF-alpha-mediated inflammation in cerebral aneurysms: A potential
link to growth and rupture. Vasc Health Risk Manag. 4:805–817.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Frösen J, Tulamo R, Heikura T, Sammalkorpi
S, Niemelä M, Hernesniemi J, Levonen AL, Hörkkö S and Ylä-Herttuala
S: Lipid accumulation, lipid oxidation and low plasma levels of
acquired antibodies against oxidized lipids associate with
degeneration and rupture of the intracranial aneurysm wall. Acta
Neuropathol Commun. 1:712013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ruigrok YM, Tan S, Medic J, Rinkel GJ and
Wijmenga C: Genes involved in the transforming growth factor beta
signalling pathway and the risk of intracranial aneurysms. J Neurol
Neurosurg Psychiatry. 79:722–724. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tan F, Morris PW, Skidgel RA and Erdos EG:
Sequencing and cloning of human prolylcarboxypeptidase
(angiotensinase C). Similarity to both serine carboxypeptidase and
prolylendopeptidase families. J Biol Chem. 268:16631–16638.
1993.PubMed/NCBI
|
|
30
|
Shoja MM, Agutter PS, Tubbs RS, Payner TD,
Ghabili K and Cohen-Gadol AA: The role of the renin-angiotensin
system in the pathogenesis of intracranial aneurysms. J Renin
Angiotensin Aldosterone Syst. 12:262–273. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dutton A, Young LS and Murray PG: The role
of cellular FLICE inhibitory protein (c-FLIP) in the pathogenesis
and treatment of cancer. Expert Opin Ther Targets. 10:27–35. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Aoki T, Kataoka H, Morimoto M, Nozaki K
and Hashimoto N: Macrophage-derived matrix metalloproteinase-2 and
−9 promote the progression of cerebral aneurysms in rats. Stroke.
38:162–169. 2007. View Article : Google Scholar : PubMed/NCBI
|