Introduction
Gastric cancer (GC) is one of the most prevalent and
lethal malignancies worldwide (1).
Although its incidence and mortality rates have markedly declined
in western countries over the last century (2,3), GC
remains a serious health burden in some parts of the world, such as
China and Japan (1,4). Surgery is currently the only
available radical treatment for GC, but patients are often
diagnosed at later stages, rendering them unfit for radical
surgery, or they experience postsurgical disease relapse or
metastasis, which worsens their prognosis (5). Comprehensive treatment for advanced
GC remains unsatisfactory. Therefore, elucidation of the molecular
mechanisms underlying GC invasion and metastasis is of vital
importance.
TAFA chemokine like family member 5 (TAFA5), also
known as Family with sequence similarity 19-member A5 C-C motif
chemokine like, belongs to the TAFA family, which includes 5
members that are predominantly expressed in the central nervous
system (6). These genes encode
small secreted proteins named TAFA1-TAFA5, which are evolutionarily
close to macrophage inflammatory protein 1A [currently known as
chemokine C-C motif ligand 3 (CCL3)], a member of the small C-C
chemokine family (6). Under
physical conditions, TAFA5 colocalizes with vasopressin and
oxytocin in the hypothalamic paraventricular nucleus, and may be
involved in fluid homeostasis regulation (7). In addition, TAFA5 is a downstream
target of Wnt family member 9 (Wnt9) b and can be activated by
interactions between c-Myc and β-catenin to induce
Wnt9b/β-catenin-mediated progenitor renewal in the developing
kidney (8,9). Furthermore, TAFA5 inhibits osteoclast
formation along with its receptor, formyl peptide receptor 2
(FPR2), by stimulating bone marrow-derived macrophages (10). Using a proteomic approach,
Janvilisri et al (11)
identified TAFA5 as one of the most increased serum tumor markers
that could distinguish human cholangiocarcinoma from benign biliary
tract diseases. In addition, Diaz de Stahl et al (12) analyzed 50 glioblastoma samples with
a high-resolution tiling-path chromosome 22 array and discovered 2
amplified regions on chromosome 22 that were characteristics for
patients with tumors. Further analysis of these regions revealed
two novel genes, including TAFA5. As no such variation was
identified in a series of normal individuals, the authors
speculated that these genes were involved in glioma tumorigenesis
(12). In a large-scale
genome-wide association study, Wu et al (13) identified 5 loci associated with
susceptibility to pancreatic cancer, including one that was located
upstream of TAFA5 at chromosome 22.
Although an accumulating body of evidence has been
suggestive of the involvement of TAFA5 in tumorigenesis, its role
in GC development and progression remains unclear. The present
study evaluated the clinical and prognostic significance of TAFA5
in 90 human GC samples and validated the results with data from two
public datasets. The present study also investigated the in
vitro activities of TAFA5 in cultured GC cells and
characterized the potential underlying mechanisms of action.
Materials and methods
Patients and specimens
A total of 18 paired human GC samples and their
matched gastric normal tissues (NTs) were collected at the time of
surgical resection at the Fifth People's Hospital of Shanghai,
Fudan University (Shanghai, China) between February 2017 and
February 2018. These samples were from 13 males and 5 females, with
a median age of 64 (range 52–86). Patients were included in the
study if they were initially diagnosed with GC, underwent the
surgery and had complete clinicopathological information. Those who
had extensively metastatic tumors, suffered from life-threatening
diseases such as severe cardiovascular disease or had other types
of tumors besides GC were excluded from the study. Samples were
snap-frozen in liquid nitrogen and stored at −80°C.
Paraffin-embedded tissues were retrieved from the Tissue Bank of
the Fifth People's Hospital of Shanghai, Fudan University, and 4-µm
tissue sections were prepared by the Department of Pathology of the
same hospital. The present study was approved by the Institutional
Ethics Committee at the Fifth People's Hospital of Shanghai, Fudan
University (ethical approval form no. 2017-097) and adhered to the
principles in the Declaration of Helsinki. Written informed consent
was obtained from each patient prior to tissue collection for
experimentation.
Tissue microarray and
immunohistochemistry (IHC)
Microarray sections of GCs and neighboring NTs were
prepared by Shanghai Outdo Biotech Co., Ltd. These sections
contained 90 paired GC and NTs from patients [the tissue microarray
(TMA) cohort] and the clinicopathological characteristics of these
patients are summarized in Table
I. Following deparaffination, rehydration in graded ethanol,
antigen retrieval with citrate buffer pH 6.0 (1:300 dilution; cat.
no. ZLI-9065; OriGene Technologies, Inc.) and blocking with goat
serum (1:20 dilution; cat. no. C0265; Beyotime Institute of
Biotechnology) at room temperature for 1 h, slides were stained
with a rabbit polyclonal antibody against human TAFA5 (1:50
dilution; cat. no. 13948-1-AP; ProteinTech Group, Inc.) at 4°C
overnight. Normal rat immunoglobulin G (1:50 dilution; cat. no.
D110504; Sangon Biotech Co., Ltd.) instead of the primary antibody
was used as a control. Subsequently, after washing with PBS, a
horseradish peroxidase (HRP)-conjugated secondary antibody
(1:2,000; goat anti-rabbit, cat. no. A0208 and goat anti-rat, cat.
no. A0192; Beyotime Institute of Biotechnology) was added and
incubated at room temperature for 1 h. Then, these sections were
stained with 3,3′-diaminobenzidine (1:25 dilution; cat. no.
GK500705; Gene Tech Co., Ltd.) at room temperature for 5 min and
counterstained with 100% hematoxylin (cat. no. C0107; Beyotime
Institute of Biotechnology) at room temperature for 2 min. A
modified H-score system was used to semi-quantitate TAFA5
expression, as previously described (14). Briefly, the maximal intensity of
staining (0, negative; 1, weak; 2, moderate; and 3, strong) was
multiplied by the percentage of positive tumor cells (0–100%) to
generate the modified H-score (range: 0-300). TAFA5 staining was
categorized as high or low expression using the median H-score.
 | Table I.Clinical and pathologic features of
patients with gastric cancera (n=90). |
Table I.
Clinical and pathologic features of
patients with gastric cancera (n=90).
| Feature | Number of patients
(%) |
|---|
| Sex |
|
|
Male | 69 (76.7) |
|
Female | 21 (23.3) |
| Age |
|
|
<70 | 45 (50) |
|
≥70 | 45 (50) |
|
Differentiation |
|
| G2 | 28 (31.1) |
| G3 | 62 (68.9) |
| Tumor size
(cm) |
|
|
<5 | 35 (38.9) |
| ≥5 | 54 (60.0) |
| NA | 1 (1.1) |
| TNM stage |
|
| I | 12 (13.3) |
| II | 27 (30.0) |
|
III | 49 (54.5) |
| IV | 2 (2.2) |
| Tumor stage |
|
| T1 | 5 (5.6) |
| T2 | 15 (16.7) |
| T3 | 46 (51.1) |
| T4 | 24 (26.6) |
| Nodal stage |
|
| N0 | 24 (26.7) |
| N1 | 15 (16.7) |
| N2 | 23 (25.5) |
| N3 | 28 (31.1) |
| Vessel
invasion |
|
| No | 74 (82.2) |
|
Yes | 16 (17.8) |
| Nerve invasion |
|
| No | 74 (82.2) |
|
Yes | 15 (16.7) |
| NA | 1 (1.1) |
| Expression of
TAFA5 |
|
|
Low | 41 (45.6) |
|
High | 49 (54.4) |
Access to public datasets
The mRNA expression of TAFA5 in GC tissues and
normal mucosae was acquired from Oncomine (www.oncomine.org) and Gene Expression Omnibus (GEO;
accession no. GSE79973). The Cancer Genome Atlas (TCGA) Stomach
Adenocarcinoma (STAD) RNA-sequence data and corresponding phenotype
data were downloaded from UCSC Xena (https://xenabrowser.net/datapages/). The results in
the present study are in whole or part based upon data generated by
the TCGA Research Network (cancergenome.nih.gov). The expression of TAFA5 in TCGA
STAD was further validated by two online analyzing tools
specifically designed for TCGA data analysis (gepia.cancer-pku.cn/detail.php?gene=FAM19A5 and
ualcan.path.uab.edu/cgi-bin/TCGAExResultNew2.pl?genenam=FAM19A5&ctype=STAD).
The original data for the prognostic analysis of TAFA5 were
downloaded from Kaplan-Meier Plotter (www.kmplot.com) and UCSC Xena (xenabrowser.net/heatmap/).
Cell lines
A gastric epithelial cell line (GES-1) and GC cell
lines (AGS, HGC27, NCI-N87, MGC803 and SNU-1) were obtained from
the Type Culture Collection of the Chinese Academy of Sciences. All
of the cells were validated by short tandem repeat DNA profiling
and were confirmed negative for Mycoplasma contamination. Cells
were cultured in RPMI-1640 (BBI Life Sciences Corporation) or F12K
medium (Zhong Qiao Xin Zhou Biotechnology Co., Ltd.) supplemented
with 10% fetal bovine serum (cat. no. E600001; Sangon Biotech Co.,
Ltd.), 100 µg/ml penicillin, and 100 mg/ml streptomycin at 37°C
with 5% CO2 in a humidified incubator (Thermo Fisher
Scientific, Inc.).
Construction of TAFA5-targeting short hairpin
(sh)-RNAs and packaging of lentiviruses. Two targeting shRNAs
(shTAFA5-1 and shTAFA5-2) and a nontargeting scrambled RNA
(scramble) were subcloned into the GV248 lentivirus vector by
Shanghai GeneChem Co., Ltd. The shTAFA5-1 target sequence was
5′-CGCAAGAATCATCAAGACCAA-3′, and the shTAFA5-2 target sequence was
5′-CACCTGTGAGATTGTGACCTT-3′. Lentiviral stocks were prepared, as
previously described (15).
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated from cell cultures or from
snap-frozen tissues from patients with GC using RNAiso Plus (Takara
Bio, Inc.), according to the manufacturer's instructions, and
reverse transcribed with HiScript Q Select RT SuperMix (cat. no.
R132-01; Vazyme Biotech Co., Ltd.), according to the manufacturer's
protocol. qPCR was performed with SYBR Green Master Mix (cat. no.
Q131-02; Vazyme Biotech Co., Ltd.) and quantified using the
2−ΔΔCq method (16).
The thermocycling conditions were as follows: 95°C for 30 sec,
followed by 40 cycles of 95°C for 10 sec, 60°C for 32 sec, 95°C for
15 sec, 60°C for 60 sec and 95°C for 15 sec. Each sample was
assayed in duplicate. All PCR products were confirmed by 2.0%
agarose gel electrophoresis. The sequences of the primers were as
follows: TAFA5, forward 5′-GTGAGTTTGGCCACTCCGTA-3′ and reverse
5′-GAATGGACAGATGGCTGGCA-3′; and GAPDH, forward
5′-GTCAAGGCTGAGAACGGGAA-3′ and reverse 5′-AAATGAGCCCCAGCCTTCTC-3′.
Experiments were repeated three times in duplicate.
Western blotting
Total protein was extracted from cell cultures or
homogenized tissues from patients with GC using
radioimmunoprecipitation assay lysis buffer (Beyotime Institute of
Biotechnology) containing phenylmethylsulfonyl fluoride (Beyotime
Institute of Biotechnology) and proteinase inhibitor cocktail
solution (Roche Diagnostics), and quantified via the bicinchoninic
acid assay method, as recommended by the manufacturer. Western
blotting was performed according to standard methods as previously
described (17). Briefly, proteins
(30 µg) were separated by 10% SDS-PAGE and then transferred onto a
polyvinylidene fluoride membrane. Subsequently, the membranes were
blocked with 5% fat-free dry milk at room temperature for 1 h.
Then, the blots were incubated with a rabbit polyclonal antibody
against TAFA5 (1:1,000 dilution; cat. no. 13948-1-AP; ProteinTech
Group, Inc.) and monoclonal antibodies against E-cadherin,
N-cadherin, Vimentin and ZEB-1 (1:1,000 dilution; cat. nos. 3195,
13116, 5741 and 3396, respectively; Cell Signaling Technology,
Inc.) overnight at 4°C. GAPDH (1:2,000; rabbit anti-human; cat. no.
AF1186; Beyotime Institute of Biotechnology) or β-actin (1:2,000;
mouse anti-human; cat. no. AF0003; Beyotime Institute of
Biotechnology) were detected as the loading controls. The membranes
were washed with TBST and incubated with the respective
HRP-conjugated secondary antibodies (1:2,000; goat anti-mouse cat.
no. A0216 and goat anti-rabbit cat. no. A0208; Beyotime Institute
of Biotechnology) at room temperature for 1 h. The proteins were
finally visualized using an enhanced chemiluminescence system (cat.
no. P0018AS; Beyotime Institute of Biotechnology). The grayscale
values of protein bands were analyzed using ImageJ software
(National Institutes of Health).
Proliferation assay
Stably infected AGS and HGC-27 cells
(2×103 cells/well) were seeded in 96-well plates and
cultured for 24, 48, 72 or 96 h. Then, 10 µl Cell Counting Kit-8
(CCK-8) reagent (10% v/v in serum-free RPMI-1640 medium; Beyotime
Institute of Biotechnology) was added to each well and incubated at
37°C for 1 h. Absorbance was measured at 450 nm using a microplate
reader (BioTek Synergy 2; BioTek Instruments, Inc.).
Migration assay
A Transwell migration assay was performed as
previously reported (18).
Briefly, cells (4×105 cells/ml) were seeded in
serum-free RPMI-1640 or F12K medium in the top chamber of the
Transwell insert. Medium containing 20% FBS in the lower chamber
served as chemoattractant. Following incubation for 24 h at 37°C,
the cells on the top side of the membrane were removed with a
cotton swab, and the migrated cells on the bottom side were fixed
with methanol for 20 min and then stained with crystal violet (0.1%
in PBS) for 15 min. A total of 6 randomly selected fields per well
were imaged under an inverted microscope (Leica Microsystems GmbH)
using the ImageScope software (Leica Microsystems GmbH) with a
magnification of ×200. Then cells were manually counted and
analyzed by Excel 2010 (Microsoft Corporation) and the number of
migrated cells was counted.
Scratch wound healing assay
A monolayer scratch wound assay was employed to
evaluate cell migration, as previously described (19). Briefly, cells (4×105
cells/well) were seeded in 12-well plates and grown to nearly 100%
confluence in 10% FBS-containing medium, before being washed with
PBS and transferred to serum-free medium for 24 h. Then, a scratch
wound was generated with a 200 µl pipette tip. Wound closure was
imaged at 0, 24 and 48 h under an inverted microscope (Leica
Microsystems GmbH) using the ImageScope software (Leica Biosystems
GmbH) with a magnification of ×40. Then the wound closure rates
were analyzed by ImageJ (v1.8.0; National Institutes of
Health).
Gene set enrichment analysis
The Gene set enrichment analysis (GSEA) is developed
by the Broad Institute website (http://www.broadinstitute.org/gsea/index.jsp). All
GSEA analyses were performed using the GSEA 2.2.2 software. The
TCGA STAD RNA-seq data were loaded into GSEA and run with the
latest version of Hallmarks gene sets. The number of permutations
was set at 1,000, and patient data were divided into TAFA5 high and
low groups by the median of TAFA5 expression. Then GESA was
performed according to the default weighted enrichment statistics
and genes were ranked by the Pearson method. Gene sets were
considered significantly enriched if P<0.05 and FDR < 0.25.
Those gene sets that were significantly enriched in the TAFA5 high
group were designated as TAFA5-positively related gene sets, while
those that were significantly enriched in the TAFA5 low group were
designated as TAFA5-negatively correlated gene sets.
Statistical analysis
Analyses were performed using Microsoft Excel 2010
(Microsoft Corporation), GraphPad Prism7 (GraphPad Software, Inc.),
and SPSS version 22 (IBM Corp.). Student's t-test was used for
statistical comparisons between two groups, while statistical
comparisons among multiple groups were performed by one-way
analysis of variance followed by Bonferroni's post hoc test.
Pearson's χ2 test and Fisher's exact test were used for
categorical comparisons. Survival analyses were conducted using the
Kaplan-Meier method. Univariate and multivariate survival analyses
were conducted with the Cox proportional hazards regression model.
All statistical tests were two-sided. P<0.05 was considered to
indicate a statistically significant difference.
Results
TAFA5 is upregulated in GC
By analyzing the TAFA5 expression profile data from
a GEO GC dataset (GSE79973), the present study revealed that TAFA5
was upregulated in GC tissues compared with NTs (n=10 pairs;
P<0.001; Fig. 1A). RNA
sequencing data from TCGA STAD also revealed an increase in TAFA5
expression in GC tissues (n=415) compared with in NTs (n=35;
P<0.001; Fig. 1B), which was
further validated by online analysis tools Gepia and Ualcan (data
not shown). Two additional GC datasets [Derrico gastric(GEO:
GSE13911), n=69; and Cho gastric(GEO: GSE13861), n=90](20,21)
with Lauren's classification in the Oncomine database indicated
that TAFA5 was generally overexpressed in each cancer group when
compared with the control group, although the differences were not
significant in three cases with smaller samples (Fig. 1C and D). To further validate these
observations, the present study measured TAFA5 expression in 18 GC
tissues and their matched NTs by RT-qPCR and western blotting. The
results indicated that TAFA5 mRNA and protein expression levels
were increased in GC tissues compared with NTs (Fig. 1E-H). Taken together, these results
indicated that TAFA5 was upregulated in GC.
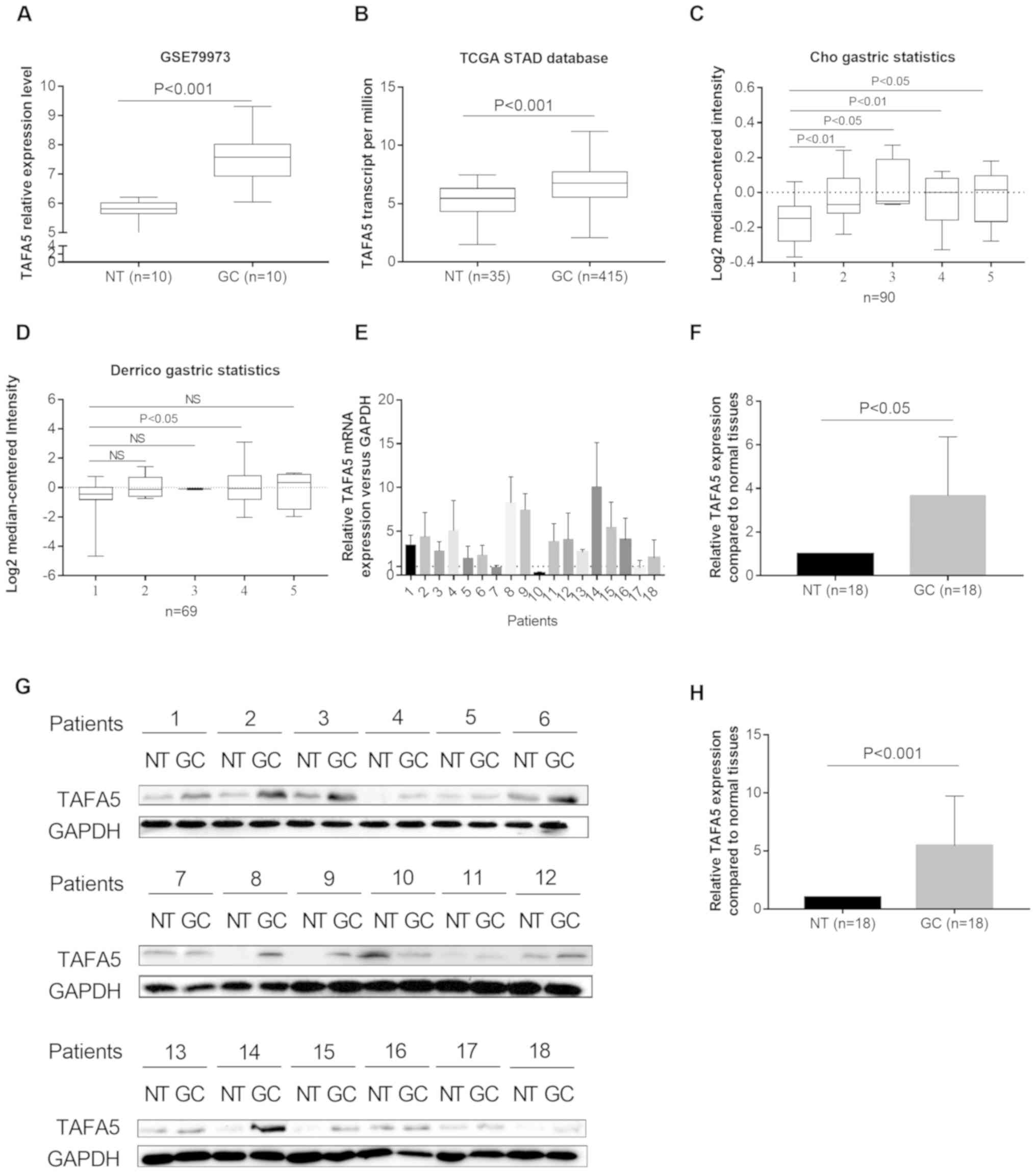 | Figure 1.TAFA5 is upregulated in GC. (A) TAFA5
expression was upregulated in GC tissues compared with NTs (n=10
per group) in the GSE79973 database. (B) TAFA5 expression was
upregulated in GC (n=415) when compared with NTs (n=35) in TCGA
cohort. (C) A logarithmic 2−ΔΔCq scale was used to
represent the fold-changes in TAFA5 mRNA expression in the
microarray datasets from the Oncomine Cho gastric and (D) Derrico
gastric databases. The cases were grouped as follows: 1, no value;
2, gastric intestinal type adenocarcinoma; 3, gastric mixed
adenocarcinoma; 4, diffuse gastric adenocarcinoma; and 5, gastric
adenocarcinoma. (E and F) TAFA5 mRNA expression levels in 18 paired
GC tissues and their adjacent NTs. (G and H) Representative blots
and quantification of TAFA5 protein expression levels from the 18
paired GC tissues and their adjacent NTs. The average TAFA5
expression was normalized to the expression of GAPDH. A total of 3
replicates were conducted for each experiment. TAFA5, TAFA
chemokine like family member 5; GC, gastric cancer; NT, normal
tissue; TCGA, The Cancer Genome Atlas; NS, not significant. |
Overexpression of TAFA5 in GC
correlates with unfavorable prognosis
To test the hypothesis that TAFA5 protein abundance
in GC contributes to increased malignancy, the present study
examined the TAFA5 expression levels in primary GC tissues of a TMA
cohort by IHC. As shown in Fig.
2A, TAFA5 was mainly expressed in the cytoplasm of cancer
cells, and its expression was stronger in GC tissues than in NTs.
The H-score was then used to quantitate TAFA5 staining in these
tissues, which revealed that TAFA5 expression was significantly
increased in GC compared with NTs (Fig. 2B). The associations between TAFA5
expression and clinicopathological variables are summarized in
Table II. High TAFA5 expression
was significantly associated with poor differentiation, and worse
tumor, nodal and metastasis stages.
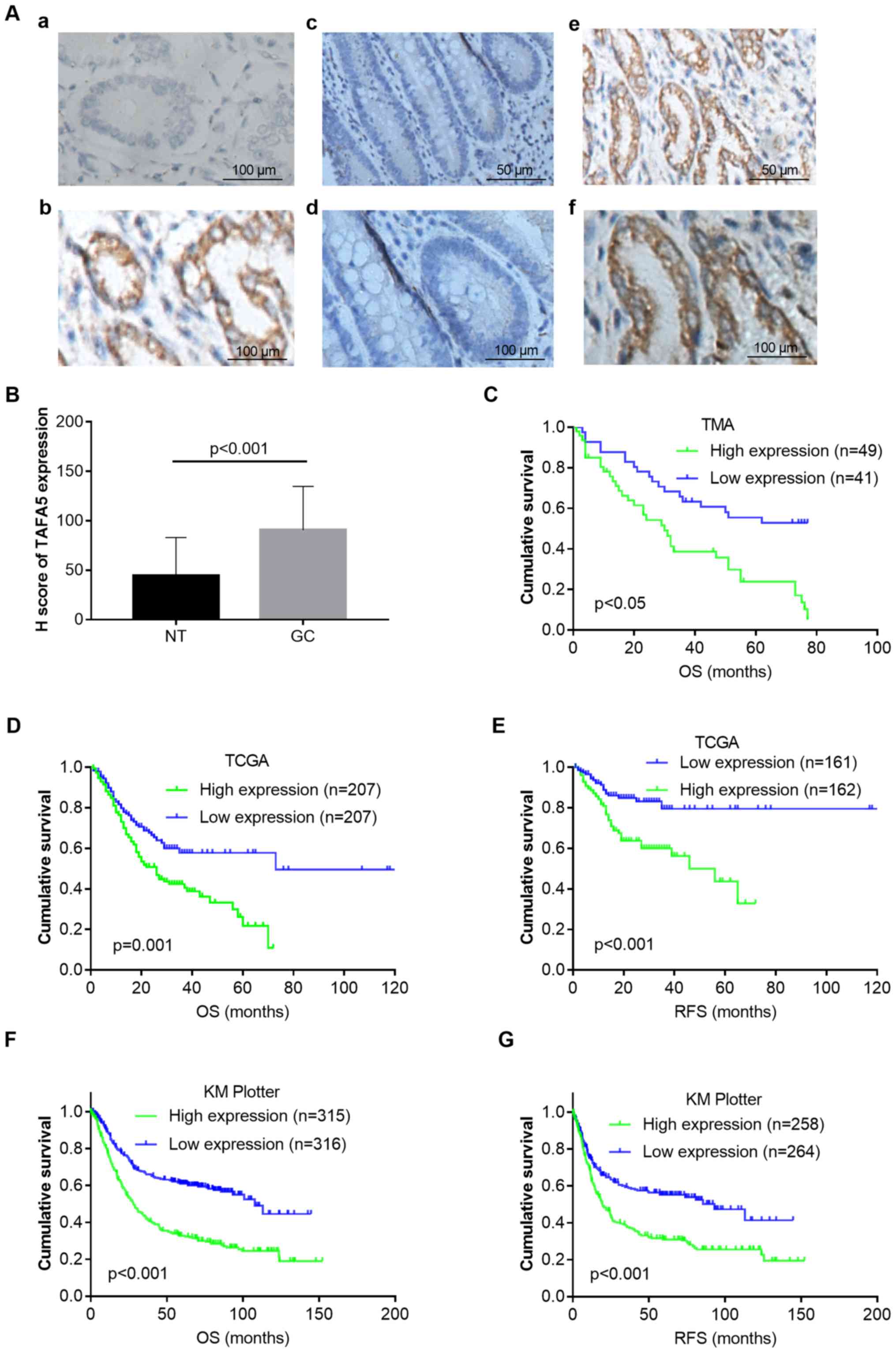 | Figure 2.TAFA5 overexpression in GC is
associated with poor patient survival. (A) Immunohistochemical
staining of TAFA5 in GC tissues and NTs. (A-a) Immunoglobulin G
negative control. (A-b) Positive staining of TAFA5 in GC tissue
(magnification, ×400). (A-c) TAFA5 staining in NT (magnification,
×200). (A-d) TAFA5 staining in NT (magnification, ×400). (A-e)
TAFA5 staining in GC tissue (magnification, ×200). (A-f) TAFA5
staining in GC tissue (magnification, ×400). (B) H-scores of TAFA5
staining in NTs and GC. (C) Kaplan-Meier plots for the OS of
patients in the TMA cohort. (D) Kaplan-Meier plots for OS and (E)
RFS in TCGA cohort. (F) Kaplan-Meier plots for OS and (G) RFS in
the Kaplan-Meier Plotter cohort. P-values were obtained using the
log-rank test. Censored data are indicated by the + symbol.
Patients were stratified into low and high TAFA5 expression groups
according to TAFA5 mRNA expressions (<median vs. ≥median) in the
TCGA and Kaplan-Meier Plotter cohorts, or H-scores of TAFA5
staining (<median vs. ≥median) in the TMA cohort. TAFA5, TAFA
chemokine like family member 5; GC, gastric cancer; NT, adjacent
normal tissue; OS, overall survival; RFS, recurrence-free survival;
TCGA, The Cancer Genome Atlas; TMA, tissue microarray. |
 | Table II.Association between TAFA5 expression
and clinicopathological variables in gastric cancer (n=90). |
Table II.
Association between TAFA5 expression
and clinicopathological variables in gastric cancer (n=90).
|
|
| TAFA5
expression |
|---|
|
|
|
|
|---|
| Clinicopathological
feature | Number of
patients | Low (41) | High (49) | P-value |
|---|
| Sex |
|
|
|
|
|
Male | 69 | 32 (46.4) | 37 (53.6) |
|
|
Female | 21 | 9 (42.9) | 12 (57.1) | 0.488 |
| Age |
|
|
|
|
|
<70 | 45 | 22 (48.9) | 23 (51.1) |
|
|
≥70 | 45 | 19 (42.2) | 26 (57.8) | 0.336 |
| Histological
grade |
|
|
|
|
| G2 | 28 | 18 (64.3) | 10 (35.7) |
|
| G3 | 62 | 23 (37.1) | 39 (62.9) | 0.015 |
| Tumor size
(cm) |
|
|
|
|
|
<5 | 35 | 18 (51.4) | 17 (48.6) |
|
| ≥5 | 54 | 22 (40.7) | 32 (59.3) | 0.220 |
| Tumor stage |
|
|
|
|
|
T1/T2 | 20 | 17 (85.0) | 3 (15.0) |
|
|
T3/T4 | 70 | 24 (34.3) | 46 (65.7) | 0.001 |
| Nodal stage |
|
|
|
|
| N0 | 24 | 16 (66.7) | 8 (33.3) |
|
|
N1-N3 | 66 | 25 (37.9) | 41 (62.1) | 0.014 |
| TNM stage |
|
|
|
|
|
I/II | 38 | 24 (63.2) | 14 (36.8) |
|
|
III/IV | 52 | 17 (32.7) | 35 (67.3) | 0.004 |
| Vessel
invasion |
|
|
|
|
| No | 74 | 34 (45.9) | 40 (54.1) |
|
|
Yes | 16 | 7 (43.8) | 9 (56.2) | 0.548 |
| Nerve invasion |
|
|
|
|
| No | 74 | 35 (47.3) | 39 (52.7) |
|
|
Yes | 15 | 6 (40.0) | 9 (60.0) | 0.410 |
Next, the present study determined the association
between TAFA5 expression and patient outcomes. Kaplan-Meier
survival analysis of the TMA cohort revealed that patients with
high TAFA5 expression exhibited a shorter overall survival (OS)
compared with those with low expression [estimated mean OS 30.5
months with 95% confidence interval (CI) 22.9–38.1 months vs. OS
53.0 months with 95% CI 44.5–61.6 months; log-rank test, P=0.001;
Fig. 2C]. To further confirm the
adverse prognostic role of TAFA5 in patients with GC, the present
study downloaded and analyzed TAFA5 transcription data from TCGA
and Kaplan-Meier Plotter datasets. The results demonstrated that
high TAFA5 expression was correlated with worse OS and
recurrence-free survival (RFS) in both patient cohorts (Fig. 2D-G). Multivariate analysis using a
Cox proportional hazards model revealed that TAFA5 overexpression
was significantly associated with shorter OS [hazard ratio (HR),
1.90; 95% CI, 1.04–3.44; P=0.036], following adjustments for tumor
size, tumor stage and nodal stage (Table III). Taken together, these
results indicated that TAFA5 was a prognostic marker in GC that was
associated with unfavorable prognoses.
 | Table III.Univariate and multivariate Cox
proportional hazard models for overall survival in patients with
gastric cancer (n=90). |
Table III.
Univariate and multivariate Cox
proportional hazard models for overall survival in patients with
gastric cancer (n=90).
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Clinicopathological
features | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Sex |
|
|
|
|
|
Male | 1 (Reference) |
|
|
|
|
Female | 0.60
(0.30–1.18) | 0.137 |
|
|
| Age |
|
|
|
|
|
<70 | 1 (Reference) |
|
|
|
|
≥70 | 1.67
(0.99–2.82) | 0.055 |
|
|
| Histological
grade |
|
|
|
|
| G2 | 1 (Reference) |
|
|
|
| G3 | 1.51
(0.85–2.69) | 0.162 |
|
|
| Tumor size
(cm) |
|
|
|
|
|
<5 | 1 (Reference) |
| 1 (Reference) |
|
| ≥5 | 3.38
(1.83–6.22) | 0.001 | 2.48
(1.28–4.77) | 0.007 |
| Tumor stage |
|
|
|
|
|
T1/T2 | 1 (Reference) |
| 1 (Reference) |
|
|
T3/T4 | 4.08
(1.75–9.56) | 0.001 | 1.61
(0.62–4.22) | 0.330 |
| Nodal stage |
|
|
|
|
| N0 | 1 (Reference) |
| 1 (Reference) |
|
|
N1-N3 | 3.27
(1.55–6.93) | 0.002 | 1.94
(0.88–4.29) | 0.100 |
| TNM stage |
|
|
|
|
|
I/II | 1 (Reference) |
|
|
|
|
III/IV | 3.46
(1.90–6.29) | 0.001 |
|
|
| Vessel
invasion |
|
|
|
|
| No | 1 (Reference) |
|
|
|
|
Yes | 1.05
(0.53–2.08) | 0.888 |
|
|
| Nerve invasion |
|
|
|
|
| No | 1 (Reference) |
|
|
|
|
Yes | 1.02
(0.50–2.07) | 0.963 |
|
|
| TAFA5
expression |
|
|
|
|
|
Low | 1 (Reference) |
| 1 (Reference) |
|
|
High | 2.63
(1.51–4.58) | 0.001 | 1.90
(1.04–3.44) | 0.036 |
TAFA5 promotes the proliferation and
migration of cultured GC cells
To gain insight into the potential role of TAFA5 in
GC progression, the present study first examined the intrinsic
expression levels of TAFA5 in a normal gastric epithelial cell
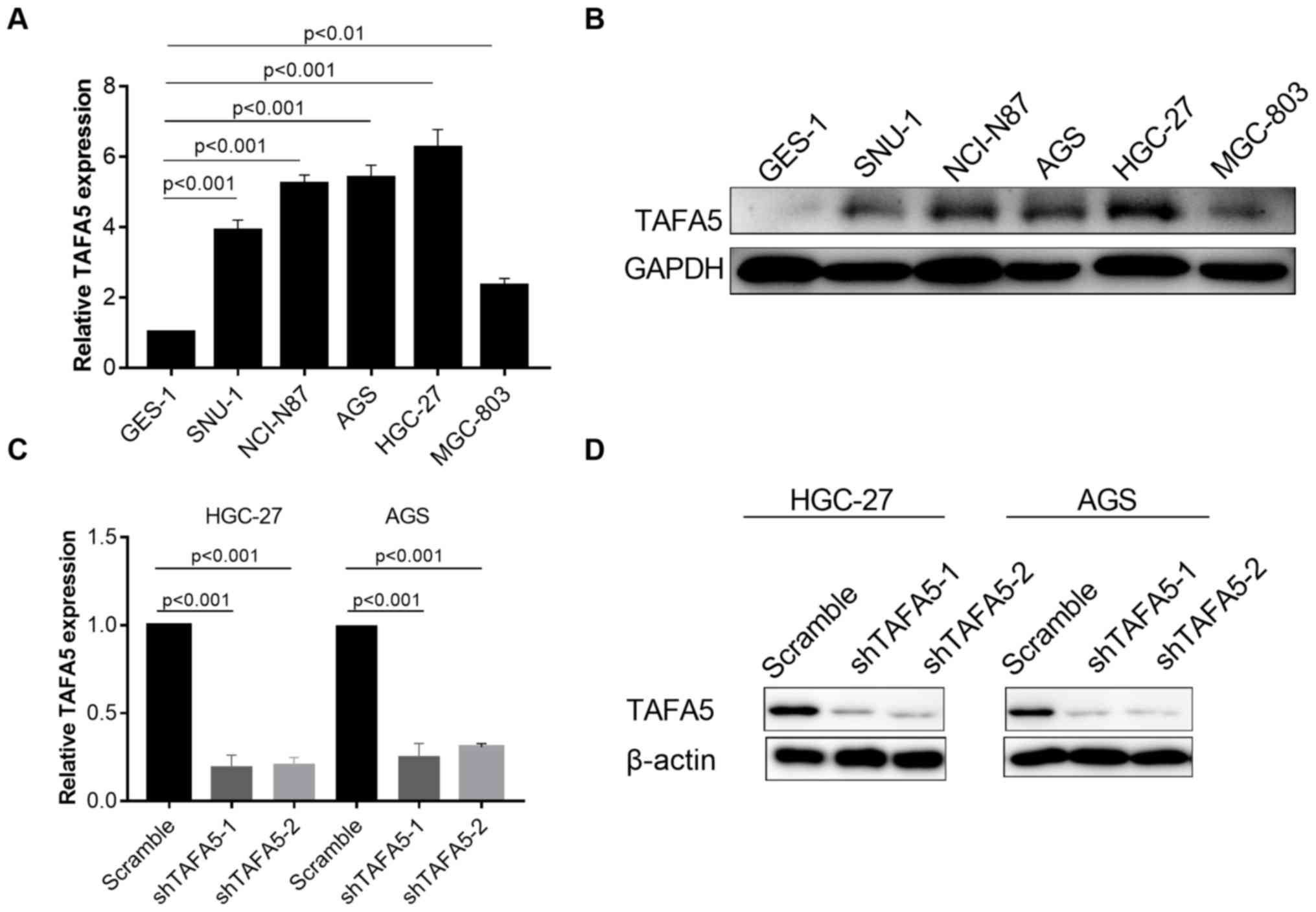
line, GES-1, and in five GC cell lines, via RT-qPCR and western
blotting. The results indicated that TAFA5 expression was markedly
increased in GC cells when compared with GES-1 cells (Fig. 3A and B). TAFA5 expression was then
silenced in HGC-27 and AGS cells with lentiviruses carrying two
TAFA5-specific shRNAs (Fig. 3C and
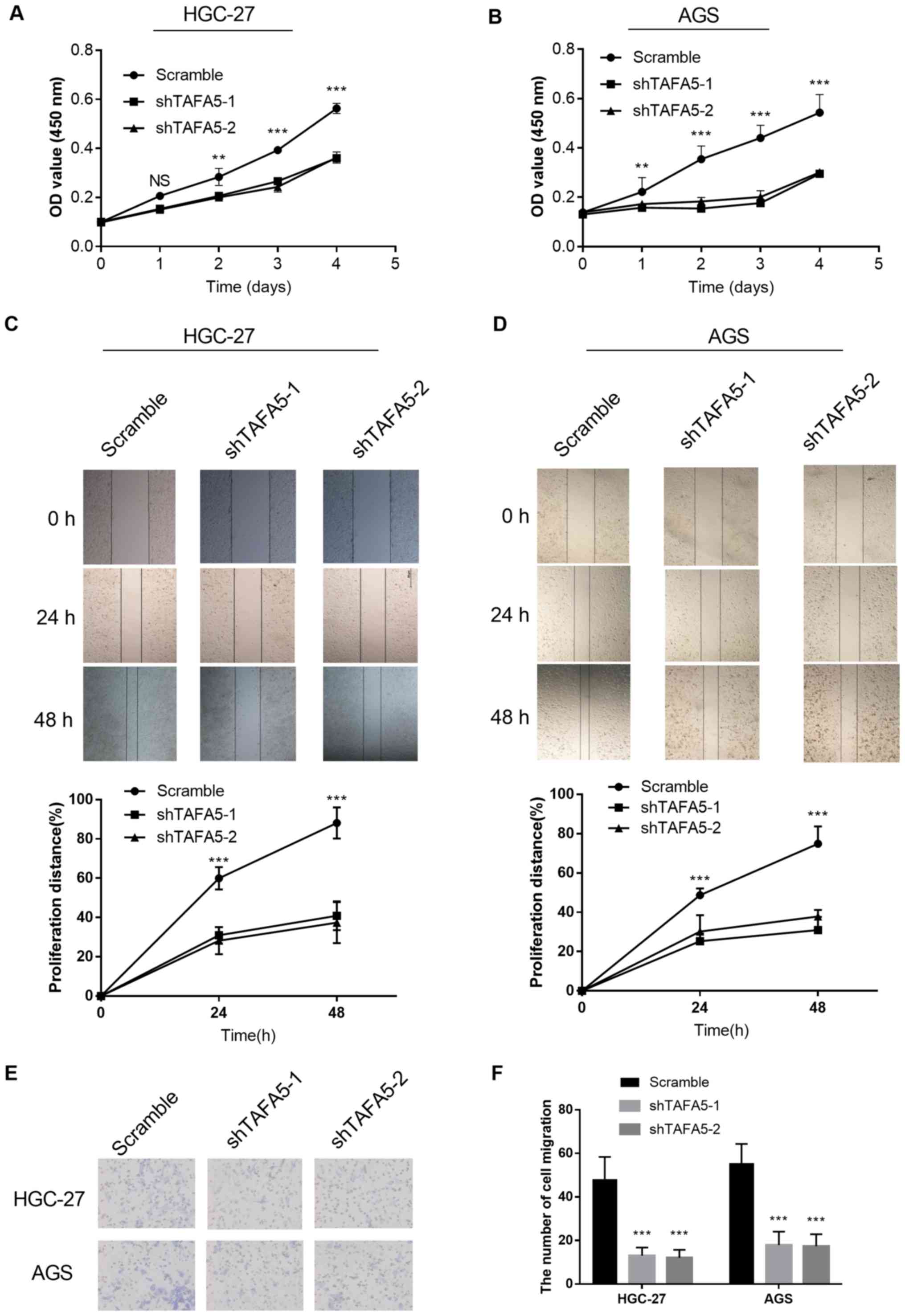
D). In cultured GC cells, CCK-8 assays revealed that knockdown
of TAFA5 expression significantly inhibited proliferation (Fig. 4A and B). In addition, knockdown of
TAFA5 expression decreased cell migration in the scratch wound
healing assay (Fig. 4C and D) and
in the Transwell migration assay (Fig.
4E and F). Collectively, these results indicated that TAFA5
promoted the proliferation and migration of GC cells.
TAFA5 may function by modulating
epithelial-mesenchymal transition (EMT) in GC
To characterize the potential mechanism of TAFA5 in
promoting tumor progression, the present study used the RNA-seq
data for GC samples from TCGA to conduct gene set enrichment
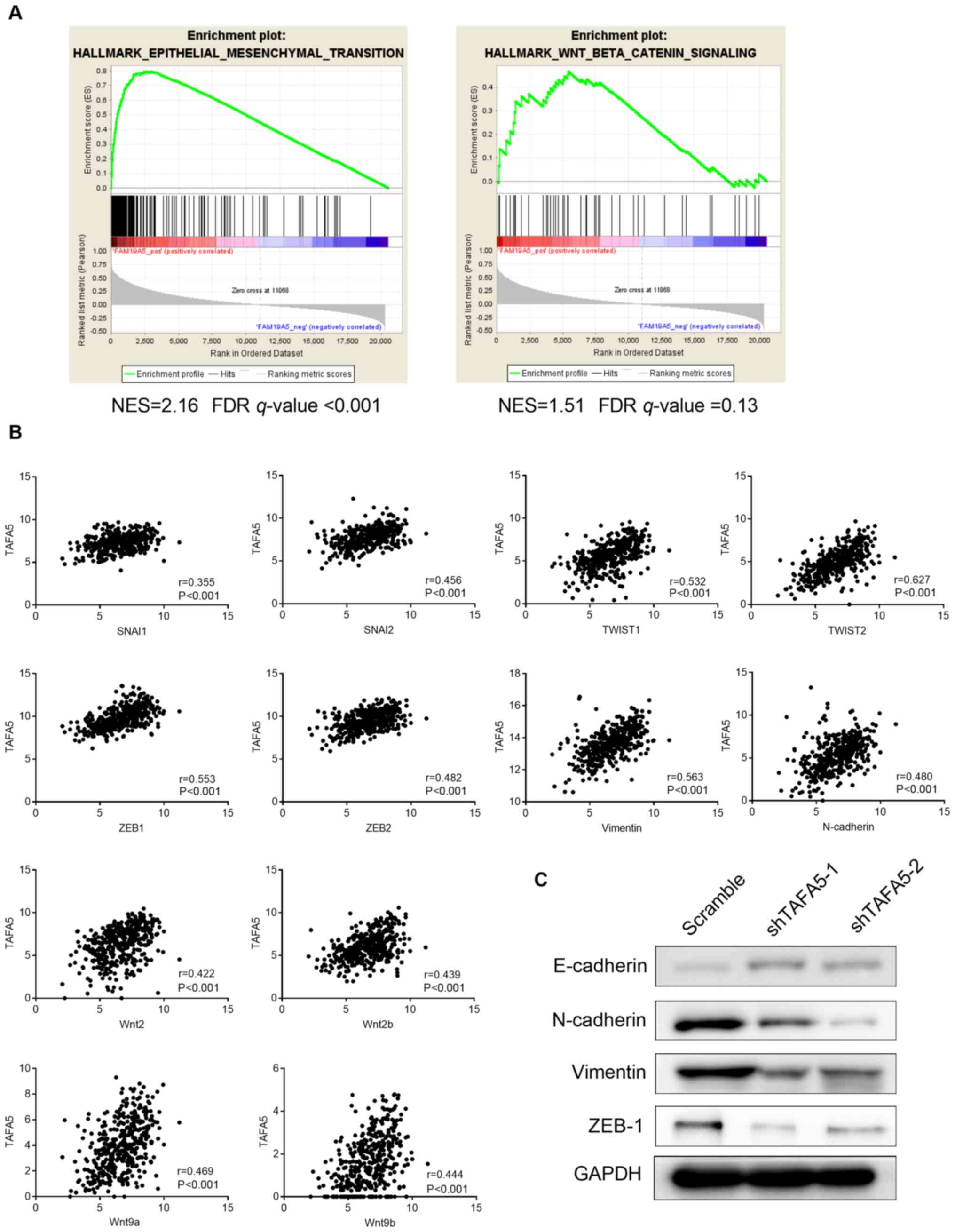
analysis (GSEA) (22,23). The results revealed that TAFA5
expression was positively correlated with genes involved in EMT,
but not with those in the Wnt/β-catenin signaling pathway (Fig. 5A). Furthermore, as shown in
Fig. 5B, TAFA5 expression was
moderately positively correlated with snail family transcriptional
repressor (SNAI) 1, SNAI2, twist family bHLH transcription factor
(TWIST) 1, TWIST2, zinc finger E-box binding homeobox (ZEB) 1,
ZEB2, Vimentin, N-cadherin, Wnt9a and Wnt9b, and weakly correlated
with SNAI3, E-cadherin and β-catenin (data not shown). In addition,
protein expression levels of the epithelial marker E-cadherin were
increased, while the mesenchymal markers N-cadherin and Vimentin,
and the EMT-associated transcription factor ZEB1 were decreased,
following TAFA5 silencing in GC cells (Fig. 5C). Taken together, these results
suggested an active role for TAFA5 in promoting tumor
aggressiveness via EMT in GC.
Discussion
Due to effective early screening and Helicobacter
pylori eradication, great achievements have been made in
controlling the prevalence and mortality rates of GC (1). However, GC remains a serious health
threat in some parts of the world, such as China, due to early
metastasis and postsurgical relapse (4,5).
Comprehensive therapy is often ineffective in patients at later
stages; thus, a better understanding of the mechanisms underlying
disease progression, and identification of potential therapeutic
targets, would be clinically and scientifically beneficial. In the
present study, TAFA5 was upregulated in GC tissues and cell lines.
High expression of TAFA5 was significantly associated with worse
clinical characteristics and unfavorable prognosis in patients with
GC. The prognostic value of TAFA5 was validated using data from
TCGA and Kaplan-Meier Plotter databases. Knockdown of TAFA5
expression inhibited the proliferation and migration of cultured GC
cells. Taken together, the present results indicated that TAFA5 may
be a novel prognostic marker in patients with GC and that it may
have a crucial role in promoting tumor progression via the
regulation of cell proliferation and migration.
As TAFA family members are mainly expressed in the
central nervous system, the majority of studies have focused on the
regulation of nerve and hormone homeostasis (24–27).
However, TAFA family members encode small secreted proteins that
are evolutionarily close to CCL3 (6), whose involvement in various
malignancies has been extensively investigated. For example, CCL3
promotes oral carcinogenesis via the induction of inflammatory and
angiogenic pathways, and eosinophil recruitment (28); it also mediates the
cyclophosphamide-induced intratumoral migration of CD4+
T cells and tumor regression in hepatic carcinomas (29), suggesting a cell type-dependent
role in carcinogenesis. Based on these results, these TAFA family
members are probably involved in carcinogenesis. In the present
study, unfavorable prognostic effects of TAFA5 in patients with GC
were reported and the results were validated with two larger
patient cohorts. Furthermore, silencing of TAFA5 in GC cells in
vitro suppressed cell proliferation and migration. These
results indicated that TAFA5 may promote progression of GC.
In total, 5 consensus Lef/Tcf binding sites are
located within intron 2 of TAFA5, and at least 2 of these sites
have been physically associated with β-catenin. Thus, TAFA5 is a
downstream target of the Wnt9b/β-catenin signaling pathway in the
maintenance of nephron progenitor cell renewal (8,9).
Furthermore, it has been reported that EMT serves an essential role
in tumor progression and is involved in the proliferation and
migration of multiple types of human tumors (30). In the present study, GSEA using
RNA-seq data from TCGA revealed that TAFA5 expression was
positively correlated with genes involved in EMT, but not the
Wnt/β-catenin signaling pathway. Further correlation analyses using
the same data revealed that TAFA5 expression was moderately
associated with Wnt9a and Wnt9b, as well as several EMT-activating
transcription factors, and only weakly associated with β-catenin.
Based on the complex cross-talk among pathways, these results
suggested that TAFA5-induced GC progression may involve multiple
pathways. Furthermore, Park et al (10) demonstrated that TAFA5 inhibited
osteoclastogenesis via the mediation of its target receptor, formyl
peptide receptor 2 (FPR2). In addition, Wang et al (31) revealed that TAFA5 was able to
inhibit postinjury neointima formation through
sphingosine-1-phosphate receptor 2 (S1PR2). Based on these studies,
it is possible that FPR2 and S1PR2 may be potential target
receptors that link TAFA5 to the proliferation and migration in GC;
this hypothesis warrants further investigation.
There are several limitations in the present study.
Firstly, since most cell lines used in the present study were
poorly differentiated, it was not possible to determine whether
TAFA5 expression is correlated with the aggressiveness of these
cell lines. Secondly, as TAFA5 is widespread in serum and has been
identified as a novel serum biomarker to differentiate
cholangiocarcinoma from benign biliary tract diseases (11), measuring its serum concentration in
GC patients may also have clinical significance. As the cases are
still accumulating, data from sera analyses were not included in
the current study. The above limitations will be addressed in
future studies.
In conclusion, the present study identified TAFA5 as
a novel prognostic marker for GC, which may accelerate tumor
progression by promoting GC cell proliferation and migration via
the modulation of EMT-associated pathways. The present results thus
highlight the importance of future investigation of this putative
marker.
Acknowledgements
The authors greatly appreciated the technical help
from the Department of Pathology of the Fifth People's Hospital of
Shanghai for the IHC staining and data analysis. The authors would
also like to thank Dr Jun Hou at Zhongshan Hospital (Shanghai,
China) for her interpretation of the IHC staining, as well as Dr
Lijie Ma at the Fifth People's Hospital of Shanghai (Shanghai,
China) for his work in packaging the lentiviral particles. In
addition, the authors give special thanks to Dr Yan Zhang at
Carillon Clinic (Roanoke, VA, USA) for his valuable work on data
mining and the processing of TCGA RNA-seq dataset.
Funding
The present study was supported by the Institutional
Grants for Newly Imported Talents of the Fifth People's Hospital of
Shanghai, Fudan University (grant no. 2016WYRC01), the Talent
Program of the Fifth People's Hospital of Shanghai, Fudan
University (grant no. 2017WYRCSG 01), the Great Discipline
Construction Project from the Medical System Shanghai Minhang
District (grant no. 2017MWDXK01), the Research Grant from Shanghai
Minhang District Health and Family Planning Commission (grant no.
2016MW03), and the Research Grant from Shanghai Minhang District
Science and Technology Commission (grant no. 2017MHZ02).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CK designed the study. ZH, GN, JR, XW, LC and RH
performed the experiments. ZH and GN wrote the manuscript. CK and
JR revised the manuscript for important intellectual content. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Ethics Committee at the Fifth People's Hospital of Shanghai, Fudan
University (ethical approval no. 2017-097) and adhered to the
principles in the Declaration of Helsinki. Written informed consent
was obtained from each patient prior to tissue collection for
experimentation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Malvezzi M, Bonifazi M, Bertuccio P, Levi
F, La Vecchia C, Decarli A and Negri E: An age-period-cohort
analysis of gastric cancer mortality from 1950 to 2007 in Europe.
Ann Epidemol. 20:898–905. 2010. View Article : Google Scholar
|
|
4
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Van Cutsem E, Sagaert X, Topal B,
Haustermans K and Prenen H: Gastric cancer. Lancet. 388:2654–2664.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tom Tang Y, Emtage P, Funk WD, Hu T,
Arterburn M, Park EE and Rupp F: TAFA: A novel secreted family with
conserved cysteine residues and restricted expression in the brain.
Genomics. 83:727–734. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Paulsen SJ, Christensen MT, Vrang N and
Larsen LK: The putative neuropeptide TAFA5 is expressed in the
hypothalamic paraventricular nucleus and is regulated by
dehydration. Brain Res. 1199:1–9. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pan X, Karner CM and Carroll TJ: Myc
cooperates with beta-catenin to drive gene expression in nephron
progenitor cells. Development. 144:4173–4182. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Karner CM, Das A, Ma Z, Self M, Chen C,
Lum L, Oliver G and Carroll TJ: Canonical Wnt9b signaling balances
progenitor cell expansion and differentiation during kidney
development. Development. 138:1247–1257. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Park MY, Kim HS, Lee M, Park B, Lee HY,
Cho EB, Seong JY and Bae YS: FAM19A5, a brain-specific chemokine,
inhibits RANKL-induced osteoclast formation through formyl peptide
receptor 2. Sci Rep. 7:155752017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Janvilisri T, Leelawat K, Roytrakul S,
Paemanee A and Tohtong R: Novel serum biomarkers to differentiate
cholangiocarcinoma from benign biliary tract diseases using a
proteomic approach. Dis Markers. 2015:1053582015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Diaz de Stahl T, Hartmann C, de Bustos C,
Piotrowski A, Benetkiewicz M, Mantripragada KK, Tykwinski T, von
Deimling A and Dumanski JP: Chromosome 22 tiling-path array-CGH
analysis identifies germ-line- and tumor-specific aberrations in
patients with glioblastoma multiforme. Genes Chromosomes Cancer.
44:161–169. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu C, Miao X, Huang L, Che X, Jiang G, Yu
D, Yang X, Cao G, Hu Z, Zhou Y, et al: Genome-wide association
study identifies five loci associated with susceptibility to
pancreatic cancer in Chinese populations. Nat Genet. 44:62–66.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Howitt BE, Sun HH, Roemer MG, Kelley A,
Chapuy B, Aviki E, Pak C, Connelly C, Gjini E, Shi Y, et al:
Genetic basis for PD-L1 expression in squamous cell carcinomas of
the cervix and vulva. JAMA Oncol. 2:518–522. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shi JY, Ma LJ, Zhang JW, Duan M, Ding ZB,
Yang LX, Cao Y, Zhou J, Fan J, Zhang X, et al: FOXP3 is a HCC
suppressor gene and Acts through regulating the TGF-beta/Smad2/3
signaling pathway. BMC Cancer. 17:6482017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao S, Zhou L, Niu G, Li Y, Zhao D and
Zeng H: Differential regulation of orphan nuclear receptor TR3
transcript variants by novel vascular growth factor signaling
pathways. FASEB J. 28:4524–4533. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hu HM, Chen Y, Liu L, Zhang CG, Wang W,
Gong K, Huang Z, Guo MX, Li WX and Li W: C1orf61 acts as a tumor
activator in human hepatocellular carcinoma and is associated with
tumorigenesis and metastasis. Faseb J. 27:163–173. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Niu G, Ye T, Qin L, Bourbon PM, Chang C,
Zhao S, Li Y, Zhou L, Cui P, Rabinovitz I, et al: Orphan nuclear
receptor TR3/Nur77 improves wound healing by upregulating the
expression of integrin β4. Faseb J. 29:131–140. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
D'Errico M, de Rinaldis E, Blasi MF, Viti
V, Falchetti M, Calcagnile A, Sera F, Saieva C, Ottini L, Palli D,
et al: Genome-wide expression profile of sporadic gastric cancers
with microsatellite instability. Eur J Cancer. 45:461–469. 2009.
View Article : Google Scholar
|
|
21
|
Cho JY, Lim JY, Cheong JH, Park YY, Yoon
SL, Kim SM, Kim SB, Kim H, Hong SW, Park YN, et al: Gene expression
signature-based prognostic risk score in gastric cancer. Clin
Cancer Res. 17:1850–1857. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mootha VK, Lindgren CM, Eriksson KF,
Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E,
Ridderstråle M, Laurila E, et al: PGC-1alpha-responsive genes
involved in oxidative phosphorylation are coordinately
downregulated in human diabetes. Nat Genet. 34:2672003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zheng C, Chen D, Zhang Y, Bai Y, Huang S,
Zheng D, Liang W, She S, Peng X, Wang P, et al: FAM19A1 is a new
ligand for GPR1 that modulates neural stem-cell proliferation and
Differentiation. Faseb J fj201800020RRR. 2018. View Article : Google Scholar
|
|
25
|
Wang X, Shen C, Chen X, Wang J, Cui X,
Wang Y, Zhang H, Tang L, Lu S, Fei J and Wang Z: Tafa-2 plays an
essential role in neuronal survival and neurobiological function in
mice. Acta Biochim Biophys Sin (Shanghai). 50:984–995. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kambrun C, Roca-Lapirot O, Salio C, Landry
M, Moqrich A and Le Feuvre Y: TAFA4 Reverses Mechanical Allodynia
through Activation of GABAergic Transmission and Microglial Process
Retraction. Cell Rep. 22:2886–2897. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shao Y, Deng T, Zhang T, Li P and Wang Y:
FAM19A3, a novel secreted protein, modulates the
microglia/macrophage polarization dynamics and ameliorates cerebral
ischemia. FEBS Lett. 589:467–475. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
da Silva JM, Moreira Dos Santos TP, Sobral
LM, Queiroz-Junior CM, Rachid MA, Proudfoot AEI, Garlet GP, Batista
AC, Teixeira MM, Leopoldino AM, et al: Relevance of CCL3/CCR5 axis
in oral carcinogenesis. Oncotarget. 8:51024–51036. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Naito T, Baba T, Takeda K, Sasaki S,
Nakamoto Y and Mukaida N: High-dose cyclophosphamide induces
specific tumor immunity with concomitant recruitment of
LAMP1/CD107a-expressing CD4-positive T cells into tumor sites.
Cancer Lett. 366:93–99. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
De Craene B and Berx G: Regulatory
networks defining EMT during cancer initiation and progression. Nat
Rev Cancer. 13:97–110. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang Y, Chen D, Zhang Y, Wang P, Zheng C,
Zhang S, Yu B, Zhang L, Zhao G, Ma B, et al: Novel Adipokine,
FAM19A5, inhibits neointima formation after injury through
sphingosine-1-phosphate receptor 2. Circulation. 138:48–63. 2018.
View Article : Google Scholar : PubMed/NCBI
|