Introduction
Coronary artery disease (CAD), including acute
myocardial infarction (AMI), is a common complex disease caused by
atherosclerosis. Dysregulated lipid metabolism and inflammation
serve critical roles in the initiation and progression of
atherosclerosis (1,2). A large number of genetic loci and
variants have been identified in CAD and AMI, accounting for ~10%
of cases (3–5). To date, the genetic causes for CAD
and AMI remain largely unknown. Previous epidemiological studies
have indicated that the incidence of CAD in patients with
congenital heart disease is markedly higher than that observed in
the healthy population (6–8). Therefore, dysregulation of cardiac
developmental genes may contribute to the pathogenesis of CAD.
The GATA transcription factor family, including GATA
binding proteins 1–6 (GATA1-6), is involved in diverse
physiological and pathological processes. GATA4-6 serve critical
functions in the differentiation and proliferation of endoderm- and
mesoderm-derived tissues (9–11).
GATA4 is essential in the development of the heart, liver,
pancreas, adrenal glands, gonads, gut, ovaries and testes (12). In experimental animals, GATA4 is
essential in ventral morphogenesis and heart tube formation, as
well as cardiomyocyte proliferation (13–15).
Neonatal GATA4 gene inactivation causes severe and lethal systolic
heart failure, indicating its autonomous function for physiological
cardiomyocyte growth (16). GATA4
and its cofactor Friend of GATA (FOG) regulate coronary vascular
development (17–19). In addition, deletion of GATA4
specifically from cardiomyocytes reduces myocardial capillary
density (18). GATA4-null embryos
exhibit an absent proepicardium and blocked epicardium formation
(20,21). Epicardium-derived cells populate
the myocardial wall and develop into vascular smooth muscle cells,
fibroblasts and endothelial cells (20). GATA4 also promotes myocardial
regeneration in neonatal mice (22). In a recent report, GATA4 was
reported to be involved in DNA damage response-induced inflammation
and senescence, and GATA4 accumulation may result in aging and
age-associated inflammation (23,24).
GATA4 gene mutations have been implicated in
familial and sporadic congenital heart diseases (25–27),
and may cause neonatal and childhood-onset diabetes (28). Human GATA4 gene polymorphisms are
also associated with plasma triglyceride levels (29,30).
In addition, GATA4 regulates cardiac morphogenesis and
cardiovascular development in a dose-dependent manner (31,32).
Therefore, it was hypothesized that altered GATA levels may
contribute to CAD and AMI through different pathways. In the
present study, the GATA4 gene promoter was genetically and
functionally analyzed in large cohorts of patients with AMI and
ethnically-matched healthy controls.
Materials and methods
Study populations
Patients with AMI (n=395; 295 male and 100 female
patients; median age, 61.48 years) were recruited between April
2014 and July 2016 from the Cardiac Care Unit, Division of
Cardiology, Affiliated Hospital of Jining Medical University,
Jining Medical University (Shandong, China). Patients with AMI were
diagnosed according to clinical symptoms, electrocardiograms,
elevated biochemical markers of myocardial necrosis or coronary
angioplasty. Ethnically-matched healthy controls (n=397; 210 male
and 187 female patients; median age, 50.39 years) were recruited
from the same hospital during the same time period. Healthy
controls with a familial history of CAD and congenital heart
diseases were excluded from this study. The research was performed
according to the principles of the Declaration of Helsinki, and the
present study protocol was approved by the Human Ethics Committee
of the Affiliated Hospital of Jining Medical University. Written
informed consent was obtained from all of the participants.
Direct DNA sequencing
Peripheral leukocytes were collected from venous
blood using the Human Leukocyte Isolation system (Haoyang
Biological Products Technology Co., Ltd., Tianjin, China),
according to the manufacturer's protocol. Genomic DNA was extracted
with the QIAamp DNA Mini kit (Qiagen, Inc., Valencia, CA, USA). Two
overlapped DNA fragments covering the GATA4 gene promoter region,
510 bp (between-961 and −451 bp) and 569 bp (between −502 and +67
bp), were generated by polymerase chain reaction (PCR) with Taq DNA
polymerase PCR master mix (Promega Corporation, Madison, WI, USA)
and directly sequenced. The thermocycling conditions were as
follows: 510 bp fragment, 35 cycles of 94°C for 30 sec, 56°C for 30
sec and 72°C for 45 sec; 569 bp fragment, 35 cycles of 94°C for 30
sec, 62°C for 30 sec and 72°C for 45 sec. PCR primers were designed
using the human GATA4 genomic sequence (National Center for
Biotechnology Information GenBank accession no. NG_008177.2;
https://www.ncbi.nlm.nih.gov/genbank/; Table I). Bidirectional sequencing of PCR
products was performed on a 3500XL genetic analyzer (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) by Sangon Biotech Co., Ltd.
(Shanghai, China). DNA sequences were then compared with the
wild-type GATA4 gene promoter using the DNAMAN program (version
5.2.2; Lynnon BioSoft, Quebec, Canada), and DNA variants were
identified. The DNA variants in the GATA4 gene promoter were first
analyzed using JASPAR program (jaspar.genereg.net) to predict their effects on
binding sites for transcription factors, which were further
experimentally confirmed.
 | Table I.PCR primers for the GATA4 gene
promoter. |
Table I.
PCR primers for the GATA4 gene
promoter.
| PCR primers | Sequence | Location | Position | Product size
(bp) |
|---|
| Sequencing |
|
|
|
|
|
GATA4-F1 |
5′-AAGTTTAACCGAAAGCGTGAG-3′ | 31,292 | −961 | 510 |
|
GATA4-R1 |
5′-CCAGACTGCCTCCTAAAATCA-3′ | 31,781 | −451 |
|
|
GATA4-F2 |
5′-GGCAAAAGGGAGGCTTCGGTC-3′ | 31,731 | −502 | 569 |
|
GATA4-R2 |
5′-CCGCCTCCAAGTCCCCAGCTC-3′ | 32,299 | +67 |
|
| Function |
|
|
|
|
|
GATA4-F |
5′-(SacI)-GCCGGCTGTTATCTGGGGCTGAAGG-3′ | 31,270 | −932 | 971 |
|
GATA4-R |
5′-(HindIII)-GGGTCCCCGGCCCAGCAACT-3′ | 32,271 | +39 |
|
Functional analysis of the GATA4 gene
promoter by dual-luciferase reporter assay
Wild-type and variant GATA4 gene promoters (971 bp,
between −932 bp and +39 bp) were generated by PCR and inserted into
the SacI and HindIII sites of a luciferase reporter
vector (pGL3-basic, Promega Corporation), in order to generate
expression constructs. The PCR primers are presented in Table I. 293 [CRL-1573; American Type
Culture Collection (ATCC), Manassas, VA, USA] and H9c2 cells (rat
cardiomyocyte line; CRL-1446; ATCC) were transiently transfected
with the designated constructs. Briefly, on the day prior to
transfection the cells were seeded in 6-well plates at 40–50 and
50–60% confluence for 293 cells and H9c2 cells, respectively.
Designated expression constructs (1.0 µg) and
Lipofectamine® (3.0 µl; Invitrogen; Thermo Fisher
Scientific, Inc.) in 500 ml serum-free medium were used to
transfect the cells in each well. The vector expressing
Renilla luciferase (pRL-TK; 25 ng, Promega Corporation) was
used as an internal control for transfection efficiency. A total of
48 h post-transfection, luciferase activity was examined using the
Promega Dual-Luciferase® Reporter Assay system on a
Promega Glomax 20/20 luminometer (both Promega Corporation). GATA4
gene promoter activity was expressed as the ratio of luciferase
activity over Renilla luciferase activity. The activity of
the wild-type GATA4 gene promoter was set as 100%.
Nuclear extract preparation and
electrophoretic mobility shift assay (EMSA)
Nuclear extracts from 293 and H9c2 cells were
prepared using NE-PER® Nuclear and Cytoplasmic
Extraction Reagent kit (Thermo Fisher Scientific, Inc.) and protein
concentrations were determined using the Bradford protein assay.
EMSA was conducted using the LightShift®
Chemiluminescent EMSA kit (Thermo Fisher Scientific, Inc.).
Biotinylated double-stranded oligonucleotides (30 bp) containing
the DNA variants identified in AMI patients were used as probes.
DNA-protein binding reactions were conducted for 20 min at room
temperature with equal amounts of probes (0.2 pM) and nuclear
extracts (3.0 µg). Subsequently, the reactions were separated on a
6% polyacrylamide gel and transferred onto a nylon membrane (Thermo
Fisher Scientific, Inc.). The oligonucleotides were cross-linked to
the membrane using the UV Stratalinker 1800 (Stratagene; Agilent
Technologies, Inc., Santa Clara, CA, USA) and were detected by
chemiluminescence using the LightShift® Chemiluminescent
EMSA kit (Thermo Fisher Scientific, Inc.).
Statistical analysis
All transfection experiments were repeated three
times independently, in triplicate. Transfection results are
expressed as the means ± standard error of the mean and were
analyzed using two-way analysis of variance followed by Dunnett's
test. The frequency of DNA variants was compared using SPSS v13.0
software (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered
to indicate a statistically significant difference.
Results
DNA variants in the GATA4 gene
promoter
A total of 14 DNA variants were identified in the
GATA4 gene promoter, including two single-nucleotide polymorphisms
(SNPs). The frequency and locations of the DNA variants and SNPs
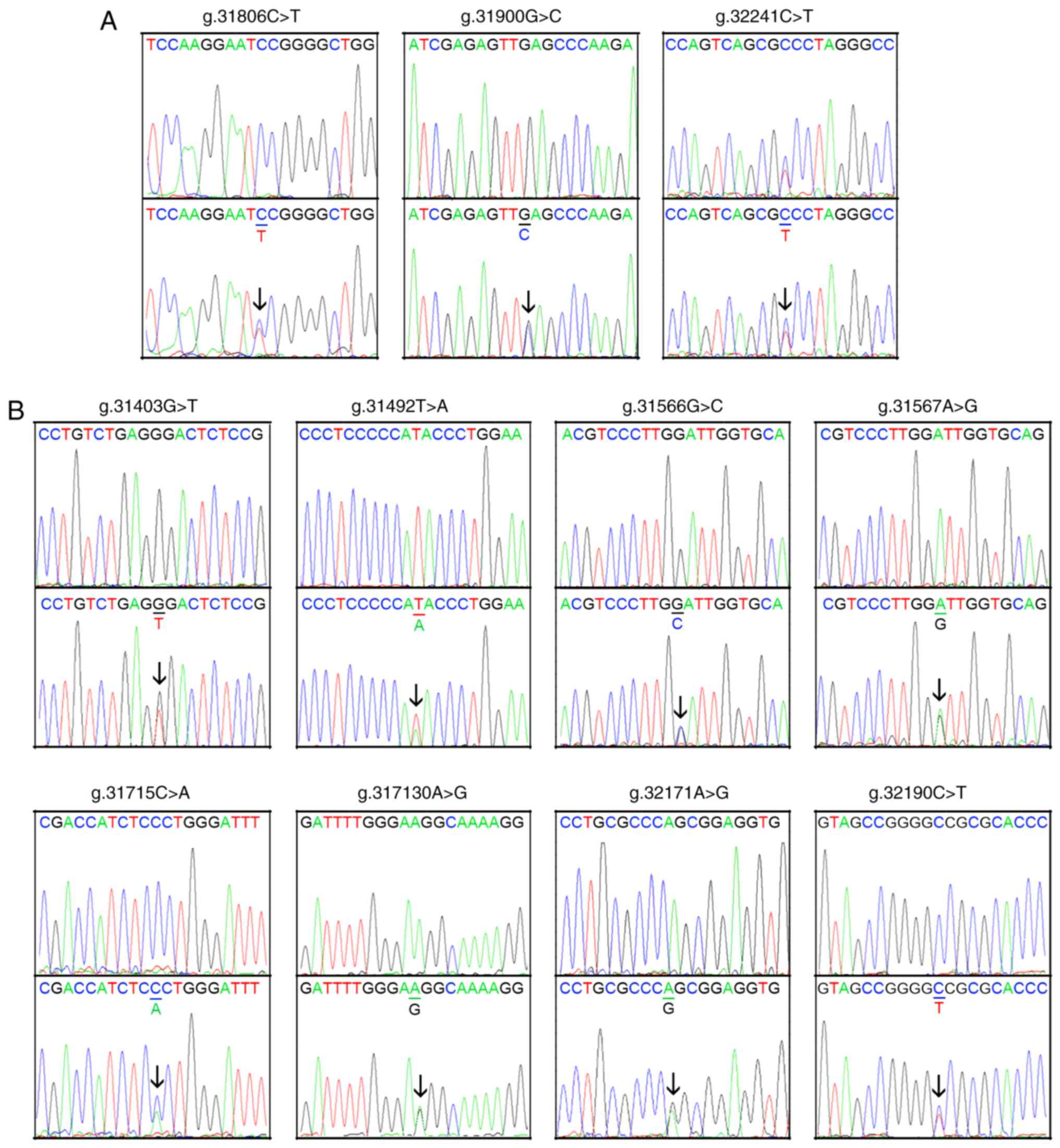
are presented in Fig. 1 and
Table II. Three novel
heterozygous DNA variants (g.31806C>T, g.31900G>C and
32241C>T) were only identified in three patients with AMI
(Fig. 2A). Eight novel
heterozygous DNA variants (g.31403G>T, g.31492T>A,
g.31566G>C, g.31567A>G, g.31715C>A, g.31730A>G,
g.32171A>G and g.32190C>T) were only found in healthy
controls (Fig. 2B). One insertion,
heterozygous DNA variant (g.31979_31980InsG) and two SNPs
[g.31360T>C (rs372004083) and g.31437C>A (rs769262495)] were
reported in patients with AMI and controls with similar frequencies
(P>0.05), the sequencing chromatograms of which were not
shown.
 | Table II.DNA variants within the GATA binding
protein 4 gene promoter in patients with AMI and controls. |
Table II.
DNA variants within the GATA binding
protein 4 gene promoter in patients with AMI and controls.
| DNA variants | Genotype |
Locationa (bp) | Controls
(n=397) | AMI (n=395) | P-value |
|---|
| g.31360T>C
(rs372004083) | TC | −837 | 1 | 1 | 1.000 |
| g.31403G>T | GT | −830 | 1 | 0 | – |
| g.31437C>A
(rs769262495) | CA | −796 | 4 | 1 | 0.373 |
| g.31492T>A | TA | −741 | 1 | 0 | – |
| g.31566G>C | GC | −667 | 1 | 0 | – |
| g.31567A>G | AG | −666 | 1 | 0 | – |
| g.31715C>A | CA | −518 | 1 | 0 | – |
| g.31730A>G | AG | −503 | 1 | 0 | – |
| g.31806C>T | CT | −427 | 0 | 1 | – |
| g.31900G>C | GC | −333 | 0 | 1 | – |
|
g.31979_31980InsG | −/G | −256 | 8 | 2 | 0.107 |
| g.32171A>G | AG |
−62 | 1 | 0 | – |
| g.32190C>T | CT |
+43 | 1 | 0 | – |
| g.32241C>T | CT |
+9 | 0 | 1 | – |
DNA variants affect the binding of
transcription factors
The GATA4 gene promoter was first analyzed using
JASPAR program (jaspar.genereg.net/). The JASPAR program predicts
whether DNA variants alter the putative binding sites for
transcription factors, and these predictions require experimental
confirmation. For the DNA variants identified in patients with AMI,
the DNA variant (g.31806C>T) abolished a putative binding site
for early growth response protein 1. The DNA variant
(g.31900G>C) disrupted a binding site for zinc finger protein
354C, and the DNA variant (g.32241C>T) altered a binding site
for the transcription factor AP-2α. For the DNA variants only
identified in controls, the DNA variants (g.31403G>T,
g.31567A>G, g.31715C>A, g.32171A>G and g.32190C>T) did
not affect the binding of transcription factors. The DNA variant
(g.31492T>A) altered a binding site for myeloid zinc finger
protein 1 (MZF1). The DNA variant (g.31566G>C) disrupted a
binding site for helicase-like transcription factor and the DNA
variant (g.31730A>G) disrupted a binding site for signal
transducer and activator of transcription 3. For the DNA variants
detected in patients with AMI and controls, the DNA variant
g.31360T>C (rs372004083) altered a binding site for homeodomain
transcription factor Distal-less 6. Furthermore, the DNA variant
g.31437C>A (rs769262495) created a binding site for MZF1.
Finally, the DNA variant (g.31979_31980insG) created a binding site
for zinc finger protein 704.
GATA4 gene promoter activity
The expression constructs containing wild-type and
variant GATA4 gene promoters: pGL3-WT (wild-type), pGL3-31715A,
pGL3-31730G, pGL3-31806T, pGL3-31900C and pGL3-32241T, were
transfected into 293 and H9c2 cells. The dual-luciferase activities
were measured and relative activity of wild-type and variant GATA4
gene promoters were examined. Three DNA variants (g.31806C>T,
g.31900G>C and 32241C>T) identified only in patients with AMI
were assessed for their effects on GATA4 gene promoter activity. In
addition, two of the eight DNA variants found only in controls were
tested as negative controls for transfection. The transcriptional
activity of pGL3-WT containing wild-type GATA4 gene promoter was
set as 100%. The transcriptional activity of variant GATA4 gene
promoters was compared to that of pGL3-WT.
293 cells were used in this study, as the cell line
has been widely used in transient transfection experiments. The
results in 293 cells are shown in Fig.
3A. Compared with pGL3-WT, the transcriptional activity of
empty pGL3-basic containing no promoter was close to zero,
confirming that transfection with the wild-type and variant GATA4
gene promoters was successful. In 293 cells, the DNA variants
(g.31806C>T, g.31900G>C and 32241C>T) identified only in
patients with AMI significantly increased transcriptional activity
of the GATA4 gene promoter (P<0.001). Conversely, the DNA
variants (31715C>A and 31730A>G) only identified in controls
did not significantly affect activity of the GATA4 gene promoter
(P>0.05; Fig. 3A).
 | Figure 3.Relative transcriptional activity of
wild-type and variant GATA4 gene promoters. The transcriptional
activity of the WT GATA4 gene promoter was set as 100%. Empty
pGL3-basic, containing no promoter, was used as a blank control for
transfection. (A) Relative activity of the WT and variant GATA4
gene promoters in 293 cells. (B) Relative activities of the WT and
variant GATA4 gene promoters in H9c2 cells. 1, pGL3-basic; 2,
pGL3-WT; 3, pGL3-31715A; 4, pGL3-31730G; 5, pGL3-31806T; 6,
pGL3-31900C; 7, pGL3-32241T. *P<0.001 compared with pGL3-WT.
GATA4, GATA transcription factor 4; WT, wild-type. |
Since human cardiomyocyte cell lines are currently
not available, the H9c2 rat cardiomyocyte cell line was used.
Similar to in 293 cells, the transcriptional activity of empty
pGL3-basic containing no promoter was close to zero in H9c2 cells,
thus indicating successful transfection. In H9c2 cells, the DNA
variants (g.31806C>T, g.31900G>C and 32241C>T) identified
only in patients with AMI significantly increased activity of the
GATA4 gene promoter (P<0.001), which was consistent with the
results observed in 293 cells. The DNA variants (31715C>A and
31730A>G) observed only in the controls did not significantly
alter the activity of the GATA4 gene promoter (P>0.05).
Collectively, these findings indicated that the effects of the DNA
variants were not tissue-specific (Fig. 3B).
Transcription factor binding as
determined by EMSA
To investigate whether the DNA variants affected the
binding of transcription factors, EMSA was performed with wild-type
or variant oligonucleotides, including the DNA variants
g.31806C>T [5′-GAACCTCCAAGGAAT(C/T)CGGGGCTGGGAGGA-3′)],
g.31900G>C [5′-ACAAGATCGAGAGTT(G/C)AGCCCAAGAGGTCA-3′] and
g.32241C>T [5′-CACAGCGAACCCAAT(C/A)GACCTCCGGCTGGG-3′]. The DNA
variant g.32241C>T markedly reduced the binding of an unknown
transcription factor in 293 and H9c2 cells (Fig. 4). The affected transcription
factor, which acted as a transcriptional activator, requires
further identification. The effects of the other two DNA variants
(g.31806C>T and g.31900G>C) on the binding of transcription
factors were not detected, likely due to the sensitivity limit of
EMSA experiments. Therefore, these DNA variants may affect the
binding of transcription factors, altering the activity of the
GATA4 gene promoter.
Discussion
Human studies and animal experiments have indicated
that GATA4 is a critical regulator in heart development, as well as
cardiac function. Insufficient or excessive GATA4 not only cause
congenital heart diseases, but are also involved in the development
of late-onset heart disease. In our previous study, five functional
DNA variants in the GATA4 gene promoter were identified in patients
with congenital heart diseases (33). In the present study, the GATA4 gene
promoter was further genetically and functionally analyzed in a
large cohort of patients with AMI and healthy controls. The results
revealed that three novel heterozygous DNA variants (g.31806C>T,
g.31900G>C and 32241C>T) were found in three patients with
AMI. In 293 cells and H9c2 cardiomyocytes, these DNA variants
significantly increased the transcriptional activity of the GATA4
gene promoter. Furthermore, EMSA experiments revealed that the DNA
variant g.32241C>T affected the binding of transcription
factors. The effects of the other two DNA variants (g.31806C>T
and g.31900G>C) were not detected by EMSA, likely due to EMSA
sensitivity. Collectively, the frequency of the DNA variants was
0.76% (3 out 395), thus suggesting that the GATA4 gene promoter DNA
sequence variants were rare. Therefore, these DNA variants may
contribute to AMI as a rare risk factor.
The human GATA4 gene has been localized to
8p23.1-p22 (34,35). The GATA4 gene is regulated in a
modular manner to control distinct temporal and spatial expression
patterns (12). The human GATA4
gene promoter is a typical TATA-less promoter with conserved
GC-boxes and an E-box. A GATA motif at ~1.0 kb upstream of the
transcription start site indicates that GATA4 gene expression may
be autoregulated (36). In
addition, the GATA4 gene is regulated by Forkhead factors, subunits
of SWI/SNF complex, bone morphogenetic protein signaling molecules
and microRNAs (miRs) (37–41). In a previous study, GATA4 gene
transcript levels were revealed to be significantly higher in the
peripheral blood mononuclear cells of patients with severe stable
CAD (42). This study provided
further supportive evidence.
During cardiac development, numerous
GATA4-interacting proteins, including transcriptional activators
and repressors, and downstream targets of GATA4 have been reported.
GATA4 serves a crucial role in cardiogenesis and α-cardiomyocyte
function by regulating cardiac-associated genes, including troponin
C, cardiac-myosin heavy chain and brain-type natriuretic factor
genes (43–46). GATA4, together with the cardiac
specific factors T-box transcription factor 5 (TBX5) and NK2
homeobox 5 (NKX2.5), directly regulates the cardiac-specific
expression of the connexin40 gene (47). GATA4 interacts and directly induces
the expression of cyclin D2 and cyclin-dependent kinase 4 genes,
which are required for cardiomyocyte proliferation (48–50).
Furthermore, GATA4 specifically cooperates with NKX2.5 to activate
atrial natriuretic factor (ANF) and other cardiac genes (43,51).
GATA4 also activates the NKX2-5 gene via a novel upstream enhancer
(52). GATA4 interacts with GATA5
and GATA6 in endocardial cushion formation and outflow tract
morphogenesis (53). Interactions
between GATA4 and GATA6 with TBX5 serve a unique role in normal
cardiac morphogenesis (54,55).
In addition, the complex interdependence of GATA4, NKX2-5 and TBX5
controls cardiac gene expression in cardiac morphogenesis (56). GATA4 physically interacts with
heart and neural crest derivatives expressed 2, a basic
helix-loop-helix transcription factor, to synergistically activate
cardiac gene expression, including ANF, the B-type natriuretic
peptide gene and the α-myosin heavy chain gene (57,58).
Therefore, altered levels of GATA4 may interfere with the cardiac
gene regulatory network, resulting in cardiomyocyte
dysfunction.
GATA4 has a role in cardiac angiogenesis and
promotes pressure overload-induced angiogenesis. GATA4 induces the
angiogenic factor, vascular endothelial growth factor A, by
directly binding to its promoter and enhancing its transcription
(18). GATA4-mediated miR-144/451
cluster exerts synergistic effects in protecting against
cardiomyocyte death (59). GATA4
and the cardiac transcription factors, nuclear factor of activated
T-cells, myocyte enhancer factor-2 and serum response factor,
synergistically activate the expression of the endothelial-specific
endothelin-1 gene (11,60–63).
Endothelin-1 causes endothelial dysfunction and inflammation,
contributing to atherosclerotic plaque formation (64). FOG-2 is also essential for cardiac
morphogenesis and coronary vessel development in the epicardium
(19). Therefore, elevated GATA4
may contribute to AMI through its function in coronary artery
formation.
In conclusion, the GATA4 gene promoter was
genetically and functionally analyzed. The DNA variants identified
in patients with AMI significantly increased GATA4 gene promoter
activity. EMSA revealed that the DNA variants affected the binding
of transcription factors, which may modify GATA4 gene promoter
activity to subsequently alter its expression levels. Therefore,
DNA variants in the GATA4 gene promoter may contribute to AMI as a
rare risk factor. The data from the present study may provide a
genetic basis for designing potential precision therapy for
patients with AMI.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81370271, 81400291
and 81670341) and the Taishan Scholar Program (grant no.
TSHW201502063), Shandong, China.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
BY and RGH designed the present study. JC, SW and YC
collected the samples and performed the experiments. SP and YC
analyzed the data. JC and SW wrote the paper. BY and RGH reviewed
and edited the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Human Ethics
Committee of the Affiliated Hospital of Jining Medical University.
Written informed consent was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Connelly MA, Shalaurova I and Otvos JD:
High-density lipoprotein and inflammation in cardiovascular
disease. Transl Res. 173:7–18. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shapiro MD and Fazio S: From lipids to
inflammation: New approaches to reducing atherosclerotic risk. Circ
Res. 118:732–749. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Assimes TL and Roberts R: Genetics:
Implications for prevention and management of coronary artery
disease. J Am Coll Cardiol. 68:2797–2818. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Björkegren JL, Kovacic JC, Dudley JT and
Schadt EE: Genome-wide significant loci: How important are they?
Systems genetics to understand heritability of coronary artery
disease and other common complex disorders. J Am Coll Cardiol.
65:830–845. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
McPherson R and Tybjaerg-Hansen A:
Genetics of coronary artery disease. Circ Res. 118:564–578. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fedchenko M, Mandalenakis Z, Rosengren A,
Lappas G, Eriksson P, Skoglund K and Dellborg M: Ischemic heart
disease in children and young adults with congenital heart disease
in Sweden. Int J Cardiol. 248:143–148. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Olsen M, Marino B, Kaltman J, Laursen H,
Jakobsen L, Mahle W, Pearson G and Madsen N: Myocardial infarction
in adults with congenital heart disease. Am J Cardiol.
120:2272–2277. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tutarel O, Kempny A, Alonso-Gonzalez R,
Jabbour R, Li W, Uebing A, Dimopoulos K, Swan L, Gatzoulis MA and
Diller GP: Congenital heart disease beyond the age of 60: Emergence
of a new population with high resource utilization, high morbidity,
and high mortality. Eur Heart J. 35:725–732. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lentjes MH, Niessen HE, Akiyama Y, de
Bruïne AP, Melotte V and van Engeland M: The emerging role of GATA
transcription factors in development and disease. Expert Rev Mol
Med. 18:e32016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Peterkin T, Gibson A, Loose M and Patient
R: The roles of GATA-4, −5 and −6 in vertebrate heart development.
Semin Cell Dev Biol. 16:83–94. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pikkarainen S, Tokola H, Kerkelä R and
Ruskoaho H: GATA transcription factors in the developing and adult
heart. Cardiovasc Res. 63:196–207. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Burch JB: Regulation of GATA gene
expression during vertebrate development. Semin Cell Dev Biol.
16:71–81. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kuo CT, Morrisey EE, Anandappa R, Sigrist
K, Lu MM, Parmacek MS, Soudais C and Leiden JM: GATA4 transcription
factor is required for ventral morphogenesis and heart tube
formation. Genes Dev. 11:1048–1060. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Molkentin JD, Lin Q, Duncan SA and Olson
EN: Requirement of the transcription factor GATA4 for heart tube
formation and ventral morphogenesis. Genes Dev. 11:1061–1072. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zeisberg EM, Ma Q, Juraszek AL, Moses K,
Schwartz RJ, Izumo S and Pu WT: Morphogenesis of the right
ventricle requires myocardial expression of Gata4. J Clin Invest.
115:1522–1531. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Prendiville TW, Guo H, Lin Z, Zhou P,
Stevens SM, He A, VanDusen N, Chen J, Zhong L, Wang DZ, et al:
Novel roles of GATA4/6 in the postnatal heart identified through
temporally controlled, cardiomyocyte-specific gene inactivation by
Adeno-associated virus delivery of Cre recombinase. PLoS One.
10:e01281052015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Crispino JD, Lodish MB, Thurberg BL,
Litovsky SH, Collins T, Molkentin JD and Orkin SH: Proper coronary
vascular development and heart morphogenesis depend on interaction
of GATA-4 with FOG cofactors. Genes Dev. 15:839–844. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Heineke J, Auger-Messier M, Xu J, Oka T,
Sargent MA, York A, Klevitsky R, Vaikunth S, Duncan SA, Aronow BJ,
et al: Cardiomyocyte GATA4 functions as a stress-responsive
regulator of angiogenesis in the murine heart. J Clin Invest.
117:3198–3210. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tevosian SG, Deconinck AE, Tanaka M,
Schinke M, Litovsky SH, Izumo S, Fujiwara Y and Orkin SH: FOG-2, a
cofactor for GATA transcription factors, is essential for heart
morphogenesis and development of coronary vessels from epicardium.
Cell. 101:729–739. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Schlueter J and Brand T: Epicardial
progenitor cells in cardiac development and regeneration. J
Cardiovasc Transl Res. 5:641–653. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Watt AJ, Battle MA, Li J and Duncan SA:
GATA4 is essential for formation of the proepicardium and regulates
cardiogenesis. Proc Natl Acad Sci USA. 101:12573–12578. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Malek Mohammadi M, Kattih B, Grund A,
Froese N, Korf-Klingebiel M, Gigina A, Schrameck U, Rudat C, Liang
Q, Kispert A, et al: The transcription factor GATA4 promotes
myocardial regeneration in neonatal mice. EMBO Mol Med. 9:265–279.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kang C, Xu Q, Martin TD, Li MZ, Demaria M,
Aron L, Lu T, Yankner BA, Campisi J and Elledge SJ: The DNA damage
response induces inflammation and senescence by inhibiting
autophagy of GATA4. Science. 349:aaa56122015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mazzucco AE, Smogorzewska A, Kang C, Luo
J, Schlabach MR, Xu Q, Patel R and Elledge SJ: Genetic
interrogation of replicative senescence uncovers a dual role for
USP28 in coordinating the p53 and GATA4 branches of the senescence
program. Genes Dev. 31:1933–1938. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Garg V, Kathiriya IS, Barnes R,
Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS,
Hirayama-Yamada K, Joo K, et al: GATA4 mutations cause human
congenital heart defects and reveal an interaction with TBX5.
Nature. 424:443–447. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rajagopal SK, Ma Q, Obler D, Shen J,
Manichaikul A, Tomita-Mitchell A, Boardman K, Briggs C, Garg V,
Srivastava D, et al: Spectrum of heart disease associated with
murine and human GATA4 mutation. J Mol Cell Cardiol. 43:677–685.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Su W, Zhu P, Wang R, Wu Q, Wang M, Zhang
X, Mei L, Tang J, Kumar M, Wang X, et al: Congenital heart diseases
and their association with the variant distribution features on
susceptibility genes. Clin Genet. 91:349–354. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shaw-Smith C, De Franco E, Lango Allen H,
Batlle M, Flanagan SE, Borowiec M, Taplin CE, van Alfen-van der
Velden J, Cruz-Rojo J, Perez de Nanclares G, et al: GATA4 mutations
are a cause of neonatal and childhood-onset diabetes. Diabetes.
63:2888–2894. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lamina C, Coassin S, Illig T and
Kronenberg F: Look beyond one's own nose: Combination of
information from publicly available sources reveals an association
of GATA4 polymorphisms with plasma triglycerides. Atherosclerosis.
219:698–703. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Muiya NP, Wakil SM, Tahir AI, Hagos S,
Najai M, Gueco D, Al-Tassan N, Andres E, Mazher N, Meyer BF and
Dzimiri N: A study of the role of GATA4 polymorphism in
cardiovascular metabolic disorders. Hum Genomics. 7:252013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xin M, Davis CA, Molkentin JD, Lien CL,
Duncan SA, Richardson JA and Olson EN: A threshold of GATA4 and
GATA6 expression is required for cardiovascular development. Proc
Natl Acad Sci USA. 103:11189–11194. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pu WT, Ishiwata T, Juraszek AL, Ma Q and
Izumo S: GATA4 is a dosage-sensitive regulator of cardiac
morphogenesis. Dev Biol. 275:235–244. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu G, Shan J, Pang S, Wei X, Zhang H and
Yan B: Genetic analysis of the promoter region of the GATA4 gene in
patients with ventricular septal defects. Transl Res. 159:376–382.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Huang WY, Heng HH and Liew CC: Assignment
of the human GATA4 gene to 8p23.1->p22 using fluorescence in
situ hybridization analysis. Cytogenet Cell Genet. 72:217–218.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
White RA, Dowler LL, Pasztor LM, Gatson
LL, Adkison LR, Angeloni SV and Wilson DB: Assignment of the
transcription factor GATA4 gene to human chromosome 8 and mouse
chromosome 14: Gata4 is a candidate gene for Ds (disorganization).
Genomics. 27:20–26. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ohara Y, Atarashi T, Ishibashi T,
Ohashi-Kobayashi A and Maeda M: GATA-4 gene organization and
analysis of its promoter. Biol Pharm Bull. 29:410–419. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liang W, Guo J, Li J, Bai C and Dong Y:
Downregulation of miR-122 attenuates hypoxia/reoxygenation
(H/R)-induced myocardial cell apoptosis by upregulating GATA-4.
Biochem Biophys Res Commun. 478:1416–1422. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mehta G, Kumarasamy S, Wu J, Walsh A, Liu
L, Williams K, Joe B and de la Serna IL: MITF interacts with the
SWI/SNF subunit, BRG1, to promote GATA4 expression in cardiac
hypertrophy. J Mol Cell Cardiol. 88:101–110. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Rojas A, De Val S, Heidt AB, Xu SM,
Bristow J and Black BL: Gata4 expression in lateral mesoderm is
downstream of BMP4 and is activated directly by Forkhead and GATA
transcription factors through a distal enhancer element.
Development. 132:3405–3417. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Si L, Shi J, Gao W, Zheng M, Liu L, Zhu J
and Tian J: Smad4 mediated BMP2 signal is essential for the
regulation of GATA4 and Nkx2.5 by affecting the histone H3
acetylation in H9c2 cells. Biochem Biophys Res Commun. 450:81–86.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhou C, Cui Q, Su G, Guo X, Liu X and
Zhang J: MicroRNA-208b alleviates post-infarction myocardial
fibrosis in a rat model by inhibiting GATA4. Med Sci Monit.
22:1808–1816. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kontaraki JE, Kochiadakis GE, Marketou ME,
Chlouverakis G, Igoumenidis NE, Saloustros IG and Vardas PE: Early
cardiac gene transcript levels in peripheral blood mononuclear
cells reflect severity in stable coronary artery disease. Hellenic
J Cardiol. 55:119–125. 2014.PubMed/NCBI
|
|
43
|
Durocher D, Charron F, Warren R, Schwartz
RJ and Nemer M: The cardiac transcription factors Nkx2-5 and GATA-4
are mutual cofactors. EMBO J. 16:5687–5696. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hasegawa K, Lee SJ, Jobe SM, Markham BE
and Kitsis RN: cis-Acting sequences that mediate induction of
beta-myosin heavy chain gene expression during left ventricular
hypertrophy due to aortic constriction. Circulation. 96:3943–3953.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Huang WY, Cukerman E and Liew CC:
Identification of a GATA motif in the cardiac alpha-myosin
heavy-chain-encoding gene and isolation of a human GATA-4 cDNA.
Gene. 155:219–223. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Molkentin JD, Kalvakolanu DV and Markham
BE: Transcription factor GATA-4 regulates cardiac muscle-specific
expression of the alpha-myosin heavy-chain gene. Mol Cell Biol.
14:4947–4957. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Linhares VL, Almeida NA, Menezes DC,
Elliott DA, Lai D, Beyer EC, Campos de Carvalho AC and Costa MW:
Transcriptional regulation of the murine Connexin40 promoter by
cardiac factors Nkx2-5, GATA4 and Tbx5. Cardiovasc Res. 64:402–411.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gallagher JM, Yamak A, Kirilenko P, Black
S, Bochtler M, Lefebvre C, Nemer M and Latinkić BV: Carboxy
terminus of GATA4 transcription factor is required for its
cardiogenic activity and interaction with CDK4. Mech Dev.
134:31–41. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Rojas A, Kong SW, Agarwal P, Gilliss B, Pu
WT and Black BL: GATA4 is a direct transcriptional activator of
cyclin D2 and Cdk4 and is required for cardiomyocyte proliferation
in anterior heart field-derived myocardium. Mol Cell Biol.
28:5420–5431. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yamak A, Latinkic BV, Dali R, Temsah R and
Nemer M: Cyclin D2 is a GATA4 cofactor in cardiogenesis. Proc Natl
Acad Sci USA. 111:1415–1420. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lee Y, Shioi T, Kasahara H, Jobe SM, Wiese
RJ, Markham BE and Izumo S: The cardiac tissue-restricted homeobox
protein Csx/Nkx2.5 physically associates with the zinc finger
protein GATA4 and cooperatively activates atrial natriuretic factor
gene expression. Mol Cell Biol. 18:3120–3129. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Brown CO III, Chi X, Garcia-Gras E, Shirai
M, Feng XH and Schwartz RJ: The cardiac determination factor,
Nkx2-5, is activated by mutual cofactors GATA-4 and Smad1/4 via a
novel upstream enhancer. J Biol Chem. 279:10659–10669. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Laforest B and Nemer M: GATA5 interacts
with GATA4 and GATA6 in outflow tract development. Dev Biol.
358:368–378. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ang YS, Rivas RN, Ribeiro AJS, Srivas R,
Rivera J, Stone NR, Pratt K, Mohamed TMA, Fu JD, Spencer CI, et al:
Disease model of GATA4 mutation reveals transcription factor
cooperativity in human cardiogenesis. Cell. 167:1734–1749.e22.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Maitra M, Schluterman MK, Nichols HA,
Richardson JA, Lo CW, Srivastava D and Garg V: Interaction of Gata4
and Gata6 with Tbx5 is critical for normal cardiac development. Dev
Biol. 326:368–377. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Luna-Zurita L, Stirnimann CU, Glatt S,
Kaynak BL, Thomas S, Baudin F, Samee MA, He D, Small EM,
Mileikovsky M, et al: Complex interdependence regulates heterotypic
transcription factor distribution and coordinates cardiogenesis.
Cell. 164:999–1014. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Dai YS, Cserjesi P, Markham BE and
Molkentin JD: The transcription factors GATA4 and dHAND physically
interact to synergistically activate cardiac gene expression
through a p300-dependent mechanism. J Biol Chem. 277:24390–24398.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Dai YS and Markham BE: p300 Functions as a
coactivator of transcription factor GATA-4. J Biol Chem.
276:37178–37185. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhang X, Wang X, Zhu H, Zhu C, Wang Y, Pu
WT, Jegga AG and Fan GC: Synergistic effects of the GATA-4-mediated
miR-144/451 cluster in protection against simulated
ischemia/reperfusion-induced cardiomyocyte death. J Mol Cell
Cardiol. 49:841–850. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Glenn DJ, Rahmutula D, Nishimoto M, Liang
F and Gardner DG: Atrial natriuretic peptide suppresses endothelin
gene expression and proliferation in cardiac fibroblasts through a
GATA4-dependent mechanism. Cardiovasc Res. 84:209–217. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Molkentin JD, Lu JR, Antos CL, Markham B,
Richardson J, Robbins J, Grant SR and Olson EN: A
calcineurin-dependent transcriptional pathway for cardiac
hypertrophy. Cell. 93:215–228. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Morimoto T, Hasegawa K, Wada H, Kakita T,
Kaburagi S, Yanazume T and Sasayama S: Calcineurin-GATA4 pathway is
involved in beta-adrenergic agonist-responsive endothelin-1
transcription in cardiac myocytes. J Biol Chem. 276:34983–34989.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Morin S, Charron F, Robitaille L and Nemer
M: GATA-dependent recruitment of MEF2 proteins to target promoters.
EMBO J. 19:2046–2055. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Kolettis TM, Barton M, Langleben D and
Matsumura Y: Endothelin in coronary artery disease and myocardial
infarction. Cardiol Rev. 21:249–256. 2013. View Article : Google Scholar : PubMed/NCBI
|