Introduction
Methicillin-resistant Staphylococcus aureus
(MRSA) is a S. aureus-associated infection that has been
reported worldwide, which has now become one of the most important
pathogens in nosocomial and communal infections (1). The mortality rates associated with
MRSA infection are as high as 63.1%, and nosocomial infections
account for >60–80% of cases (2,3).
Previous research has revealed that the bacterial efflux system is
the main cause of multidrug resistance (MDR), and therefore
identifying effective efflux pump inhibitors that prevent the
bacteria from expelling the drugs out of the cells is a valid
method of targeting bacterial MDR (4). A number of efflux pump inhibitors
have been reported, including reserpine, LY35979, glyburide and
phenothiazine-metal complexes, cyanide chlorobenzene hydrazone and
verapamil, all of which inhibited the bacterial efflux system and
restored bacterial sensitivity to antibiotics and other drugs
(5,6). However, these drugs exhibit several
side effects that limit their use for clinical application.
Medicinal herbs used in traditional Chinese medicine tend to have
fewer side effects, and therefore recent research has focused on
the screening of efflux pump inhibitors from these herbs. Extracts
from certain herbs have been demonstrated to inhibit the efflux
pump of S. aureus, including Fritillaria, Belamcanda
chinensis, Andrographis paniculata (7). Silybin is a flavonolignan component
of the extract from the milk thistle seed, termed silymarin.
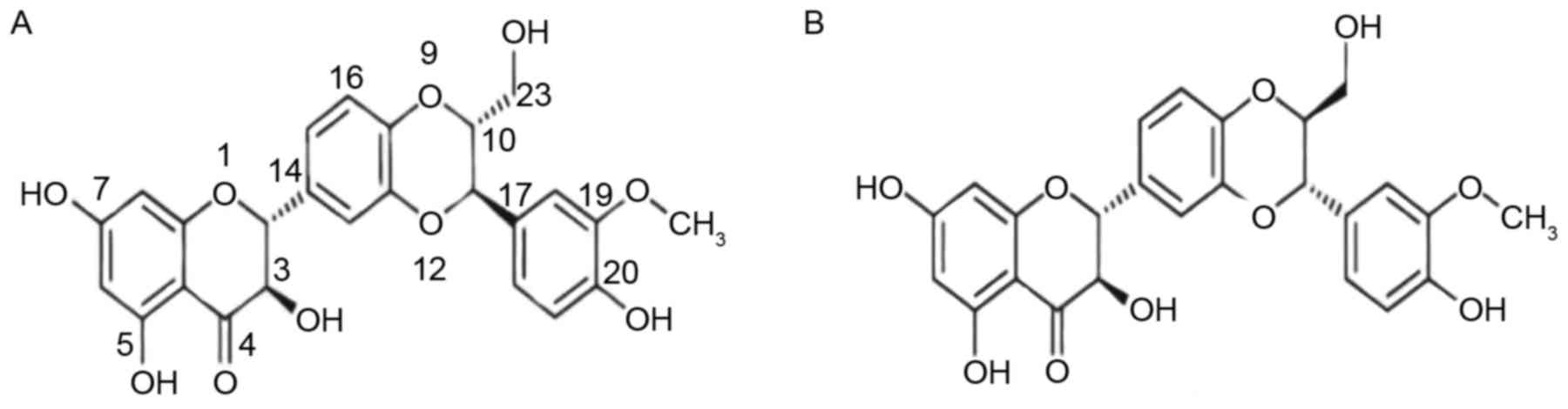
Silybin is present in silymarin as an approximately equal-molar
mixture of two diastereoisomers: silybinA and silybinB (Fig. 1). To date, silybin has been
reported to have anti-oxidative, anticancer, antibacterial and
anti-osteoporosis activities (8);
however, the exact mechanism by which silybin inhibits MDR to
improve the effectiveness of antibiotics remains unclear. Molnar
et al (9) demonstrated that
silybin acted as an efflux pump inhibitor as it inhibited the
efflux pump of S. aureus. In the present study, the
mechanism by which silybin inhibits the MRSA efflux system was
investigated by performing antimicrobial susceptibility testing,
the double-plate method, fluorescence spectrophotometry, plasmid
elimination and polymerase chain reaction (PCR) assays.
Materials and methods
Materials
MRSA41577 was provided by the Dalian Municipal
Central Hospital (Dalian, Liaoning, China). The quality control
strain, S. aureus ATCC25923 was provided by the Chinese
Medical Culture Collection Center, National Center for Medical
Culture Collections (Beijing, China; www.cmccb.org.cn/cmccbnew). Ciprofloxacin (CIP) and
benzalkonium chloride (BAC; 40%) were purchased from Dalian Sanxing
Wuzhou Chemical Co., Ltd. (Dalian, China; sanxingwuzhou.1688.com). Cefoxitin paper (30
µg/piece), reserpine and silybin were purchased from Novozymes
Biotechnology Co., Ltd. (Shenyang, China). Primers and PCR reagents
were purchased from Takara Biotechnology Co., Ltd. (Dalian, China).
2,3,5-Triphenyltetrazolium chloride (TTC) was purchased from Sangon
Biotech Co., Ltd. (Shanghai, China).
Detection of drug resistance in
bacterial strains
Drug resistant strains of bacteria were detected
using the cefoxitin paper method of the American Association of
Clinical and Laboratory Standards Institute (CLSI2014) (10). S. aureus ATCC25923 was used
as the quality control strain.
PCR amplification of the drug
resistance and efflux pump genes
MRSA41577 cells were cultured in lysogeny broth (LB;
Sangon Biotech Co., Ltd.) to the logarithmic phase and aliquots
(3–6 ml) of the culture were then centrifuged at 3,000 × g for 5
min at 4°C, and the supernatant was discarded. Genomic DNA was
extracted from the cell pellets according to previously described
methods (11,12). Primers specific for the penicillin
binding protein 2 (mecA), an MRSA-inherent gene, quinolone
resistance protein NorA (norA), a surrogate for measuring
resistance to fluoroquinolone antibiotics, and quaternary ammonium
resistance proteins A/B (qacA/B) genes, representative of
MRSA resistance to quarternary ammonium antibiotics (Table I), were designed according to the
sequences published in GenBank (www.ncbi.nlm.nih.gov/genbank) using Primer Premier 5.0
(Premier Biosoft International, Palo Alto, CA, USA; www.premierbiosoft.com) software and reference
documents. Premix Taq (Takara Taq Version 2.0 plus dye; Takara
Biotechnology Co., Ltd.) was used for the PCR analysis. PCR was
performed with the following thermocycling conditions: Initial
denaturation at 94°C for 5 min, followed by 30 cycles of 94°C for
30 sec, 55°C 30 sec and 72°C for 45 sec, and a final extension at
72°C for 10 min. The amplified products were visualized by 1%
agarose gel electrophoresis. The DNA samples were loaded onto a 1%
agarose gel and visualized using ethidium bromide and a
transilluminator.
 | Table I.Sequences of the primers used for
reverse transcription-quantitative polymerase chain reaction. |
Table I.
Sequences of the primers used for
reverse transcription-quantitative polymerase chain reaction.
| Primer | Sequence (5′-3′) | Product size
(bp) |
|---|
| mecA | F:
TTAGATTGGGATCATAGCGT | 813 |
|
| R:
GGAACTTGTTGAGCAGAGGT |
|
| norA | F:
GTTACTTGTTGCTGCTTTTG | 435 |
|
| R:
GCTTGTCGTAGACTTTTTCG |
|
| qacA/B | F:
ATGCCAACTACTCTTTCA | 304 |
|
| R:
TCCATACCAGTCCAATCA |
|
Antimicrobial susceptibility
testing
The minimum inhibitory concentrations (MICs) of CIP
and BAC were determined using a two-fold dilution method according
to the National Committee for Clinical Laboratory Standards
guidelines (13). A total of 100
µl MRSA41577 (105 cfu/ml) was mixed with different
concentrations of CIP (16, 32, 64 and 128 µg/ml) and BAC (0.5, 1, 2
and 4 µg/ml) in a 96-well plate and incubated at 37°C on a shaking
plate (120 rpm) for 24 h. MRSA41577 cultured with 0 µg/ml CIP and
BAC were used as a control. Following incubation, 20 µl 20% TTC was
added to each sample and the plate was further incubated at 37°C in
the dark for 4 h.
In order to elucidate the effect of CIP and BAC
activity in the presence of silybin, various concentrations of
silybin (16, 32, 64 and 128 µg/ml) were applied to 100 µl MRSA41577
(105 cfu/ml) cells alone or in combination with either
CIP (16, 32, 64 and 128 µg/ml) or BAC (0.5, 1, 2 and 4 µg/ml), in
96-well plates. The plates were incubated at 37°C for 24 h.
Following incubation, 20 µl 20% TTC was added to each sample and
the plate was further incubated at 37°C in the dark for 4 h. A
blank control and a positive control were also prepared. The blank
control was comprised of MRSA41577 cells cultured in culture media
only without CIP, BAC or silybin treatment. Positive control
MRSA4157 cells were cultured with reserpine only. The MIC of CIP to
MRSA 41577 was 12 µg/ml. When the silybin was added, the MIC of CIP
decreased notably, demonstrating that the sensitivity of MRSA41577
to CIP was improved. Each experiment was repeated three times.
Determination of the synergistic
effect of silybin
Silybin, CIP and BAC were added alone or in
combination (CIP or BAC with silybin) to a sample of 20 ml
MRSA41577 (105 cfu/ml). The final concentrations of
silybin, CIP and BAC were 32, 16 and 0.24 µg/ml, respectively. The
samples were incubated at 37°C on a shaking plate set at 120 rpm.
The optical density values of the samples were continuously
measured at 580 nm over a 12-h period. The control was comprised of
MRSA41577 cells cultured in culture media only without CIP, BAC or
silybin treatment. The experiment was repeated three times.
Effect of silybin on the efflux pump
of MRSA
The double-plate method (14) was used to evaluate the effect of
silybin on the efflux plump in MRSA. Briefly, four LB-agar plates
were prepared, each containing 20 ml of the LB solid medium. To two
of the plates, 200 µl of the MRSA41577 culture containing 1% agar
was poured on top and allowed to solidify. To the other two plates,
the same volume of MRSA41577 culture containing 1% agar was added,
with the addition of either 32 µg/ml CIP or 2 µg/ml BAC. Following
solidification, four holes, each with a diameter of 6 mm, were
punched into the plate. Two of the holes were filled with 100 µl of
a solution containing silybin (60 and 80 µg, respectively), while
the other two holes were filled with 100 µl of a solution
containing reserpine (60 and 80 µg, respectively). The plates were
incubated at 37°C for 24 h, and the zone of bacteriostatic activity
around each hole was then observed. A vernier caliper was used to
accurately measure bacteriostatic circle, with the expected result
that a higher concentration of the drug leads to an increased
diameter of the bacteriostatic circle.
Accumulation of CIP dynamics test
Following treatment with 32 µg/ml silybin for 16 h,
20 ml MRSA41577 (105 cfu/ml) cells were harvested and
washed with phosphate-buffered saline (PBS; pH 7.4). The cells were
then resuspended in PBS (pH 7.4) and incubated at 37°C for 10 min.
CIP was added to the cells to 20 µg/ml, which was followed by
further incubation at 37°C. Aliquots (0.5 ml) of the cell
suspension were removed at 3, 8, 15 and 20 min intervals during
incubation. The samples were centrifuged (at 4°C and 3,000 × g for
5 min) and the cell pellets were each resuspended in 1 ml
glycine-hydrochloric acid (0.1 mol/l; pH 3.0). All samples were
incubated at 25°C for 2 h, followed by centrifugation (at 4°C and
3,000 × g for 5 min). The supernatants were removed for
fluorescence analysis using emission and excitation wavelengths of
450 and 382 nm, respectively. The positive control in this
experiment contained 20 µg/ml reserpine as an efflux pump inhibitor
and the negative control contained no drugs.
Assessing the effect of silybin on
MRSA41577 norA and qacA/B expression levels
20 ml MRSA41577 (105 cfu/ml) was cultured
in the absence or presence of 32 µg/ml silybin at 37°C to the
logarithmic phase. Aliquots (4–6 ml) of the culture were then
centrifuged at 10,000 × g and 4°C for 1 min to harvest the cells.
Total RNA was extracted from the cells using the TRIzol method
(Takara Biotechnology Co., Ltd.). RNA concentration was measured
with a nucleic acid protein analysis instrument (Cole-Palmer Ltd.,
Stone, UK). The RNA was used as a template to synthesize the cDNAs
of the norA and qacA/B genes by two-step reverse
transcription, using RNA to cDNA EcoDry™ Premix (Oligo dT) (Takara
Biotechnology Co., Ltd.). The 16S RNA was used as the internal
reference gene. The temperature protocol was as follows: 42°C for
60 min, followed by heating at 70°C for 10 min. Amplification was
performed using the following conditions: Initial denaturation at
95°C for 10 min, followed by 40 cycles of 95°C for 30 sec, 95°C for
5 sec and 53°C for 30 sec. At the end of amplification, the samples
were then subjected to a further cycle of 95°C for 15 sec, 60°C for
30 sec and 95°C for 15 sec with continuous fluorescence monitoring
using Thermal Cycler Dice Real Time System II (Takara Biotechnology
Co., Ltd.). The primers used were: norA (F: 5′GAG TGC TGG
TAT GGT AAT GCC3′; R: 5′CCCTGGTCCTAAAATGAATCC3′); qacA/B (F:
5′TTGAGCAATTTTCATGGCACTC3′; R: 5′CCCACGAGTGAGACTTTTCTTTT3′); and
16S RNA (F: 5′GCTCGTGTCGTGAGATGTTGG3′; R:
5′TTTCGCTGCCCTTTGTATTGT3′). The relative expression levels of
norA and qacA/B genes were calculated using the
equation 2−ΔΔCq (15),
where ΔCq was measured by subtracting the Cq of the 16S RNA value
from the Cq value of norA or qacA/B. The experiment
was repeated three times.
Silybin elimination MRSA41577 plasmid
assay
MRSA41577 cells were cultured at 37°C to the
logarithmic phase and aliquots (3–4 ml) of the culture were
centrifuged at 3,000 × g for 1 min at 4°C to harvest the cells for
plasmid DNA extraction, according to the method described by Chen
et al (16). The extracted
plasmid DNA was subjected to 1% agarose gel electrophoresis and
visualized using ethidium bromide and a transilluminator.
MRSA41577 cells were cultured at 37°C in LB medium
containing 32 µg/ml silybin to the logarithmic phase. A total of
200 µl cell suspension (109 cfu/ml) was spread onto an
LB agar plate and then incubated at 37°C for 24 h. Single colonies
(~300) were randomly picked and replica-plated onto new plates
containing 0 or 2 µg/ml BAC. The two plates were incubated at 37°C
for 24 h and the number of colonies observed on the plates were
counted to calculate the plasmid elimination rate. Plasmid
elimination rate (%)=(A/B) × 100; where A was the number of
colonies observed on the plate containing common culture medium
without antibiotics, but, could not grow in the selective medium
containing antibiotics and B is the number of colonies that were
observed on the plate containing common culture medium without
antibiotics (17).
Statistical analysis
Statistical analysis was performed using SPSS
software version 13.0 (SPSS, Inc., Chicago, IL, USA). Data are
expressed as the mean ± standard deviation and were analyzed by
one-way analysis of variance followed by least significant
difference post hoc test (equal variances). P<0.05 was
considered to indicate a statistically significant difference.
Results
Drug resistance in the strains
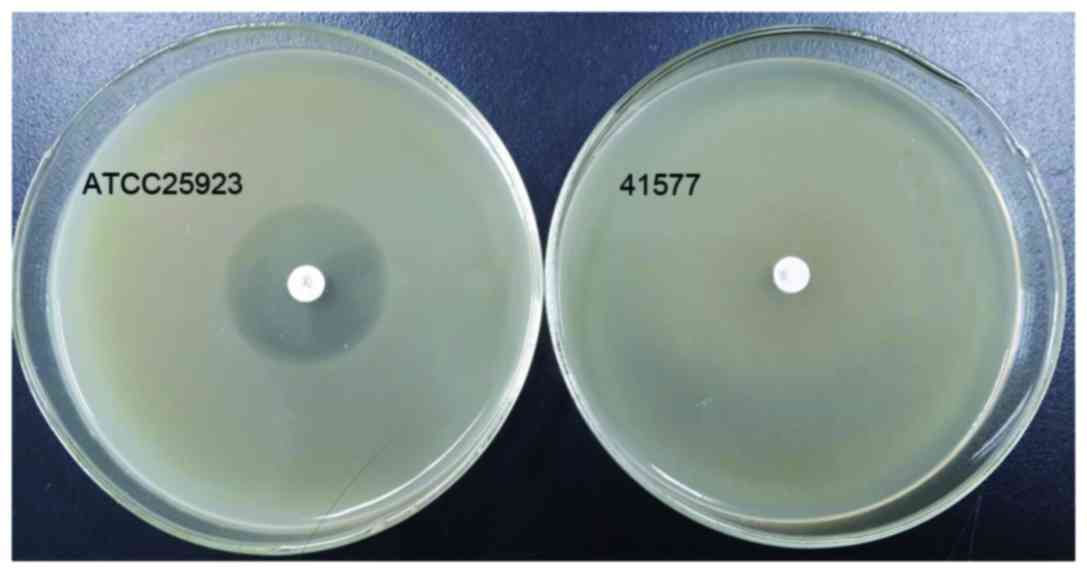
The quality control strain ATCC25923 was sensitive
to cefoxitin, however, MRSA41577 was resistant to it. The diameter
of the inhibition zone of ATCC25923 was 29 mm (Fig. 2). According to the CLSI2014
criteria, bacterial strains that produce a bacteriostatic zone of
<21 mm are considered to be resistant MRSA strains, while
strains that produce a bacteriostatic zone of >22 mm are
regarded as sensitive strains. Thus, the MRSA41577 strain used in
the present study was regarded as a MRSA resistant strain.
Screening of MRSA41577 resistant and
efflux pump genes
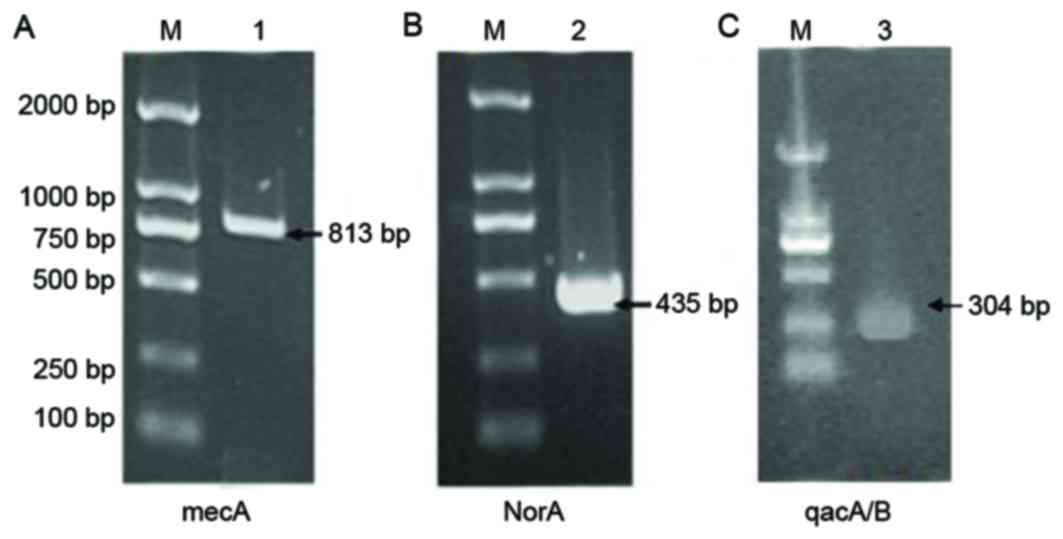
The mecA gene has been become a molecular
index for detecting MRSA, and the norA and qacA/B
genes are considered to be significant contributors to antibiotic
resistance (18). The mecA
gene was detected in MRSA41577 cells (Fig. 3A), further confirming that it was a
MDR bacterial strain. In addition, the efflux pump genes
norA (Fig. 3B) and
qacA/B (Fig. 3C) were also
detected in these cells.
Synergistic effect of silybin with CIP
and BAC
Silybin had no bacteriostatic effect on MRSA41577 at
the range of concentrations tested (16 to 128 µg/ml). The MICs of
CIP and BAC were 128 and 4 µg/ml, respectively. When silybin was
combined with CIP and BAC, the resulting mixture reduced the MIC of
CIP and BAC. Following treatment with 32 µg/ml silybin, the MIC of
CIP was reduced by 4-fold and the MIC of BAC was reduced by 2-fold
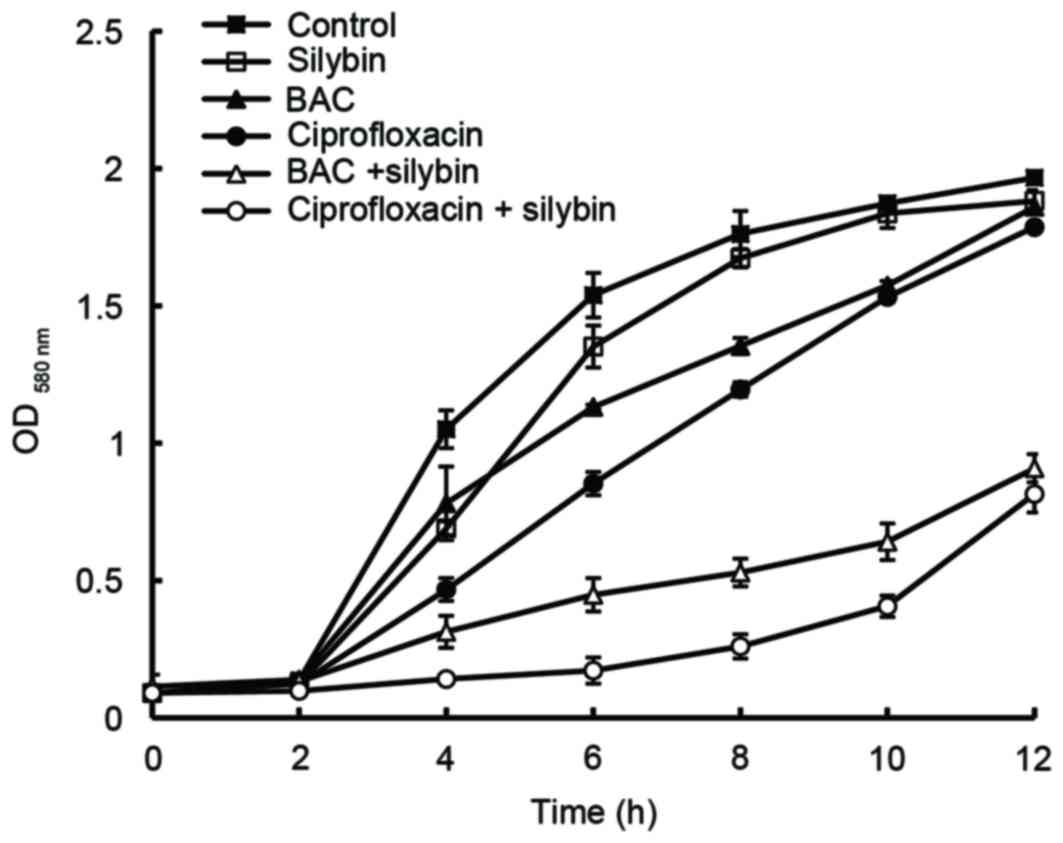
when compared with treatment without silybin (Table II). When these drugs were used
individually, the growth curves obtained for MRSA41577 were
comparable to that produced by the control (Fig. 4). However, when silybin was used in
combination with BAC or CIP, growth was markedly inhibited when
compared to the control; treatment with silybin+CIP was more
inhibitory than silybin+BAC. These results indicated that silybin
may be an efflux pump inhibitor of the MRSA41577 efflux pump
system.
 | Table II.Synergistic effect of CIP and BAC in
combination with silybin. |
Table II.
Synergistic effect of CIP and BAC in
combination with silybin.
|
| BAC (µg/ml) | CIP (µg/ml) |
|---|
|
|
|
|
|---|
| EPI (µg/ml) | 4 | 2 | 1 | 0.5 | 0 | 128 | 64 | 32 | 16 | 0 |
|---|
| Silybin |
|
|
|
|
|
|
|
|
|
|
| 0 | − | + | + | + | + | − | + | + | + | + |
|
128 | − | − | − | + | + | − | − | − | − | + |
| 64 | − | − | + | + | + | − | − | − | + | + |
| 32 | − | − | + | + | + | − | − | − | + | + |
| 16 | − | + | + | + | + | − | − | + | + | + |
| Reserpine |
|
|
|
|
|
|
|
|
|
|
| 20 | − | − | + | + | + | − | − | − | + | + |
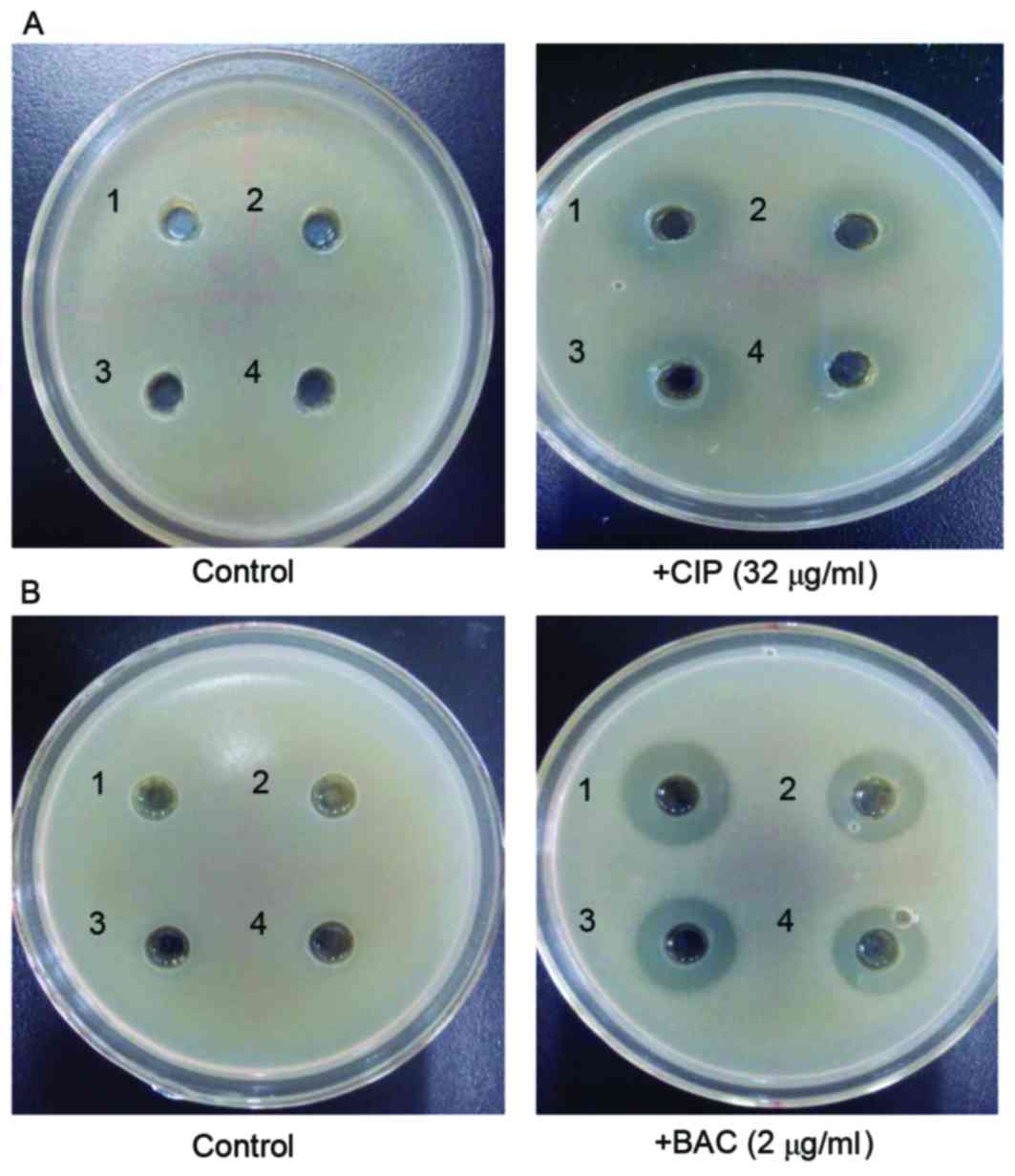
Silybin inhibits the MRSA41577 efflux
pump
The results of the double-plate assay suggested that
in the control group (no CIP and BAC in the culture), silybin had
no effect on MRSA41577. However, when CIP (Fig. 5A) and BAC (Fig. 5B) were included in the culture,
positive bacteriostatic zones were observed, indicating that
silybin may inhibit MRSA41577. In addition, its inhibitory effect
was consistent with the positive control reserpine.
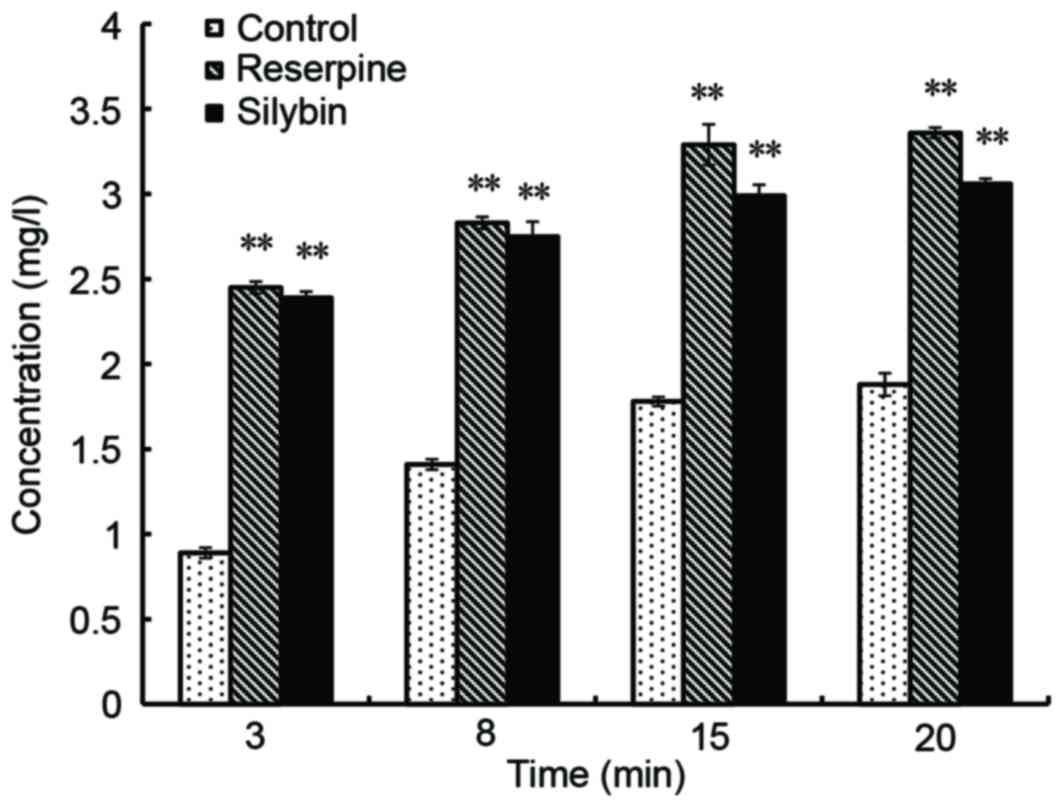
Effect of silybin on CIP accumulation
in the bacteria
According to the CIP standard curve, a linear
relationship between fluorescence intensity and CIP concentration
was observed for the range of 0.2 to 20 µg/ml, with linear
regression y=4.8418×+2.7472, where R2=0.9974. When
compared with the control, MRSA41577 treated with silybin exhibited
increased fluorescence, which also increased over time, which
indicated that the concentration of CIP entering the bacteria
increased. After 15 min, the level of was CIP increased by ~70% in
the silybin-treated culture compared with those treated with 32
µg/ml CIP only (P<0.01; Fig.
6). This further illustrated that silybin inhibits the release
of CIP out of cells via the efflux pump, indicating that silybin
may function as a efflux pump inhibitor.
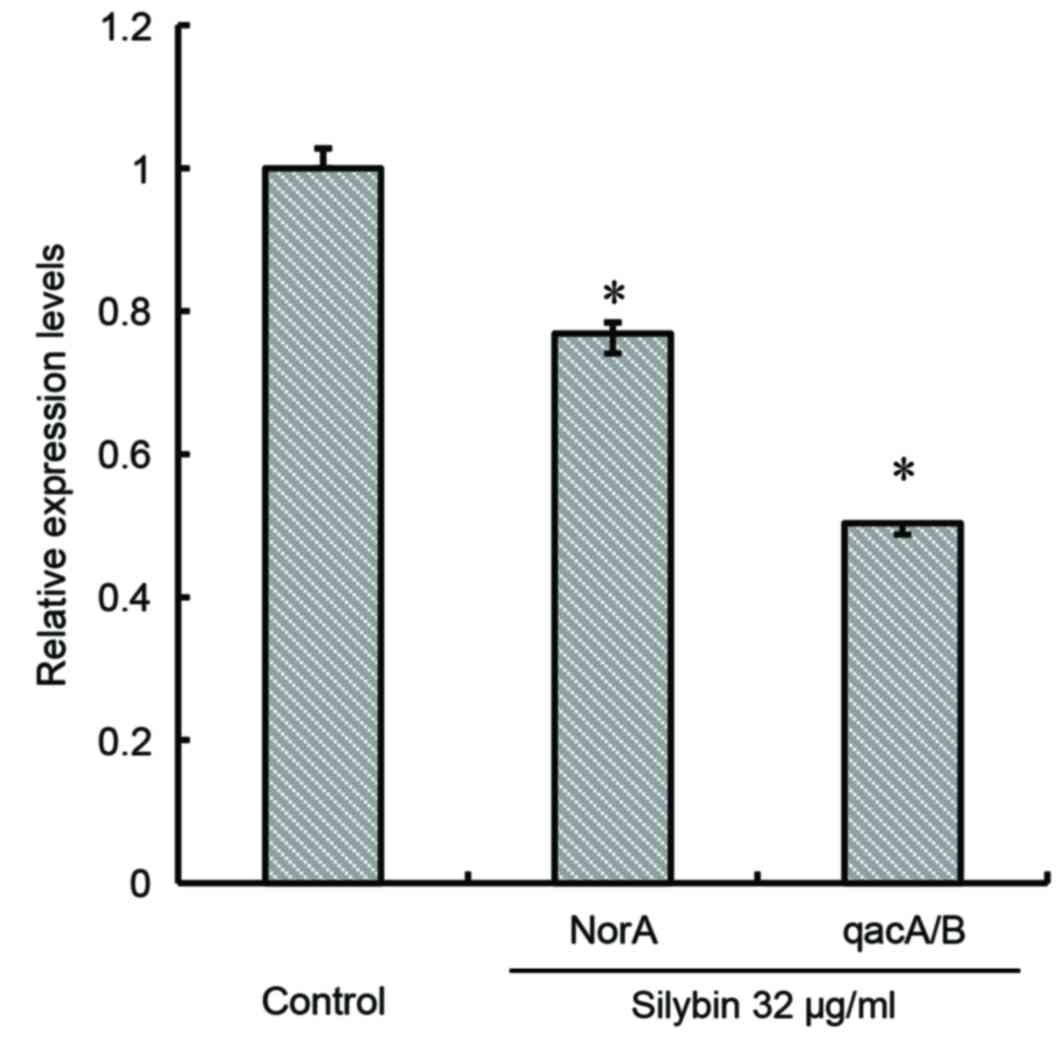
norA and qacA/B expression
Silybin appeared to significantly inhibit the
expression of the efflux gene norA (P<0.05). Following 16
h of treatment, silybin (32 µg/ml) reduced the expression of
norA by 36% and that of qacA/B by 49% compared with
the respective controls (P<0.05; Fig. 7).
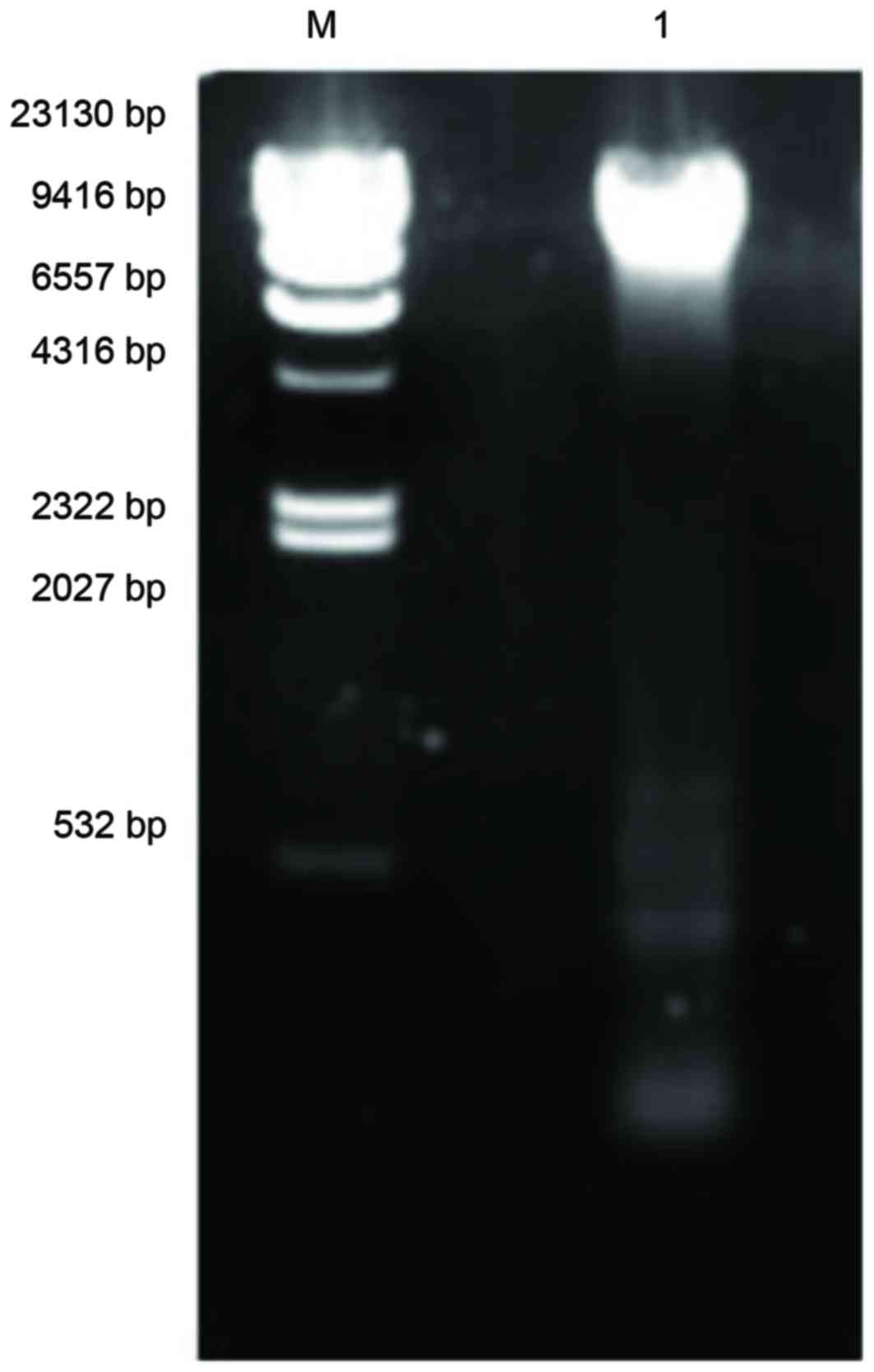
Emission of plasmid
Three different DNA plasmids were detected in
MRSA41577; a major one of ~23,130 bp, and two minor ones of ~500
and 100 bp, respectively (Fig. 8).
The identification of these plasmids in MRSA41577 suggested that
the qacA/B gene may be carried by these plasmids, as it is
known to be a plasmid-encoded gene.
The plasmid elimination experiment demonstrated that
300 MRSA41577 single colonies were able to grow on plates without
BAC, however, when grown with BAC only 29 single colonies were
produced. The plasmid elimination rate reached 90%.
Discussion
Recent evidence have suggested that efflux pumps may
be used by the cell as a first-line defense mechanism, preventing a
drug from reaching lethal concentrations inside the cell (19). Thus, using efflux pump inhibitors
with high-efficiency and low toxicity may be an effective strategy
to improve the status of clinical therapies. The effectiveness of
an efflux pump inhibitor can be judged by the reduction in the MIC
associated with an antibiotic. In order to detect the effect of
silybin on the MRSA efflux system, the present study measured the
synergistic effect of silybin alone and in combination with CIP or
BAC. Silybin alone was not bacteriostatic in MRSA41577, however, in
combination with either CIP or BAC, the MICs of CIP and BAC were
reduced and the sensitivity of MRSA41577 to the antibiotic was
greatly enhanced, suggesting that silybin may be an efflux pump
inhibitor. The double-plate method and fluorescence
spectrophotometry further verified this result, revealing that
silybin blocked or prevented the release of CIP by MRSA41577,
resulting in CIP accumulation inside the cells. This would
eventually lead to cell death or an enhancement in the
bacteriostatic activity of CIP in a time-dependent manner.
The bacterial efflux system is mediated by membrane
transporters. In general, these membrane transporters are comprised
of outer membrane channel proteins, fusion proteins and efflux
protein. To date, >10 multidrug efflux pumps have been described
for S. aureus, including NorA, NorB, MedA, QacA/B and Smr
(19). The NorA efflux pump is
considered to be a significant contributor to antibiotic resistance
as it can expel an array of chemically and structurally dissimilar
compounds from the cell, namely hydrophilic fluoroquinolones,
including norfloxacin, CIP, dyes such as ethidium bromide and
biocides such as quaternary ammonium compounds. NorA is a multiple
drug-resistant S. aureus transporter and is a 42-kDa protein
encoded by the norA (20)
The expression of norA can be induced by the presence of
antibiotics, resulting in a significant increase in expression
levels that in turn improves the ability of the bacteria to expel
the antibiotics, thereby allowing them to acquire MDR (21). Silybin significantly reduced the
expression of norA (Fig.
8), resulting in the enhanced sensitivity of MRSA41577 to
antibiotics (Fig. 4). This
suggested that by suppressing the expression of the efflux
proteins, silybin was able to reverse the MDR phenotype in
MRSA41577.
Currently, the primary method for controlling the
MRSA infection in clinical settings is to use a class of quaternary
ammonium salt disinfectants (such as BAC and benzalkonium bromide)
and double guanidine kind disinfectant (chlorhexidine). However,
following their mass overuse, MRSA has now become resistant to
these disinfectants (22). A
previous study indicated that in MRSA, resistance to disinfectants
is predominantly induced by the presence of a qac resistance
gene, particularly the qacA/B gene (23). The qacA/B gene is encoded by
a plasmid and an increase in its expression would eventually
enhance the resistance of the bacteria to disinfectants as a result
of increased expression of the MDR protein (24). The present study has demonstrated
that silybin significantly reduced the expression of the
qacA/B gene, revealing its ability to inhibit qacA/B
transcription. This suppression of the synthesis of the efflux pump
protein may be an important mechanism by which silybin could
restore the sensitivity of the bacteria to disinfectants.
A previous study has revealed that bacteria
resistant to disinfectants may also be resistant to antibiotics,
which may be associated with the existence of plasmid in the
bacteria. The MRSA plasmids contain the efflux genes and also the
qac gene (25). For
example, MRSA qacA genes are located in the pSK1 plasmid,
which carries the antibiotic resistant genes TMγ,
Kγ and Gmγ (26). However, it also carries the gene
qacA, which provides resistance against ethidium bromide and
quaternary amine disinfectant. Thus, plasmids appear to serve an
important role in bacterial resistance to disinfectants and
antibiotics (27). Fan et
al (28) revealed that the
complete elimination of plasmid from a drug-resistant strain can
transform it into a drug-sensitive strain. The results of plasmid
elimination in the present study demonstrated that silybin
eliminated the plasmid of MRSA41577, achieving a plasmid
elimination ratio of ~90%. Thus, silybin may restore the
sensitivity of MRSA41577 to antibiotics and disinfectants by
eliminating the plasmid carried by MRSA41577.
In conclusion, the results of the present study
demonstrated protein expression alterations which may indicate that
silybin may be an effective efflux pump inhibitor. It reduced the
expression levels of norA and qacA/B, thereby
decreasing the expression of efflux protein, and it also eliminated
the plasmid from MRSA41577, restoring bacterial sensitivity to
antibiotics and disinfectants. Due to the complexity of the MRSA
efflux system, inhibition of this efflux system may include a
number of aspects; for example, the interaction of a wide variety
of efflux genes and corresponding efflux proteins. Further study is
required to elucidate the precise mechanism by which silybin
inhibited the efflux system of MRSA, in order to provides novel
therapeutic strategies and means for the treatment of MRSA
infection.
Acknowledgements
The present study was supported by the Scientific
Research Projects of the Education Department of Liaoning Province
(grant no. L201683675) and the Natural Science Foundation of
Liaoning Province (grant no. 201602462).
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zou D, Xie KM, Wang H, Chen Y and Xie M:
Inhibitory effects of biochanin A on the efflux pump of
methicillin-resistant Staphylococcus aureus (MRSA). Wei Sheng Wu
Xue Bao. 54:1204–1211. 2014.(In Chinese). PubMed/NCBI
|
|
2
|
Feng M: Research progress of detection and
drug resistance of methicillin resistant Staphylococcus aureus.
China Med Herald. 9:8–9. 2012.
|
|
3
|
Yu XH, Wang D and Wang R: Progress in
methicillin-resistant Staphylococcus aureus. Chin J Clin Pharmacol.
27:306–310. 2011.
|
|
4
|
Garvev MI, Rahman MM, Gibbon S and Piddock
LJ: Medicinal plant extracts with efflux inhibitory activity
against Gram-negative bacteria. Int J Antimicrob Agents.
37:145–151. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dantziq AH, Law KL, Cao J and Starling JJ:
Reversal of multidrug resistance by the P-glycoprotein modulator,
LY335979 from the bench to the clinic. Curr Med Chem. 8:39–50.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gamier-Suillerot A, Marbeuf-Gueve C,
Salemo M, Loetchutinat C, Fokt I, Krawaczyk M, Kowalczyk T and
Priebe W: Analysis of drug transport kinetics in mutidrug-resistant
cells:implications for drug action. Curr Med Chem. 8:51–64. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Song ZJ, Feng X, Han WY, Ding Z, Chen AD
and Lei LC: Selection of Chinese herbal medicine Staphylococcus
aureus norA efflux pump inhibitor. J Jilin Agricultural University.
29:329–333. 2007.
|
|
8
|
Wlcek K, Koller F, Ferenci P and Stieger
B: Hepatocellular organic anion-transporting polypeptides (OATPs)
and multidrug resistance-associated protein 2(MRP2) are inhibited
by silibinin. Drug Metab Dispos. 41:1522–1528. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Molnár J, Engi H, Hohmann J, Molnár P,
Deli J, Wesolowska O, Michalak K and Wang Q: Reversal of multidrug
resitance by natural substances from plants. Curr Top Med Chem.
10:1757–1768. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Clinical and Laboratory Standards
InstitutePerformance standards for antimicrobial susceptibility
testing; 16th informational supplement clinical and laboratory
standards lnstitnte. Wayne, PA:
|
|
11
|
Noquchi N, Hase M, Kitta M, Sasatsu M,
Dequchi K and Kono M: Antiseptics susceptibility and distribution
of antiseptic-resistance genes in Methicillin-resistant
Staphylococcus aureus. FEMS Microbiol Lett. 172:247–253. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Su LT, Liang JL, Guo XJ and Feng F:
Comparison of methods for DNA extraction in Staphylococcis aureus.
J Zhong Kai Univ Agric Eng. 24:5–19. 2011.
|
|
13
|
Wu GB, Zhou JH and Zhang J: Study on
antibacterial activity of 5 kinds of quindones in vitro. Chin J
Clin Pharmacol. 30:129–130. 2014.
|
|
14
|
Yang ZC, Yang XS and Niu YL: Screening for
and bioassay-guided isolation of bacterial efflux pump inhibitors
by control method based on double plates. Nat Prod Res Dev.
22:277–280. 2010.
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen XJ, Xu XM, Niu B, Jie J, Yang Q,
Zhang YH and Zhang DC: Improving the technique to prepare plasmid
DNA from Escherichia coil on a large scale. Chin Remed Clin.
3:103–105. 2003.
|
|
17
|
Chen SB: Three kinds of Traditional
Chinese medicine (TCM) on a beta lactamase Staphylococcus aureus
plasmid elimination process. Anim Husband Vet Med. 42:81–84.
2010.
|
|
18
|
Yang CS and Liu WE: Drug resistance
mechanism of MRSA and molecular biological detection methods: A new
research progress. Chin J Nosocomiol. 17:356–358. 2007.
|
|
19
|
Costa SS, Viveiros M, Amaral L and Couto
I: Multidrug efflux pumps in Staphlococcus aureus: An update. Open
Microbiol J. 7:59–71. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chan BC, Ip M, Lau CB, Lui SL, Jolivalt C,
Ganem-Elbaz C, Litaudon M, Reiner NE, Gong H, See RH, et al:
Synergistic effects of baicalein with ciprofloxacin against NorA
over-expressed methicillinresistant Staphylococcus aureus (MRSA)
and inhibition of MRSA Pyruvate kinase. J Ethnopharmacol.
137:767–773. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kaatz GW, Thyagarajan RV and Seo SM:
Effect of promoter region mutations and mgrA overexpression on
transcription of norA, which encodes a Staphylococcis aureus
multidrug efflux transporter. Antimicrob Agents Chemother.
49:161–169. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ho J and Branlev J: Prevalence of
antiseptic resistance genes qacA/B and specific sequence types of
methicillin-resistant Staphylococcus aureus in the era of hand
hygiene. Antimicrob Chemother. 67:1549–1550. 2012. View Article : Google Scholar
|
|
23
|
Noguchi N, Suwa J, Narui K, Sasatsu M, Ito
T, Hiramatsu K and Song JH: Susceptibilities to antiseptic agents
and distribution of antiseptic-resistance genes qacA/B and smr of
methicillin-resistant Staphylococcus aureus isolated in Asia during
1998 and 1999. J Med Microbiol. 54:557–565. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Su Y, Shen W and Ju LW: Progress in
Staphylococcus aureus disinfectant resistance genes. Chin J
Disinfect. 31:1084–1087. 2014.
|
|
25
|
Irizarry L, Merlin T, Rupp J and Griffith
J: Reduced susceptibility of methicillin-resistant Staphylococcus
aureus to cetylpyridinium chloride and chlorhexidine. Chemotherapy.
42:248–252. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li S: Study on the epidemiological of
disinfectant-resistant gene qacA/B in MRSA and it's resistance of
disinfectant and mechanisims research (unpublished thesis). The
Fourth Military Medical University. 2012.
|
|
27
|
Hua DX and Qian YS: Progress in Bacterial
disinfectant resistance gene. Chin J Anibiot. 33:513–518. 2008.
|
|
28
|
Fan JC, Bai YN, Shi FY and Pei HB:
Drug-resistance and plasmid homology analysis of SA in medical
environment. Chin J Public Health. 24:816–817. 2008.
|