Introduction
Osteoporosis, or porous bone, is a serious condition
that impacts the health of hundreds of millions of people. In 2010,
>158 million people suffered from osteoporotic fractures
worldwide (1). Osteoporosis is
typically prevalent in older populations, but can also occur in
children and teenagers. Osteoporosis is characterized by a loss of
bone mass or a reduction in bone mineral density (BMD) (2). Due to the complexity and number of
causes of osteoporosis; however, the prevention and treatment of
osteoporosis is a challenging process and existing therapies have a
limited efficiency.
Estrogen deficiency (3), gene polymorphisms (4), menopause (5) and environmental factors, including
smoking (6) may contribute to
osteoporosis pathogenesis. Postmenopausal osteoporosis or reduction
of BMD is partially due to estrogen deficiency (7). Previous studies focusing on
osteoporosis drugs have demonstrated the risk factors or
side-effects involved in osteoporosis treatment, such as increasing
the risk of bone neoplasms, breast cancer and embolisms (8,9).
Novel drugs with fewer adverse side effects are required for
effective osteoporosis management.
Icariin (ICA) is a flavonol glycoside isolated from
a traditional Chinese medicinal herb Epimedium sagittatum,
the Epimedium genus (10).
Previous studies focusing on the molecular mechanisms of ICA have
demonstrated its anti-osteoporotic and osteogenic differentiation
effects (11,12), as well as its involvement in
estrogen biosynthesis, in vivo and in vitro (13,14).
It is of note that ICA may regulate the expression of
osteoporosis-associated factors, including the Wnt/β-catenin
pathway, peroxisome proliferator-activated receptor γ (PPARγ), and
bone morphogenetic protein (BMP) (15–17).
Chen et al (15)
demonstrated that the administration of ICA to an ovariectomized
rat model of osteoporosis increases the expression of β-catenin
pathway-associated proteins, including runt related transcription
factor 2 and low-density lipoprotein receptor related protein 6.
Proteomics, transcriptomics, and metabolomics analyses have
identified the dysregulation of mRNAs, proteins and metabolites
associated with osteoporosis. However, there are limited studies
that focus on the proteomics associated with the protective
activity of ICA against osteoporosis, particularly the underlying
mechanisms of ICA activity (18).
To further investigate the mechanisms of ICA against
osteoporosis, the proteomics of ICA-treated calvaria osteoblasts
were analyzed using matrix-assisted laser desorption/ionization
time-of-flight mass spectrometry (MALDI-TOF-MS) analysis.
Differentially expressed proteins (DEPs) in ICA-treated osteoblasts
were identified and further investigated. The present study aimed
to provide more information on the protective mechanisms of ICA
against osteoporosis.
Materials and methods
Animals
All animal experimental protocols were approved by
the Animal Care Committee of Nanjing University of Chinese Medicine
(Nanjing, China) and were performed in accordance with the Guide
for Care and Use of Laboratory Animals. A total of 10
Sprague-Dawley male rats (weighing 8–10 g), were obtained from
Experimental Animal Center of Nanjing University of Chinese
Medicine (Nanjing, China) within 24 h post-birth and kept in an
incubator under 12-h light/dark cycle with a humidity of 45–75% at
19–27°C, with free access to food and water.
Osteoblast isolation and cell culture
procedure
The calvaria was dissected from surface-sterilized
rats and subsequently soaked in 75% ethanol for 5–10 min at 4°C.
Isolation of calvaria osteoblasts was performed as previously
described (19,20). Briefly, the frontal and parietal
bone was separated, cut into fragments (1 mm3), digested
in 0.25% trypsin (Gibco; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) at 37°C for 15 min and the precipitates following
centrifugation at 750 × g and 4°C for 5 min were treated with
collagenase II (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for
5 min for further digestion (19).
Precipitated cells were subsequently suspended in Dulbecco's
modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 15% fetal bovine serum (FBS; Gibco; Thermo
Fisher Scientific, Inc.) at 37°C in 5% CO2 for 24 h.
Cells were then transferred to DMEM with 20% FBS. Medium was
replaced every 2 days. For ICA (95.4%; cat. no I1286;
Sigma-Aldrich; Merck KGaA) treatment, ICA was added into the cell
cultures prior to incubation at concentration of 0, 10, and 20 µg/l
at 37°C in 5% CO2 for 24 h. Each experiment was
performed in triplicate.
Cell viability analysis
An MTT (Sigma-Aldrich; Merck KGaA) assay was
performed to detect cell viability as previously described
(16). Osteoblasts (50 cells/well)
were seeded into 96-well plates and maintained in DMEM supplemented
with 20% FBS (Gibco; Thermo Fisher Scientific) at 37°C in 5%
CO2 for 24 h prior to treatment with 10 µg/l and 20 µg/l
ICA for 5 days. The supernatant of cell cultures was subsequently
discarded and 20 µl MTT solution (5 mg/ml) was added into each well
and cells were incubated for a further 4 h. 150 µl DMSO was added
and the optical density at an absorbance of 490 nm was determined
using a microplate reader (Molecular Devices, LLC, Sunnyvale, CA,
USA).
Cell cycle assay
Osteoblasts (5×104 cells/ml) were seeded
into 6-well culture plates until 90% confluence. Cells were then
treated with ICA for 48 h and harvested, fixed with 4%
paraformaldehyde at 4°C for 4 h, and stained with propidium iodide
(PI) for 30 min in the dark at room temperature. For cell cycle
analysis, the at G0/G1, S and G2/M phase distribution of 10,000
cells was determined using the BD FACS Calibur™ flow cytometer
equipped with CellQuest Pro 5.1 software (BD Biosciences, San Jose,
CA, USA). Each experiment was performed in triplicate.
Apoptotic analysis
Osteoblasts were seeded into 6-well plates
(1×105 cells/well) and cultured in DMEM at 37°C in 5%
CO2 for 48 h and treated as aforementioned. Cells were
harvested with 0.25% trypsin (Sigma-Aldrich; Merck KGaA) at 37°C in
5% CO2 for 20 min for apoptotic analysis using the
Annexin V apoptosis detection kit (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. Cells were stained with
Annexin V and PI for 30 min in the dark at room temperature
followed by BD FACS Calibur™ flow cytometry analysis using
CellQuest Pro 5.1 software (BD Biosciences). Annexin V positive and
PI negative stained cells indicated early apoptosis.
Alkaline phosphatase (ALP) staining
and enzyme-linked immunosorbent assay (ELISA)
Following incubation with ICA (10 µg/l) for 1, 2, 3
and 4 days, calvaria osteoblasts were prepared for ALP staining. A
total of 1×104 cells/well were placed into 24-well
plates and incubated in the aforementioned conditions until 85%
confluence. Cells were subsequently harvested, fixed using 4%
paraformaldehyde at 4°C for 30 min and stained for the analysis of
ALP activity with an ALP activity assay kit (BioVision, Inc.,
Milpitas, CA, USA) according to manufacturer's protocol. Images of
stained cells were captured using an Olympus BX51 inverted
fluorescent microscope (magnification, ×40; Olympus Corporation,
Tokyo, Japan). Additionally, the ALP activity of cell cultures was
analyzed using an ELISA kit (cat. no. K422; BioVision, Inc.)
according to manufacturers' instruction.
Alizarin red staining
For the in vitro visualization of nodular
patterns and calcium deposition, osteoblasts were stained with
Alizarin red S after 21 days of culture in DMEM as previously
described (20,21). Briefly, 1,000 cells/well were
placed into 24-well plates to reach to 80% confluence. Cells were
subsequently harvested, fixed using 4% paraformaldehyde at 4°C and
stained with 2% Alizarin red S (Sigma-Aldrich; Merck KGaA) for 30
min at room temperature. Cells were washed with distilled water
prior to observation of plate samples for calcium deposition, which
indicates bone nodule formation or osteoblast mineralization, using
an Olympus BX51 inverted fluorescent microscope (magnification,
×40; Olympus Corporation).
Preparation of proteomics sample
For proteomics analysis, osteoblasts
(1×106 cells/ml) were transferred to a flask and allowed
to reach 80% confluence prior to the addition of ICA (10 µg/l) at
37°C in 5% CO2 for 48 h. The secretory proteins from
conditioned cell cultures of osteoblasts were prepared for
proteomics as previously described (22,23).
In brief, the supernatants of the ICA-treated osteoblasts were
collected and gathered using TCA-acetone solution (1:3) at −20°C
overnight. Precipitates from centrifugation (750 × g at 4°C for 5
min) were subsequently washed with acetone, air-dried, quantified
using the Bradford assay method (Applygen Technologies, Inc.,
Beijing, China) (24) and stored
at −80°C.
Two-dimensional electrophoresis (2-DE)
and gel image scanning
2-DE was conducted as described previously (23). Immobilized pH gradient (IPG) strips
(17 cm; pH 3–10; GE Healthcare Life Sciences, Uppsala, Sweden) were
rehydrated with 150 µg protein for 12–16 h at room temperature,
followed by isoelectric focusing (IEF, 60,000 Vh) in a Protean IEF
cell (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The IPG
strips were subsequently equilibrated as previously described
(25) and transferred onto a 12%
polyacrylamide gel (Beyotime Institute of Biotechnology, Nanjing,
China), which was subjected to the second dimension (200 V, room
temperature) in a Protean II XI cell (Bio-Rad Laboratories, Inc.)
for 7 h at 6°C. Polyacrylamide gels were washed and silver stained
using a Fast Silver Stain kit according to the manufacturer's
protocols (Beyotime Institute of Biotechnology). Finally, the gels
were scanned and analyzed using an ImageScanner™ (GE
Healthcare Life Sciences) transmission scan with ImageMaster
version 5.0 gel image analysis (GE Healthcare Life Sciences). The
silver-stained spots with more than 2-fold change in spot intensity
and novel spots among groups were considered to be upregulated
DEPs. Spots with less than 1-fold change were considered
downregulated DEPs. Each experiment was performed in
triplicate.
MALDI-TOF/MS
A total of 60 DEP spots were manually cut from gels
and digested with trypsin (Gibco; Thermo Fisher Scientific, Inc.)
in 96-well plates, according to previously described methods
(18). DEP spots were subsequently
excised from the polyacrylamide gels. Excised bands were then
destained using 50% acetonitrile and 50 mM ammonium bicarbonate at
37°C for 30 min and digested using trypsin (Gibco; Thermo Fisher
Scientific, Inc.) at 37°C for 16 h. DEPs were subsequently
extracted with 50% acetonitrile and 0.1% trifluoroacetic acid 3
times. Gel spot extracts (1.5 µl) were placed into new 96-well
plates, vacuum dried and stored at −80°C prior to MS analysis using
a MALDI-TOF/MS system (Bruker Corporation, Ettlingen, Germany)
according to instructions as previously described (22,26).
MS database search and alignment of
peptide sequences
MS peptide mass fingerprinting (PMF) data was
obtained using Mascot Distiller software (v.2.3.2; Matrix Science
Ltd., London, UK) (22), and
searches of PMF peptide sequences were performed based on the
international protein index (IPI) rat FASTA database (v3.31; 41,251
sequences; 21,545,744 residues; ftp://ftp.ebi.ac.uk/pub/databases/IPI). Sequence
alignments were generated using the CLC Free Workbench software
package (version 4.0.3; Qiagen, Inc., Valencia, CA, USA), and
protein properties were identified as previously described
(27).
Bioinformatics analysis
In order to investigate the DEP-associated pathways
and Gene Ontology (GO) Consortiums, DEP sequences (peptides) were
analyzed with the Protein ANalysis THrough Evolutionary
Relationships (PANTHER) pathway database (http://www.pantherdb.org/) (28,29).
Proteins were classified according to their function and were
annotated with ontology terms (PANTHER pathways, GO terms and
PANTHER protein class) and sequences are assigned to PANTHER
pathways.
Western blot analysis
Western blot analysis was performed to validate
expression of several identified potential DEPs. Cellular proteins
of calvaria osteoblasts were extracted, quantified as
aforementioned. Proteins were subsequently separated by 10%
SDS-PAGE (Beyotime Institute of Biotechnology) and
electro-transferred onto polyvinylidene difluoride (PVDF) membranes
(Invitrogen; Thermo Fisher Scientific, Inc.). Membranes were
blocked with 5% milk for 1 h at 4°C, probed with the primary
antibodies against β-actin (cat. no. ab8227; 1:1,000; Abcam,
Cambridge, MA, USA), serpin family F member 1 (cat. no. ab14993;
PEDF; 1:1,000; Abcam), signal transducing adaptor molecule (cat.
no. ab48015; STAM; 1:1,000; Abcam), c-Myc (cat. no. ab32072;
1:1,000: Abcam), protein disulfide isomerase family A, member 3
(cat. no. ab13506; PDIA3; 1:1,000; Abcam), heat shock protein
family A member 4 (cat. no. ab2787; HSP70; 1:1,000; Abcam), and
nuclear protein, co-activator of histone transcription (cat. no.
ab70595; NPAT; 1:1,000; Abcam) overnight at 4°C. Membranes were
subsequently incubated with the appropriate horseradish peroxidase
(HRP)-conjugated secondary antibodies (cat. no. ab6721; 1:10,000,
Abcam) for 1 h at room temperature. Bands were subsequently
visualized using an enhanced chemiluminescence detection reagent
(Beyotime Institute of Biotechnology) and were subjected to
densitometric analysis with QuantiScan software (v.3.0; BIOSOFT,
Cambridge, UK).
Statistical analysis
Experimental data was analyzed using SPSS version
17.0 (SPSS, Inc., Chicago, IL, USA). and is expressed as the mean ±
standard deviation. Differences among groups were determined using
one-way analysis of variance (ANOVA) followed by Fisher's Least
Significant Difference for multiple comparisons. P<0.05 was
considered to indicate a statistically significant difference.
Results
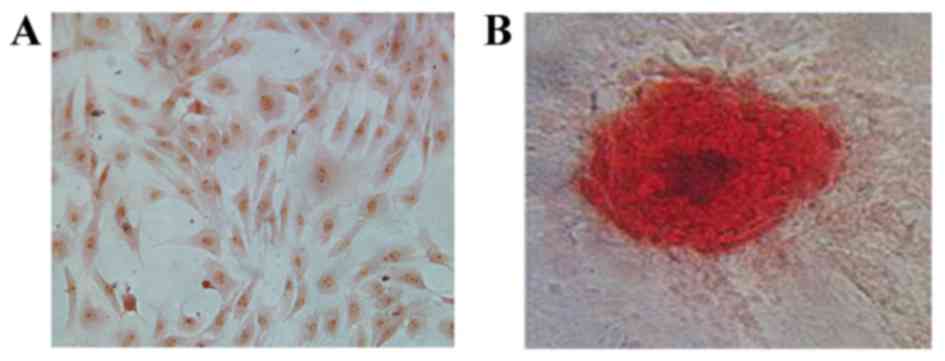
Identification of osteoblasts
Prior to ICA treatment, isolated calvaria
osteoblasts were characterized by ALP and Alizarin red S staining
(Fig. 1). At day 3, cells were
stained with an ALP assay kit and were observed to have purple
stained nuclei accompanied by red-brown stained membrane and
cytoplasm (Fig. 1A). Cells were
stained with Alizarin red S at day 21 to detect calcium and an
irregular red-orange stain was observed, indicating the formation
of mineralized bone nodules and the deposition of calcium-rich
hydroxyapatite (Fig. 1B).
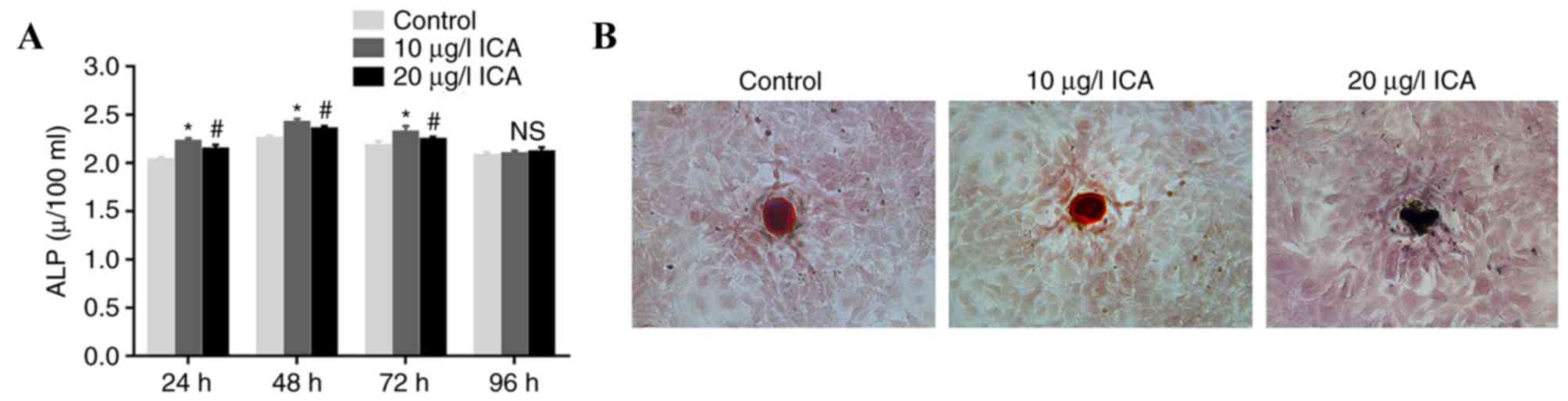
Effect of ICA on ALP activity and
calcium deposition
Osteoblasts were treated with ICA to evaluate the
effect of ICA on cell function. Osteoblasts treated with 10 µg/l
ICA for 48 h exhibited the highest ALP activity (P<0.05;
Fig. 2A). Cells treated with 10
µg/l ICA demonstrated higher ALP activity compared with cells
treated with 20 µg/l ICA at 24, 48 and 72 h time intervals
(P<0.05). For Alizarin red S staining analysis, it was confirmed
that osteoblasts treated with 10 µg/l ICA had brighter irregular
red-orange staining than 20 µg/l ICA-treated cells (Fig. 2B). These results suggested that 10
µg/l ICA concentration was the optimum treatment compared with 20
µg/l for ICA-mediated ALP activation and calcium deposition in
osteoblasts.
ICA promotes cell viability and
inhibits apoptosis
The MTT assay confirmed that ICA promoted the cell
viability of osteoblast in a dose- and time-dependent manner
(Fig. 3A). Cells treated with 10
µg/l ICA had significantly increased cell viability at 72 h
following ICA incubation (P<0.05). Notably, osteoblasts treated
with 20 µg/l ICA had decreased cell viability compared with the 10
µg/l ICA treatment (P<0.05).
Cell cycle distribution analysis revealed that ICA
treatment increased the percentage of cells in the S and G2/M
phases, and reduced the percentage in the G0/G1 phase (Fig. 3B). As expected, there was also an
increase in the percentage of cells in the S and G2/M phase and a
decrease in the G0/G1 phase in osteoblasts treated with 10 µg/l ICA
compared with those treated with 20 µg/l ICA (P<0.05). Cell
apoptosis analysis revealed that ICA inhibited cell apoptosis
(Fig. 3C). Osteoblasts treated
with 10 µg/l ICA had a significantly lower apoptotic rate when
compared with the control and 20 µg/l ICA treated cells
(P<0.05). Taken together, it was concluded that ICA increased
cell viability and inhibited cell apoptosis. Furthermore, it was
determined that the optimum ICA concentration for osteoblast
viability was 10 µg/l.
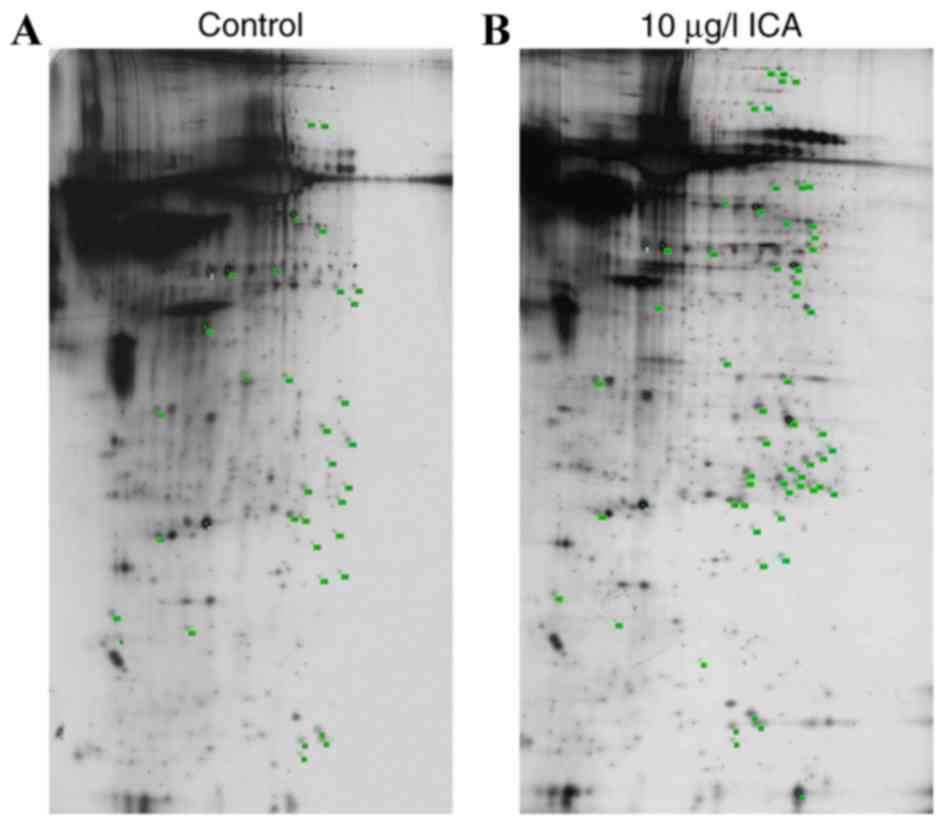
2-DE/MALDI-TOF/MS analysis
Proteomic analysis revealed 60 proteins points that
were differentially expressed (with more than two-fold change in
spot intensity) or specifically expressed (expression specifically
induced by ICA) in osteoblasts treated with 10 µg/l ICA, compared
with the control with >750 silver-stained spots (Fig. 4). Bioinformatics analysis
identified 56 DEP sequences, which were matched to reference
sequences in databases using the CLC Free Workbench software
package. Identified DEPS included 22 upregulated DEPs, including
STAM binding protein, insulin-like growth factor-binding protein 2
(IGFBP-2) precursor and PEDF. Eight downregulated DEPs were
identified, including phosphoglycerate mutase 1 and myosin-1 and
succinate dehydrogenase subunit A. A total of 24 specially
expressed proteins were identified in ICA-treated osteoblasts,
including heat shock proteins [heat shock cognate 71 kDa protein
(HSPA8) and heat shock protein, mitochondrial precursor],
myc-associated factor X (Max), nuclear protein
ataxia-telangiectasia (NPAT) and PDIA3 precursor. Glutaredoxin 5
and translationally controlled tumor protein were DEPs that were
lost in ICA treated cells (Table
I). A total of four downregulated DEPs in ICA treated
osteoblasts were not aligned in the CLC Free Workbench software
package.
 | Table I.DEPs in ICA-treated osteoblasts. |
Table I.
DEPs in ICA-treated osteoblasts.
| A, Upregulated
DEPs |
|---|
|
| Protein point | Sequence number of
protein warehouse | Name of
protein | Fold-change |
|---|
| 59 | IPI00231275 | Galectin-1 | 5.7 |
| 62 | IPI00230937 |
Phosphatidylethanol-amine-binding
protein | 5.5 |
| 65 | IPI00196994 | GDP
dissociation | 6.7 |
| 72 | IPI00231260 |
Peroxiredoxin-6 | 6.8 |
| 112 | IPI00765011 | Follistatin-related
protein 1 precursor | 7.6 |
| 118 | IPI00207063 | Follistatin-related
protein 1 precursor | 11.8 |
| 123 | IPI00464670 | Macrophage-capping
protein | 5.2 |
| 127 | IPI00778493 | Serpinf1 41 kDa
protein | 9.2 |
| 135 | IPI00189819 | β-actin | 8.6 |
| 149 | IPI00553950 | Prolactinregulatory
element-binding protein | 10.7 |
| 155 | IPI00200044 | STAM binding
protein | 12.4 |
| 186 | IPI00370815 | T-complex protein 1
subunit theta | 11.2 |
| 219 | IPI00778705 | Ephrin type-A
receptor 8 | 7.3 |
| 271 | IPI00777582 | −69 kDa
protein | 13.0 |
| 284 | IPI00188921 | Collagen alpha-2
(I) chain precursor | 17.5 |
| 285 | IPI00326412 | Gamma-enolase | 5.7 |
| 357 | IPI00201060 | Lamin-A | 5.7 |
| 358 | IPI00421947 | DPPY | 5.2 |
| 136 | IPI00201573 | Insulin-like growth
factor-binding protein 2 precursor | 2.1 |
| 159 | IPI00209148 | Isoform 1 of
Heterogeneous nuclear ribonucleoprotein M | 3.5 |
| 330 | IPI00765011 | Actin, cytoplasmic
2 | 4.3 |
| 169 | IPI00212901 | Uncharacterized
protein C18 or f19 homolog | 5.2 |
|
| B, Downregulated
DEPs |
|
| Protein point | Sequence number of
protein warehouse | Name of
protein | Fold-change |
|
| 29 | IPI00764455 | Myosin-1 | 0.1 |
| 101 | IPI00421428 | Phosphoglycerate
mutase 1 | 0.3 |
| 251 | IPI00567268 | Sdha 72 kDa
protein | 0.3 |
| 105 | IPI00362469 |
6-phosphogluconolactonase | 0.7 |
| 129 | IPI00189819 | β-actin | 0.6 |
| 227 | IPI00187662 | Cyfip1_predicted 51
kDa protein | 0.8 |
| 296 | IPI00230941 | Vimentin | 0.6 |
| 306 | IPI00195929 | CAP-Gly
domain-containing linker protein 2 | 0.7 |
|
| C, DEPs
specifically expressed in control |
|
| Protein point | Sequence number of
protein warehouse | Name of
protein | Fold-change |
|
| 8 | IPI00365904 | Glutaredoxin 5 | – |
| 20 | IPI00208306 | Translation
ally-controlled tumor protein | – |
| 45 | IPI00558185 | Max (18 kDa
protein) | – |
| 91 | IPI00213667 | Hemiferrin | – |
| 143 | IPI00464815 | Alpha enolase | – |
| 145 | IPI00464815 | Alpha enolase | – |
| 147 | IPI00464815 | Alpha enolase | – |
| 151 | IPI00566018 | Npat | – |
|
| D, DEPs
specifically expressed in response to ICA |
|
| Protein point | Sequence number of
protein warehouse | Name of
protein | Fold-change |
|
| 153 | IPI00767505 | γ-actin | – |
| 158 | IPI00780207 | Dlec1 39 kDa
protein | – |
| 174 | IPI00212810 | Sfrs2 29 kDa
protein | – |
| 176 | IPI00364170 | Spetex-2E | – |
| 180 | IPI00192078 | Biphenyl
hydrolase-like | – |
| 248 | IPI00845891 | transglutaminase
4 | – |
| 263 | IPI00208188 | Syntaxin-binding
protein | – |
| 303 | IPI00230941 | Vimentin | – |
| 314 | IPI00551812 | ATP synthase
subunit beta, mitochondrial precursor | – |
| 323 | IPI00324741 | Protein
disulfide-isomerase A3 precursor | – |
| 325 | IPI00324741 | Protein
disulfide-isomerase A3 precursor | – |
| 339 | IPI00192984 | Argbp2 78 kDa
protein | – |
| 340 | IPI00339148 | heat shock protein,
mitochondrial precursor | – |
| 342 | IPI00208205 | Heat shock cognate
71 kDa protein | – |
| 364 | IPI00366944 | Collagen alpha-1
(III) chain precursor | – |
| 365 | IPI00409539 | Filamin-A | – |
| 368 | IPI00369732 | Serine-protein
kinase ATM | – |
| 369 | IPI00360916 | GRIP and
coiled-coil domain-containing 2 | – |
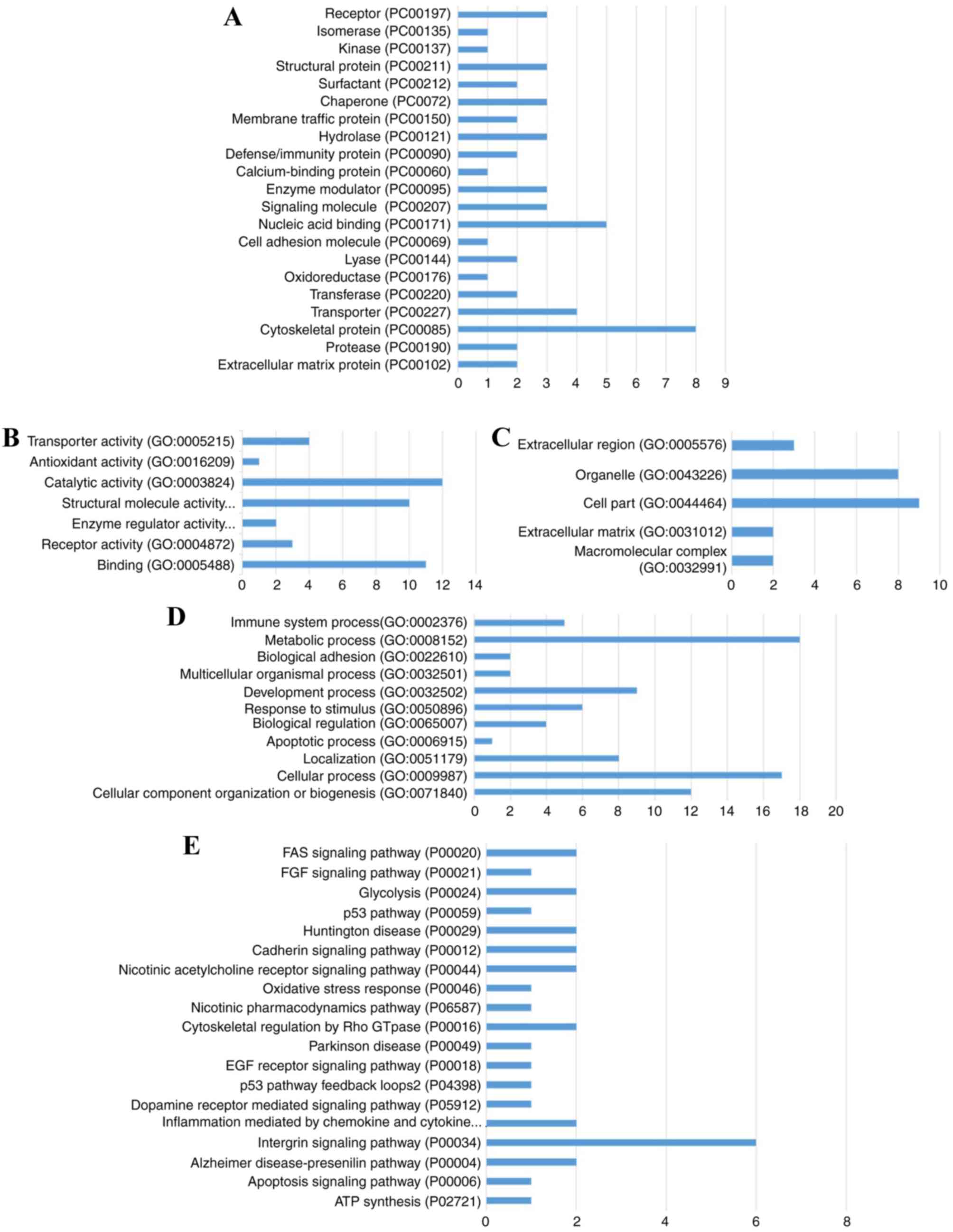
Bioinformatics analysis
Sequences of these 56 DEPs were subjected to PANTHER
pathway database analysis (29,30).
DEP enrichment analysis for PANTHER pathways, GO terms (biological
processes, molecular function and cellular component) and PANTHER
protein classification were performed (Fig. 5). Results revealed that these DEPs
were associated with PANTHER pathways, including receptor
(PC00197), isomerase (PC00135), calcium-binding protein (PC00060)
and signaling molecule (PC00207); enriched into GO terms including
‘transporter activity’ (GO: 0005215), ‘catalytic activity’ (GO:
0003824), ‘immune system process’ (GO: 0002376) and ‘cell part’
(GO: 0044464); annotated into pathways including Fas (P00020),
fibroblast growth factor (FGF; P00021), p53 (P00059) and apoptosis
(P04398).
Validation of DEP expression by
western blot analysis
Previous reports have demonstrated the association
of gene dysregulation with osteoporosis or bone mineral content
accumulation, including PEDF (30,31)
and PDIA3 (32). Furthermore,
NPAT, c-Myc and several other proteins are thought to be involved
in the regulation of histone transcription (32,33),
calcium metabolism (34) and cell
proliferation (35,36). Max associates and interacts with
c-Myc, thus forming the c-Myc-Max complex, which has core
activating roles in gene expression. Inhibition of c-Myc-Max
dimerization formation results in the inhibition of cancer cell
proliferation (37,38). HSPA8 is a gene that encodes for an
important member of HSP70 protein family (39). Therefore, the expression of HSP70
was investigated. In the present study, Max was identified as a
protein that was specifically expressed in response to ICA
administration. The expression of c-Myc protein and several
identified DEPs in ICA (10 and 20 µg/l) treated osteoblasts was
subsequently detected. ICA treatment was demonstrated to upregulate
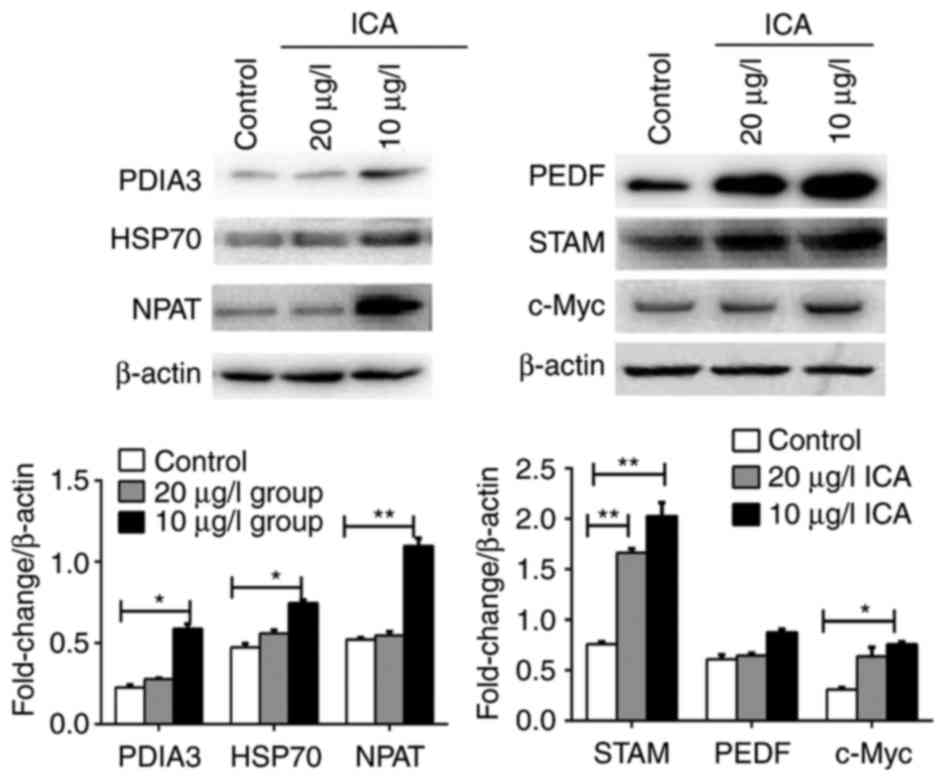
the expression of PEDF, STAM, c-Myc, PDIA3, HSP70 and NPAT.
Expression of these DEPs were higher in cells treated with 10 µg/l
ICA compared with control cells (P<0.05; Fig. 6). In addition, the expression
levels of PEDF, PDIA3, HSP70 and NPAT proteins exhibited by the
control cells were not significantly different compared with cells
treated with 20 µg/l ICA. These findings were in accordance with
the bioinformatics analysis results.
Discussion
The antiosteoporotic activity of ICA has been
previously reported (11,12) and was further confirmed in the
present study. ICA treatment promoted the viability, ALP activity
and calcium deposition of osteoblasts in a dose- and time-dependent
manner. Proteomics analysis revealed that the identified proteins
were involved in osteogenesis and bone mineral content accumulation
via several signaling pathways.
The proliferation inducing effect of ICA was
identified in our previous study (40) and other previous research (12,41).
ICA is a flavonoid that exhibits anti-apoptosis, cell
differentiation and proliferation inducing effects (42,43).
In the present study, ICA treatment was demonstrated to promote the
proliferation of osteoblasts in a dose-dependent manner. ICA (10
µg/l) demonstrated a more pronounced effect on cell viability, ALP
activity and formation of osteoblast mineralized bone nodule, with
lower apoptosis percentage compared with 20 µg/l ICA treatment
(Figs. 2 and 3). It was confirmed that 10 µg/l ICA was
the most effective concentration for osteoblast proliferation and
antiosteoporotic activity. This was in accordance with our previous
study (40).
Based on the proteomics analysis, 56 proteins were
identified as differentially expressed in the 10 µg/l ICA treated
osteoblasts compared with the control cells, including PEDF, HSPs,
NPAT, PDIA3 and STAM. These proteins were enriched in pathways, GO
function terms and PANTHER protein classes including
calcium-binding protein, signaling molecule, immune system process
and signaling pathways such as Fas, FGF, p53, and apoptosis. As
previously reported, p53-B-cell lymphoma 2 (Bcl-2) /Bcl-2
associated protein X-Fas/Fas ligand and FGF-2-p53 are important
signaling pathways that modulate apoptosis and proliferation
(44,45), demonstrating that the DEPs induced
by ICA treatment may be associated with cell differentiation,
proliferation and apoptosis. Further validation by western blot
analysis revealed that the expression of PEDF, PDIA3, NPAT, NPAT,
STAM, HSP70 and c-Myc proteins were upregulated by 10 µg/l ICA
administration. This was in accordance with cellular function
alterations observed in cells treated with ICA, demonstrating that
ICA-induced alterations may be mediated by these proteins.
Previous reports have revealed that the expression
of PEDF (30,31), NPAT (32,33)
and PDIA3 (32) may directly or
indirectly contribute to osteoporosis. PEDF, a 50-kDa secreted
glycoprotein encoded by SERPINF1, is an endogenous
anti-inflammatory factor (46),
which also acts as an anti-tumor factor (47,48).
PEDF can induce p53- and Fas-mediated cell apoptosis and the
expression of anti-inflammatory factors through PPARγ signaling
(49). PPARγ is an essential
factor for adipocyte differentiation and suppression of its
expression has been determined to be crucial for the promotion of
osteogenesis and the inhibition of adipogenesis (50,51).
NPAT is an essential factor in histone transcription
regulation and acts directly downstream of cyclin
E/cyclin-dependent kinase 2 (52).
The phosphorylation or upregulation of NPAT is required for the
expression of histone genes, which modulate DNA synthesis and cell
cycle proliferation (53).
Additionally, NPAT expression correlates with the S phase (53). However, to the best of our
knowledge, no reports have investigated the association of PEDF and
NPAT with osteoporosis. In the present study, it was confirmed that
PEDF and NPAT expression, as well as cell viability in 10 µg/l
ICA-treated osteoblasts were upregulated, suggesting that PEDF and
NPAT may contribute to osteogenesis via signaling pathways,
including Fas and p53 in vitro.
Max and c-Myc forms the c-Myc-Max complex, which has
a core role in trans-activating gene expression (54,55),
whereas the inhibition of c-Myc-Max complex has been previously
reported to lead to cell cycle arrest, apoptosis and the inhibition
of cancer cell proliferation (37,39).
Indo et al (56) reported
that the inhibition of c-Myc reduces bone-resorbing activity in
mature osteoclasts and suppresses the expression of solute carrier
family, neutral amino acid transporter (B0), which in turn results
in the suppression of osteoclastogenesis. In the present study, it
was demonstrated that the expression of Max and c-Myc was
significantly upregulated by ICA treatment. Osteoblast apoptosis
was inhibited and cell viability and proliferation was enhanced by
ICA, suggesting that the formation of the c-Myc-Max complex and
subsequent trans-activated gene expression was promoted by ICA
treatment.
The PDIA3/phospholipase A2 activating protein
receptor complex was reported to be necessary for the Wnt/β-catenin
pathway, particularly Wnt5a calcium-dependent pathways in
osteoblasts and chondrocytes (32,57).
Expression of HSP70 promotes osteoblast differentiation via
activation of the mitogen-activated protein kinase signaling
pathway (58). Additionally, c-Myc
gene products promote HSP70 expression (59) and both c-Myc and HSP70 contribute
to cell cycle regulation (60). In
the present study, c-Myc, Max, PDIA3 and HSP70 expression was
significantly upregulated by ICA treatment. Bioinformatics analysis
revealed that these proteins were associated with pathways that may
mediate calcium metabolism, suggesting a link between c-Myc, Max,
PDIA3 and HSP70 overexpression and osteoporosis, as well as the
therapeutic effects of ICA in osteoporosis.
In conclusion, it was demonstrated that ICA
treatment promoted proliferation, calcium deposition and inhibited
osteoblast apoptosis. Using proteomics analysis, 56 DEPs were
identified between cells treated with 10 µg/l ICA and control
groups, including PEDF, PDIA3, NPAT, STAM, HSP70 and c-Myc, which
may be associated with ICA-mediated osteogenesis differentiation.
However, additional experiments should be performed to further
elucidate the interaction of these DEPs with ICA-induced
osteogenesis and the involvement of these proteins in ICA-induced
cell proliferation, calcium accumulation and osteogenesis.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Science
Foundation of Traditional Chinese Medicine Administration, Jiangsu
Province (grant no. LB09083).
Availability of data and materials
All the data and materials in this manuscript are
available from the corresponding author on reasonable request.
Authors' contributions
HY and XZ conceived and designed the experiments.
Cell experiments were performed by WQ, YS and YZ. Secretome and
data analysis were performed by XZ and NG. Statistical analysis was
performed by NY. All authors participated in the writing of the
manuscript.
Ethics approval and consent to
participate
Animal experimental protocol was approved by the
Animal Care Committee of Nanjing University of Chinese Medicine
(Nanjing, China). No human subjects were enrolled in this
study.
Consent for publication
Not applicable.
Competing interests
The authors declare that there are no competing
interests.
References
|
1
|
Odén A, McCloskey EV, Kanis JA, Harvey NC
and Johansson H: Burden of high fracture probability worldwide:
Secular increases 2010–2040. Osteoporos Int. 26:2243–2248. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zambelli PY, Tercier S, Newman CJ and
Bregou A: Targeted approach to osteoporosis for children and
teenagers. Rev Med Suisse. 10:116–118. 2014.(In French). PubMed/NCBI
|
|
3
|
Tyagi AM, Srivastava K, Mansoori MN,
Trivedi R, Chattopadhyay N and Singh D: Estrogen deficiency induces
the differentiation of IL-17 secreting Th17 cells: A new candidate
in the pathogenesis of osteoporosis. PLoS One. 7:e445522012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kurt-Sirin O, Yilmaz-Aydogan H, Uyar M,
Seyhan MF, Isbir T and Can A: Combined effects of collagen type I
alpha1 (COL1A1) Sp1 polymorphism and osteoporosis risk factors on
bone mineral density in Turkish postmenopausal women. Gene.
540:226–231. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cheung A, Papaioannou A and Morin S:
Osteoporosis Canada Scientific Advisory Council: Postmenopausal
osteoporosis. N Engl J Med. 374:20962016.PubMed/NCBI
|
|
6
|
Chen H, Liu N, Xu X, Qu X and Lu E:
Smoking, radiotherapy, diabetes and osteoporosis as risk factors
for dental implant failure: A meta-analysis. PLoS One.
8:e719552013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Miyamoto T: Mechanism underlying
post-menopausal osteoporosis: HIF1α is required for osteoclast
activation by estrogen deficiency. Keio J Med. 64:44–47. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ahn HJ, Kim HJ, Kim YS, Kim MS, Huh KH,
Kim JH, Lee JH, Jeon KO and Kim SI: Risk factors for changes in
bone mineral density and the effect of antiosteoporosis management
after renal transplantation. Transplant Proc. 38:pp. 2074–2076.
2006; View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Díez-Pérez A, Adachi JD, Adami S, Anderson
FA Jr, Boonen S, Chapurlat R, Compston JE, Cooper C, Gehlbach SH,
Greenspan SL, et al: Risk factors for treatment failure with
antiosteoporosis medication: The global longitudinal study of
osteoporosis in women (GLOW). J Bone Miner Res. 29:260–267. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee MK, Choi YJ, Sung SH, Shin DI, Kim JW
and Kim YC: Antihepatotoxic activity of icariin, a major
constituent of Epimedium koreanum. Planta Med. 61:523–526. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen KM, Ge BF, Ma HP, Liu XY, Bai MH and
Wang Y: Icariin, a flavonoid from the herb Epimedium enhances the
osteogenic differentiation of rat primary bone marrow stromal
cells. Pharmazie. 60:939–942. 2005.PubMed/NCBI
|
|
12
|
Meng FH, Li YB, Xiong ZL, Jiang ZM and Li
FM: Osteoblastic proliferative activity of Epimedium brevicornum
Maxim. Phytomedicine. 12:189–193. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xue L, Wang Y, Jiang Y, Han T, Nie Y, Zhao
L, Zhang Q and Qin L: Comparative effects of er-xian decoction,
epimedium herbs, and icariin with estrogen on bone and reproductive
tissue in ovariectomized rats. Evid Based Complementary Altern Med.
2012:2414162012. View Article : Google Scholar
|
|
14
|
Yang L, Lu D, Guo J, Meng X, Zhang G and
Wang F: Icariin from Epimedium brevicornum Maxim promotes the
biosynthesis of estrogen by aromatase (CYP19). J Ethnopharmacol.
145:715–721. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen G, Wang C, Wang J, Yin S, Gao H,
Xiang LU, Liu H, Xiong Y, Wang P, Zhu X, et al: Antiosteoporotic
effect of icariin in ovariectomized rats is mediated via the
Wnt/β-catenin pathway. Exp Ther Med. 12:279–287. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu H, Xiong Y, Zhu X, Gao H, Yin S, Wang
J, Chen G, Wang C, Xiang L, Wang P, et al: Icariin improves
osteoporosis, inhibits the expression of PPARγ, C/EBPα, FABP4 mRNA,
N1ICD and jagged1 proteins, and increases Notch2 mRNA in
ovariectomized rats. Exp Ther Med. 13:1360–1368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li XF, Xu H, Zhao YJ, Tang DZ, Xu GH, Holz
J, Wang J, Cheng SD, Shi Q and Wang YJ: Icariin augments bone
formation and reverses the phenotypes of osteoprotegerin-deficient
mice through the activation of Wnt/β-catenin-BMP signaling. Evid
Based Complementary Altern Med. 2013:6523172013. View Article : Google Scholar
|
|
18
|
Xue L, Jiang Y, Han T, Zhang N, Qin L, Xin
H and Zhang Q: Comparative proteomic and metabolomic analysis
reveal the antiosteoporotic molecular mechanism of icariin from
Epimedium brevicornu maxim. J Ethnopharmacol. 192:370–381. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bakker A and Klein-Nulend J: Osteoblast
isolation from murine calvariae and long bones. Methods Mol Med.
80:19–28. 2013.
|
|
20
|
Chang H, Jin TY, Jin WF, Gu SZ and Zhou
YF: Modulation of isoflavones on bone-nodule formation in rat
calvaria osteoblasts in vitro. Biomed Environ Sci. 16:83–89.
2003.PubMed/NCBI
|
|
21
|
Honda H, Tamai N, Naka N, Yoshikawa H and
Myoui A: Bone tissue engineering with bone marrow-derived stromal
cells integrated with concentrated growth factor in Rattus
norvegicus calvaria defect model. J Artif Organs. 16:305–315. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Coutu DL, Wu JH, Monette A, Rivard GE,
Blostein MD and Galipeau J: Periostin, a member of a novel family
of vitamin K-dependent proteins, is expressed by mesenchymal
stromal cells. J Biol Chem. 283:17991–18001. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao Y, Ju F, Zhao Y, Wang L, Sun Z, Liu M
and Gao L: The expression of αA- and βB1-crystallin during normal
development and regeneration, and proteomic analysis for the
regenerating lens in Xenopus laevi. Mol Vis. 17:768–778.
2011.PubMed/NCBI
|
|
24
|
Bradford MM: A rapid and sensitive method
for the quantitation of microgram quantities of protein utilizing
the principle of protein-dye binding. Anal Biochem. 72:248–254.
1976. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xing X, Lai M, Gartner W, Xu E, Huang Q,
Li H and Chen G: Identification of differentially expressed
proteins in colorectal cancer by proteomics: Down-regulation of
secretagogin. Proteomics. 6:2916–2923. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ma X, Liu G, Wang S, Chen Z, Lai M, Liu Z
and Yang J: Evaluation of sphingolipids changes in brain tissues of
rats with pentylenetetrazol-induced kindled seizures using
MALDI-TOF-MS. J Chromatogr B Analyt Technol Biomed Life Sci.
859:170–177. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Powell TJ, Strutt T, Reome J, Hollenbaugh
JA, Roberts AD, Woodland DL, Swain SL and Dutton RW: Priming with
cold-adapted influenza A does not prevent infection but elicits
long-lived protection against supralethal challenge with
heterosubtypic virus. J Immunol. 178:1030–1038. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mi H, Muruganujan A, Casagrande JT and
Thomas PD: Large-scale gene function analysis with the PANTHER
classification system. Nat Protoc. 8:1551–1566. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mi H and Thomas P: PANTHER pathway: An
ontology-based pathway database coupled with data analysis tools.
Methods Mol Biol. 563:123–140. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bogan R, Riddle RC, Li Z, Kumar S, Nandal
A, Faugere MC, Boskey A, Crawford SE and Clemens TL: A mouse model
for human osteogenesis imperfecta type VI. J Bone Miner Res.
28:1531–1536. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gattu AK, Swenson ES, Iwakiri Y, Samuel
VT, Troiano N, Berry R, Church CD, Rodeheffer MS, Carpenter TO and
Chung CH: Determination of mesenchymal stem cell fate by pigment
epithelium-derived factor (PEDF) results in increased adiposity and
reduced bone mineral content. FASEB J. 27:4384–4394. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Doroudi M: Essential roles of Pdia3/PLAA
receptor complex and CaMKII IN 1α,25(OH)2D3
and Wnt5a calcium-dependent signaling pathways in osteoblasts and
chondrocytes. 2014.
|
|
33
|
Frank SR, Schroeder M, Fernandez P,
Taubert S and Amati B: Binding of c-Myc to chromatin mediates
mitogen-induced acetylation of histone H4 and gene activation.
Genes Dev. 15:2069–2082. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kaibuchi K, Tsuda T, Kikuchi A, Tanimoto
T, Yamashita T and Takai Y: Possible involvement of protein kinase
C and calcium ion in growth factor-induced expression of c-myc
oncogene in Swiss 3T3 fibroblasts. J Biol Chem. 261:1187–1192.
1986.PubMed/NCBI
|
|
35
|
Vadde R, Radhakrishnan S, Kurundu HEK,
Reddivari L and Vanamala JK: Indian gooseberry (Emblica officinalis
Gaertn.) suppresses cell proliferation and induces apoptosis in
human colon cancer stem cells independent of p53 status via
suppression of c-Myc and cyclin D1. J Funct Foods. 25:267–278.
2016. View Article : Google Scholar
|
|
36
|
Zhang W, Shen X, Wan C, Zhao Q, Zhang L,
Zhou Q and Deng L: Effects of insulin and insulin-like growth
factor 1 on osteoblast proliferation and differentiation:
differential signalling via Akt and ERK. Cell Biochem Funct.
30:297–302. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen BJ, Wu YL, Tanaka Y and Zhang W:
Small molecules targeting c-Myc oncogene: promising anti-cancer
therapeutics. Int J Biol Sci. 10:1084–1096. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zirath H, Frenzel A, Oliynyk G, Segerström
L, Westermark UK, Larsson K, Munksgaard Persson M, Hultenby K,
Lehtiö J, Einvik C, et al: MYC inhibition induces metabolic changes
leading to accumulation of lipid droplets in tumor cells. Proc Natl
Acad Sci USA. 110:pp. 10258–10263. 2013; View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Banerjee D, Upadhyay RC, Chaudhary UB,
Kumar R, Singh S, Ashutosh G JM, Polley S, Mukherjee A, Das TK and
De S: Seasonal variation in expression pattern of genes under
HSP70: Seasonal variation in expression pattern of genes underHSP70
family in heat- and cold-adapted goats (Capra hircus). Cell Stress
Chaperones. 19:401–408. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Qian W, Yin H and Sun H: Influence of
different concentrations of Icariin on osteoblast metabolism of
rats. China Med Herald. 8:23–25. 2011.(In Chinese).
|
|
41
|
Song L, Zhao J, Zhang X, Li H and Zhou Y:
Icariin induces osteoblast proliferation, differentiation and
mineralization through estrogen receptor-mediated ERK and JNK
signal activation. Eur J Pharmacol. 714:15–22. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cao H, Ke Y, Zhang Y, Zhang CJ, Qian W and
Zhang GL: Icariin stimulates MC3T3-E1 cell proliferation and
differentiation through up-regulation of bone morphogenetic
protein-2. Int J Mol Med. 29:435–439. 2012.PubMed/NCBI
|
|
43
|
Zhang DW, Cheng Y, Wang NL, Zhang JC, Yang
MS and Yao XS: Effects of total flavonoids and flavonol glycosides
from Epimedium koreanum Nakai on the proliferation and
differentiation of primary osteoblasts. Phytomedicine. 15:55–61.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Duan P, Hu C, Butler HJ, Quan C, Chen W,
Huang W, Tang S, Zhou W, Yuan M, Shi Y, et al: 4-Nonylphenol
induces disruption of spermatogenesis associated with oxidative
stress-related apoptosis by targeting p53-Bcl-2/Bax-Fas/FasL
signaling. Environ Toxicol. 32:739–753. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Han Y, Jiang Q, Wang Y, Li W, Geng M, Han
Z and Chen X: The anti-proliferative effects of oleanolic acid on
A7r5 cells-Role of UCP2 and downstream FGF-2/p53/TSP-1. Cell Biol
Int. 41:1296–1306. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang SX, Wang JJ, Gao G, Shao C, Mott R
and Ma JX: Pigment epithelium-derived factor (PEDF) is an
endogenous antiinflammatory factor. FASEB J. 20:323–325. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jarvis CL, Nelius T, Martinez-Marin D,
Cheerla S and Filleur S: Low-dose cabazitaxel inhibits the growth
of prostate cancer cells and enhances the anti-tumor properties of
PEDF with greater efficacy than docetaxel. Cancer Res. 76:2882016.
View Article : Google Scholar
|
|
48
|
Nelius T, Hirsch J and Filleur S: PEDF
inhibits bone metastases formation, prolongs survival and enhances
the antitumor efficacy of low-dose chemotherapy in
castration-refractory prostate cancer. Cancer Res. 73:50972013.
View Article : Google Scholar
|
|
49
|
Ho TC, Chen SL, Yang YC, Liao CL, Cheng HC
and Tsao YP: PEDF induces p53-mediated apoptosis through PPAR gamma
signaling in human umbilical vein endothelial cells. Cardiovasc
Res. 76:213–223. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Sun H, Kim JK, Mortensen R, Mutyaba LP,
Hankenson KD and Krebsvach PH: Osteoblast-targeted suppression of
PPARγ increases osteogenesis through activation of mTOR signaling.
Stem Cells. 31:2183–2192. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Brusotti G, Montanari R, Capelli D,
Cattaneo G, Laghezza A, Tortorella P, Loiodice F, Peiretti F,
Bonardo B, Paiardini A, et al: Betulinic acid is a PPARγ antagonist
that improves glucose uptake, promotes osteogenesis and inhibits
adipogenesis. Scienti Rep. 7:57772017. View Article : Google Scholar
|
|
52
|
Ling Zheng L, Wang FY, Cong XX, Shen Y,
Rao XS, Huang DS, Fan W, Yi P, Wang XB, Zheng L, et al: Interaction
of heat shock protein Cpn10 with the Cyclin E/Cdk2 substrate
nuclear protein ataxia-telangiectasia (NPAT) is involved in
regulating histone transcription. J Biol Chem. 290:29290–29300.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhao J: Coordination of DNA synthesis and
histone gene expression during normal cell cycle progression and
after DNA damage. Cell Cycle. 3:693–695. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lin CY, Lovén J, Rahl PB, Paranal RM,
Burge CB, Bradner JE, Lee TI and Young RA: Transcriptional
amplification in tumor cells with elevated c-Myc. Cell. 151:56–67.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Miller DM, Thomas SD, Islam A, Muench D
and Sedoris K: c-Myc and cancer metabolism. Clin Cancer Res.
18:5546–5553. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Indo Y, Takeshita S, Ishii KA, Hoshii T,
Aburatani H, Hirao A and Ikeda K: Metabolic regulation of
osteoclast differentiation and function. J Bone Miner Res.
28:2392–2399. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chen J, Olivares-Navarrete R, Wang Y,
Herman TR, Boyan BD and Schwartz Z: Protein-disulfide
isomerase-associated 3 (Pdia3) mediates the membrane response to
1,25-dihydroxyvitamin D3 in osteoblasts. J Biol Chem.
285:37041–37050. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Chen E, Xue D, Zhang W, Lin F and Pan Z:
Extracellular heat shock protein 70 promotes osteogenesis of human
mesenchymal stem cells through activation of the ERK signaling
pathway. FEBS Lett. 589:4088–4096. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kingston RE, Baldwin AS Jr and Sharp PA:
Regulation of heat shock protein 70 gene expression by c-myc.
Nature. 312:280–282. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Taira T, Sawai M, Ikeda M, Tamai K,
Iguchi-Ariga SM and Ariga H: Cell cycle-dependent switch of up-and
down-regulation of human hsp70 gene expression by interaction
between c-Myc and CBF/NF-Y. J Biol Chem. 274:24270–24279. 1999.
View Article : Google Scholar : PubMed/NCBI
|