Introduction
microRNAs (miRNAs) are important effector molecules
of RNA interference. They regulate gene expression at the
post-transcriptional level and thereby control several key
processes, including development, differentiation, proliferation,
apoptosis and organogenesis. Previous studies have demonstrated
that miRNAs are pivotal in various types of human cancer (1), for example, the miR-17–92 cluster
(2), miR-21 (3–4),
miR-221 (5), miR-372/373 (6) and miR-375 (7) have been demonstrated to be oncogenic
or anti-oncogenic. miR-21 was clearly upregulated in various types
of cancer, including breast (8)
and colorectal cancer (9). miRNAs
are generally downregulated in numerous types of cancer, although,
certain miRNAs are aberrantly overexpressed (10–11).
Let-7 is a member of a small family of miRNAs, including miR-84 and
miR-48, in C. elegans and let-7a to let-7h in humans (12), which are downregulated in lung
cancer. In addition, miR-26a is downregulated in hepatocellular
carcinoma, a finding associated with overall survival and response
to interferon therapy for these patients (13). Furthermore, it has been reported
that miRNAs regulate a diverse range of important regulators in
cancer, including cell cycle components, signal-transduction
factors and transcription factors.
The expression of miRNAs is also affected by
cellular stress, for example radiation and chemotherapy drugs,
which leads to the manifestation of numerous biological effects,
including radiation-induced DNA damage, signal transduction, gene
transcription and enzyme recruitment activation (activated in the
cell) (14–15). Previous studies have suggested that
DNA damage in response to radiation is mediated via miRNAs that
control complex regulatory pathways involved in p53 and is followed
by the induction of cell cycle arrest and/or the promotion of
apoptosis, further demonstrating that miRNA alterations in several
types of tumor possibly affect DNA damage response directly and
thus impact the effects of several types of tumor to cancer
therapy.
The present study investigated the expression levels
of several miRNAs by using the stem-loop real-time polymerase chain
reaction (PCR) in HepG2 cells treated with ultraviolet (UV)
irradiation (16–17). The examination of molecular
processes responsible for modulating the signaling pathways by
downregulating various genes which are involved in response to DNA
damage, may lead to an improved understanding of the effects of
radiation. The assessment of miRNA expression patterns that control
diverse cellular functions may provide a rationale for their roles
in promoting these processes following radiation exposure.
Materials and methods
Cell culture and radiation treatment
The human liver cancer cell lines (HepG2) from the
Cell Bank of the Shanghai Institute of Biochemistry and Cell
Biology, Chinese Academy of Sciences (Shanghai, China) were
cultured in DMEM (HyClone, Thermo Scientific, Waltham, MA, USA)
supplemented with 10% fetal bovine serum (Hyclone, Thermo
Scientific) and 1% penicillin-streptomycin (Gibco, Carlsbad, CA,
USA) in a 60 mm culture dish with 5% CO2 at 37°C. UV
irradiation was performed using modified methods previously
described (18). Briefly,
following growing to confluence, the culture medium was decanted
out and the cells were washed twice with PBS buffer and cultured in
PBS. Then, cells were treated immediately as follows in parallel:
i) cells were treated with different UV-irradiation dosages (0, 20,
50, 70, 100, 120 and 150 J/m2) and then cultured in
complete culture medium for 4 h. ii) Cells were incubated for 0, 2,
4, 7, 12, 24 and 36 h after exposure to 50 J/m2 of
UV-irradiation.
Cell viability assay
An MTT assay was used to evaluate cell viability.
Following the process above, MTT (Sigma-Aldrich, Munich, Germany)
was added to the complete DMEM culture medium to the final
concentration of 0.5 mg/ml and cultured for 4 h in the
CO2 incubator at 37°C. Then, the culture medium
containing MTT was decanted out and DMSO (AMRESCO, Solon, OH, USA)
was added to dissolve the formazan. Finally, the solution was read
at 570 nm using a microplate reader (Synergy HT; BioTek, Winooski,
VT, USA). All experiments were repeated six times.
RNA isolation and miRNA expression
assay
Total RNA was extracted using TRIzol (Invitrogen,
Carlsbad, CA, USA) reagent from HepG2 cells irradiated with UV (0,
2, 4 and 12 h post-irradiation) and the control group. In total,
seven miRNA genes were selected from the Sanger Center miRNA
Registry (http://www.mirbase.org) according to
studies reported previously, the expression level of these miRNAs
was normalized with endogenous control U6 snRNA. p53 and
phosphatase and tensin homolog (PTEN), genes involved in
irradiation damage response, were selected to examine the
association between miRNA and their expression, and the
sequence-specific primers for these genes are listed in Table I. Real-time quantitative polymerase
chain reaction (qRT-PCR) analysis was performed on an ABI 7500
real-time PCR system (Applied Biosystems, Foster City, CA, USA).
After cDNA was synthesized and amplified the product levels were
detected by real-time monitoring of Evagreen dye fluorescence. The
reaction conditions were as follows: 42°C for 60 min, 85°C for 5
min for reverse transcription, 95°C for 10 min, followed by 40
cycles of 95°C for 15 sec and 60°C for 1 min for the amplification.
The threshold cycle (Ct) was defined as the fractional cycle number
at which the fluorescence passes the fixed threshold. The gene
expression ΔCt values of miRNAs from each sample were calculated by
normalizing with the internal control U6 snRNA. The relative
expression of miRNA targets was calculated by the comparative
2−ΔΔCt method (19).
All experiments were repeated in triplicate.
 | Table IPrimer sequences used for miRNA
expression analysis with gene names. |
Table I
Primer sequences used for miRNA
expression analysis with gene names.
| Gene | Primer name | Primer sequence
(5′-3′) |
|---|
| miR-21 | RT primer |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCAACATC |
| Forward primer |
ACACTCCAGCTGGGTAGCTTATCAGACTGA |
| miR-26a | RT primer |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAGCCTATC |
| Forward primer |
ACACTCCAGCTGGGTTCAAGTAATCCAGGA |
| miR-34a | RT primer |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACAACCAG |
| Forward primer |
ACACTCCAGCTGGGTTGGCAGTGTCTTAGC |
| miR-146a | RT primer |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAACCCATG |
| Forward primer |
ACACTCCAGCTGGGTGAGAACTGAATTCCA |
| miR-181b | RT primer |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACCCACCG |
| Forward primer |
ACACTCCAGCTGGGAACATTCATTGCTGTCG |
| miR-221 | RT primer |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGAAACCCA |
| Forward primer |
ACACTCCAGCTGGGAGCTACATTGTCTGCT |
| miR-224 | RT primer |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAACGGAAC |
| Forward primer |
ACACTCCAGCTGGGCAAGTCACTAGTGGT |
| U6 | Forward primer |
CGCTTCGGCAGCACATATAC |
| Reverse primer |
TTCACGAATTTGCGTGTCAT |
| qRT-PCR | Universal | TGGTGTCGTGGAGTC |
Functional analysis of miRNA
Since a single miRNA is able to regulate hundreds of
target genes, three web-based programs [TargetScan (http://www.targetscan.org), PicTar (http://pictar.bio.nyu.edu) and DIANA-micro v3.0
(http://diana.cslab.ece.ntua.gr/microT] were employed
for the prediction of miRNA targets. Only the targets shared by all
three softwares were considered as candidate targets. Furthermore,
validated targets of the miRNA by experiments were searched in
TarBase (http://diana.cslab.ece.ntua.gr/tarbase). In addition,
a pathway analysis of the targets was performed using DIANA
Bioinformatics Resources (http://diana.cslab.ece.ntua.gr/pathways) (20).
Results
Cell viability assay
Exponentially growing HepG2 cells were irradiated
with UV which decreased cell viability in a dose-dependent manner.
The survival ratio of HepG2 cells decreased gradually with the
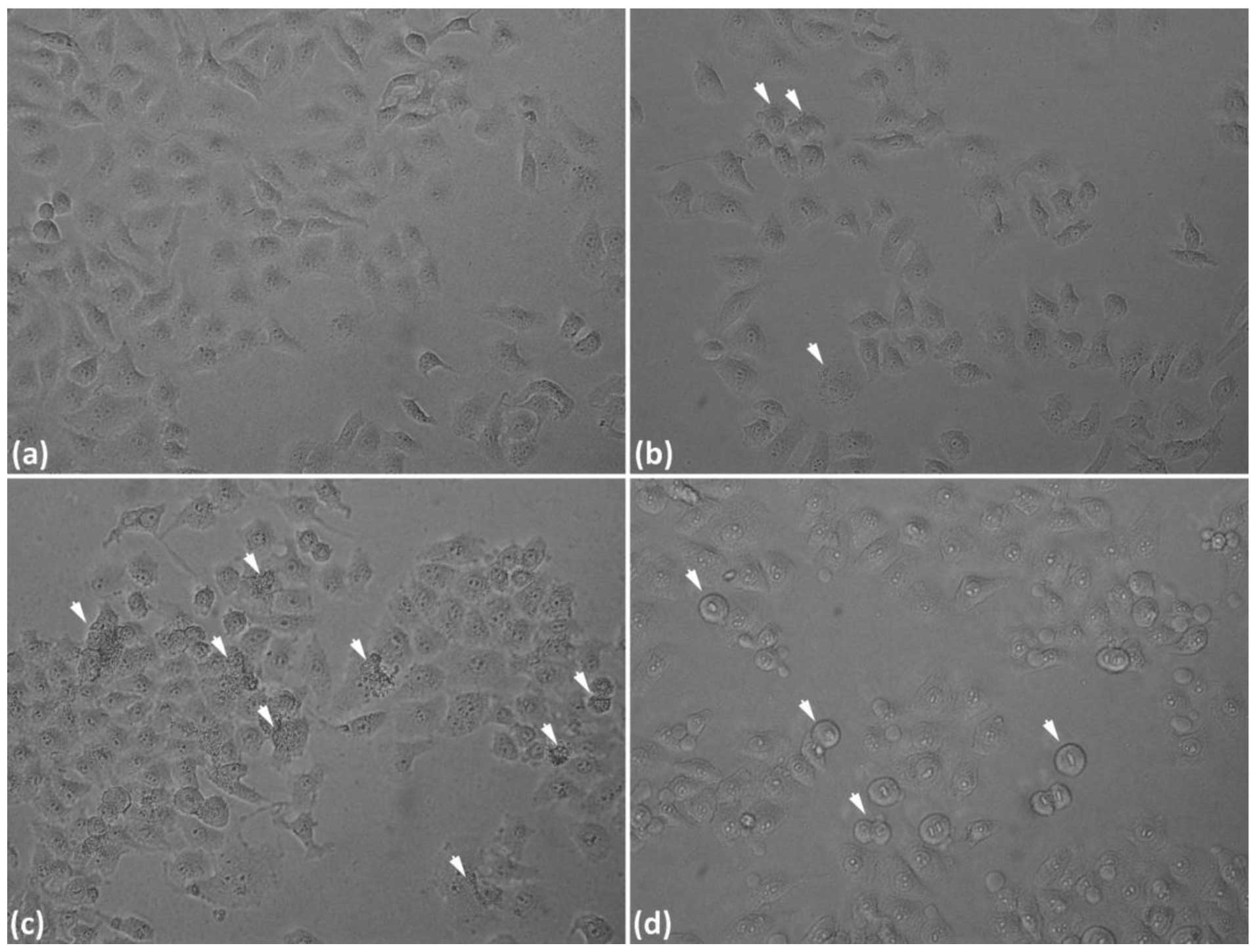
increased UV dosages. Fig. 1
demonstrated that UV exposure, with increasing dosage, resulted in
decreased cell viability and increased apoptosis. When the dosage
was increased to 100 J/m2, the mortality markedly
increased and, until 150 J/m2, the mortality continued
to increase, however, the increase was slower than that at a dosage
of 100 J/m2 (Fig. 1A).
Combining the above results, together with our morphological data
(Fig. 2), UV irradiation of 50
J/m2 was selected for the next series of experiments.
When cells were exposed to UV (50 J/m2) and then
incubated with complete culture medium for 2, 4, 7, 12, 24 and 36
h, the results demonstrated a larger change in cell viability at 2,
4 and 7 h (Fig. 1B). Fig. 2 demonstrated that UV-induced DNA
damage, thereby led to cell apoptosis with increasing UV
irradiation dosage.
Assessment of gene expression by
real-time qRT-PCR
A real-time qRT-PCR assay was used to study the
expression of seven miRNA genes, p53 and PTEN responses to DNA
damage. These miRNAs were selected based on two reasons. One was
that these miRNAs are associated with that in human liver; the
other is that their function in the progress of cell cycle and
proliferation is relatively clear (4,5,21,22).
Furthermore, p53 and PTEN are involved in the response to radiation
damage. Fig. 3A demonstrated an
upregulated expression of miR-26a, miR-34a and miR-146a in HepG2
cells treated with UV irradiation at various dosages. Among which
the expression of miR-26a and miR-146a were induced and remained
upregulated until a dosage of 100 J/m2 was examined in
the experiments (Fig. 3A), and
were upregulated 2.9-fold and 5.3-fold compared with the untreated
cells, respectively. However, the expression of miR-21 was
downregulated regardless of different radiation dosages or
different incubation times following 50 J/m2 of
UV-irradiation.
 | Figure 3UV radiation-induced gene expression
in HepG2 cells. The expression levels of various target genes and
endogenous U6 snRNA/GAPDH were determined by real-time PCR and
quantified with the comparative 2−ΔΔCt method. (A)
Relative expression of miR-21, miR-26a, miR-34a, miR-146a,
miR-181b, miR-221 and miR-224 at 50, 70 and 100 J/m2 of
UV irradiation, compared with the unirradiated sham control. (B)
Relative expression of miR-21, miR-26a, miR-34a, miR-146a,
miR-181b, miR-221 and miR-224 at 2, 4 and 12 h after 50
J/m2 of UV irradiation, compared with the unirradiated
sham control. (C and D) p53 and PTEN relative expression in HepG2
cells treated with different UV radiation dosages, respectively.
miR, microRNA; UV, ultraviolet; PTEN, phosphatase and tensin
homolog. |
The expression of miR-26a and miR-146a (Fig. 3B) was significantly induced 4 h
after the radiation exposure and then reduced to 4.9-fold and
7.3-fold of baseline levels 12 h post-irradiation. miR-34a was
upregulated at 2 h and its expression gradually decreased at 4 h,
followed by a mild increase at 12 h. The expression of miR-181b and
miR-221 was only moderately enhanced or decreased following
irradiation. The expression of miR-224 was downregulated at 2 h and
peaked at 8 h in HepG2 cells, then was followed by a gradual
downregulation 12 h post-irradiation (Fig. 3B). Furthermore, the level of p53
demonstrated an increase in a dose-dependent manner (Fig. 3C), while, PTEN expression was
slightly upregulated in 70 J/m2 followed by
downregulation in 50 J/m2, then, again downregulated in
100 J/m2 (Fig. 3D).
Bioinformatics analysis of target genes
of differentially expressed miRNAs
Studies suggested that PicTar and Targetscan had an
excellent recovery rate in target genes prediction for miRNA
compared with numerous other softwares previously developed
(20). In the present study,
potential target genes of the differentially expressed miRNAs were
predicted by miRGen. The coincidence results of the PicTar database
and TargetScanS database were selected as candidate targets, and
518 potential target genes were obtained. KEGG analysis
demonstrated that the predicted target genes were involved in
numerous signaling pathways involved in tumorigenesis and tumor
metastasis, including the Wnt signaling pathway, cell cycle
progression, apoptosis and the p53 signaling pathway, and were
selected and listed in Table
II.
 | Table IIPathway analysis of target genes of
UVB-responsive miRNAs on DIANA bioinformatics resources. |
Table II
Pathway analysis of target genes of
UVB-responsive miRNAs on DIANA bioinformatics resources.
| miRNA | KEGG pathway | Target genes | Count | P-value |
|---|
| miR-21 | Amyotrophic lateral
sclerosis | BCL2,
PPP3CA | 2 | 8.47E-05 |
| Huntington’s
disease | HIP2,
RASA1 | 2 | 2.42E-03 |
| Apoptosis | BCL2, FASLG,
PPP3CA PTEN | 4 | 5.95E-03 |
| Jak-STAT signaling
pathway | STAT3, CNTFR,
SPRY1, SPRY2 | 4 | 7.96E-03 |
| TGF-β signaling
pathway | SMAD7, ACVR2A,
PITX2 | 3 | 9.92E-03 |
| miR-26a | Adherens
junction | LEF1, YES1,
CREBBP, SSX2IP
NLK, ACVR1C, EP300, SMAD4 | 8 | 3.92E-05 |
| Cell cycle | CCNE2, CCNE1,
GSK3B, CDK6
YWHAE, CREBBP, ATM, CCND2
EP300, SMAD4 | 10 | 1.04E-04 |
| Wnt signaling
pathway | CTNNBIP1, GSK3B,
LEF1
CREBBP, NLK, SOX17, PLCB1
CCND2, EP300, SMAD4, PPP3CB | 11 | 4.22E-04 |
| p53 signaling
pathway | CCNE2, CCNE1,
CDK6
PTEN, PTENP1, ATM, CCND2 | 6 | 5.18E-04 |
| miR-34a | Non-small cell lung
cancer | RARB, E2F3,
MAP2K1, CDK6
PLCG1, CCND1 | 6 | 4.88E-04 |
| Notch signaling
pathway | APH1A, NOTCH2,
NUMBL
DLL1, JAG1 | 5 | 6.27E-04 |
| Glioma | E2F3, PDGFRA,
MAP2K1, CDK6
PLCG1, CCND1 | 6 | 2.33E-03 |
| Galactose
metabolism | HK1, RDH11,
B4GALT2, PGM1 | 4 | 2.52E-03 |
| p53 signaling
pathway | CCNE2, IGFBP3,
CDK6, EI24, p53 | 6 | 4.07E-03 |
| miR-146a | Small cell lung
cancer | RARB, TRAF6,
MAX | 3 | 7.49E-03 |
| Toll-like receptor
signaling pathway | TRAF6, MAP3K8,
IRAK1 | 3 | 1.91E-02 |
| Inositol phosphate
metabolism | PIP5K1B,
PIP4K2B | 2 | 2.98E-02 |
| Axon guidance | EFNB2, SEMA3G,
NFAT5 | 3 | 4.82E-02 |
Discussion
A wide variety of biological effects are induced in
cells by exposure to irradiation, including DNA damage, signal
transduction, mutations, altered gene expression, cell cycle arrest
and others (23,24). Cellular mechanisms exist to repair
the DNA damage or to induce apoptosis to remove severely damaged
cells. However, the majority of studies have focused on examining
protein-coding genes. Previously, studies have suggested that
miRNAs are involved in the regulation of proliferation,
differentiation, apoptosis and cell cycle progression (25,26).
It appears that miRNAs may contribute to understanding the cellular
mechanisms of response to radiation. Our present results
demonstrated that UV radiation significantly altered miRNA
expression in HepG2 cells. As participants in cell processes,
miRNAs have the potential to regulate various target genes involved
in the cell cycle and apoptosis. Therefore, it is intriguing to
hypothesize that these miRNAs may represent a type of non-coding
gene involved in apoptosis induced by UV irradiation. The theory
was supported by the study on the regulation of PIK3R1 and BCL-2 by
miR-21. The available data demonstrated that the overexpression of
miR-21 results in the downregulation of PTEN and more active
survival signaling through the PI3K signaling pathway rendering the
cells less susceptible to apoptosis (27). miR-21, reported in the present
study is downregulated during the process of HepG2 cells apoptosis
induced by various irradiation dosages, which support these
previous studies.
The diversity and abundance of miRNA targets present
multi-level regulatory network interaction with other cellular
networks. Thus, it is necessary to understand what functions miRNAs
have in cellular processes at a system level. For this reason,
bioinformatics methods were used to predict the target genes of the
differentially expressed miRNAs and then analyze the possible
mechanisms of its role in response to UV irradiation. The most
commonly used target gene predicting database, PicTar and
Targetscan, were selected from the website of miRGen. To narrow
down the scope of target gene and improve the specificity of the
prediction, the shared results were selected by the two databases
and subjected to functional analysis. The analysis of KEGG pathways
demonstrated that, the target genes obtained from our prediction
participated in numerous pathways involved in tumorigenesis, DNA
damage and the cell cycle. For example, the p53 signaling pathway
was important in DNA damage and liver tumorigenesis.
Among the selected miRNAs, a significant increased
expression in the tumor suppressors miR-26a and miR-34a was
observed in HepG2 cells treated with different UV-irradiation
doses, which is downregulated in various types of liver cancer
(21,28). The expression changes of miR-34a
and miR-26a may be involved in important protective mechanisms
counteracting UV-radiation damage. Additionally, these cellular
changes may constitute an attempt to counteract radiation-induced
DNA damage. Bioinformatics analysis suggests that this is partly
through controlling the p53 signaling pathway and genes involved in
the DNA damage response pathway (29) (Fig.
4). The expression of p53 has been increased in HepG2 cells
treated with 50 J/m2, 70 J/m2 and 100
J/m2 of UV-irradiation. Furthermore, PTEN, a target of
miR-26a, was upregulated following firstly being downregulated in
50 J/m2, then, again downregulated in 100
J/m2. It remains quite possible that miR-26a mediates
pro-apoptotic effects by regulating a number of targets not limited
to PTEN, a potential interaction with the p53 transcript is
particularly intriguing, although as of yet unverified.
Furthermore, miR-26a may possibly contribute to pro-apoptotic
pathways as a downstream regulator, including in p53-induced
apoptosis (30). Therefore, the
present study hypothesized that miR-26a has a similar function in
the p53 signaling pathway as miR-34a. Collectively, these findings
may be direct evidence that miRNAs are able to suppress resistance
to anticancer cytotoxic therapy, a common feature of cancer cells.
With respect to carcinogenesis and cancer therapy, radiation
effects may be conveyed, modified or associated with differential
regulations of miRNAs. Therefore, the modulation of miRNAs may have
implications for anticancer treatments (31).
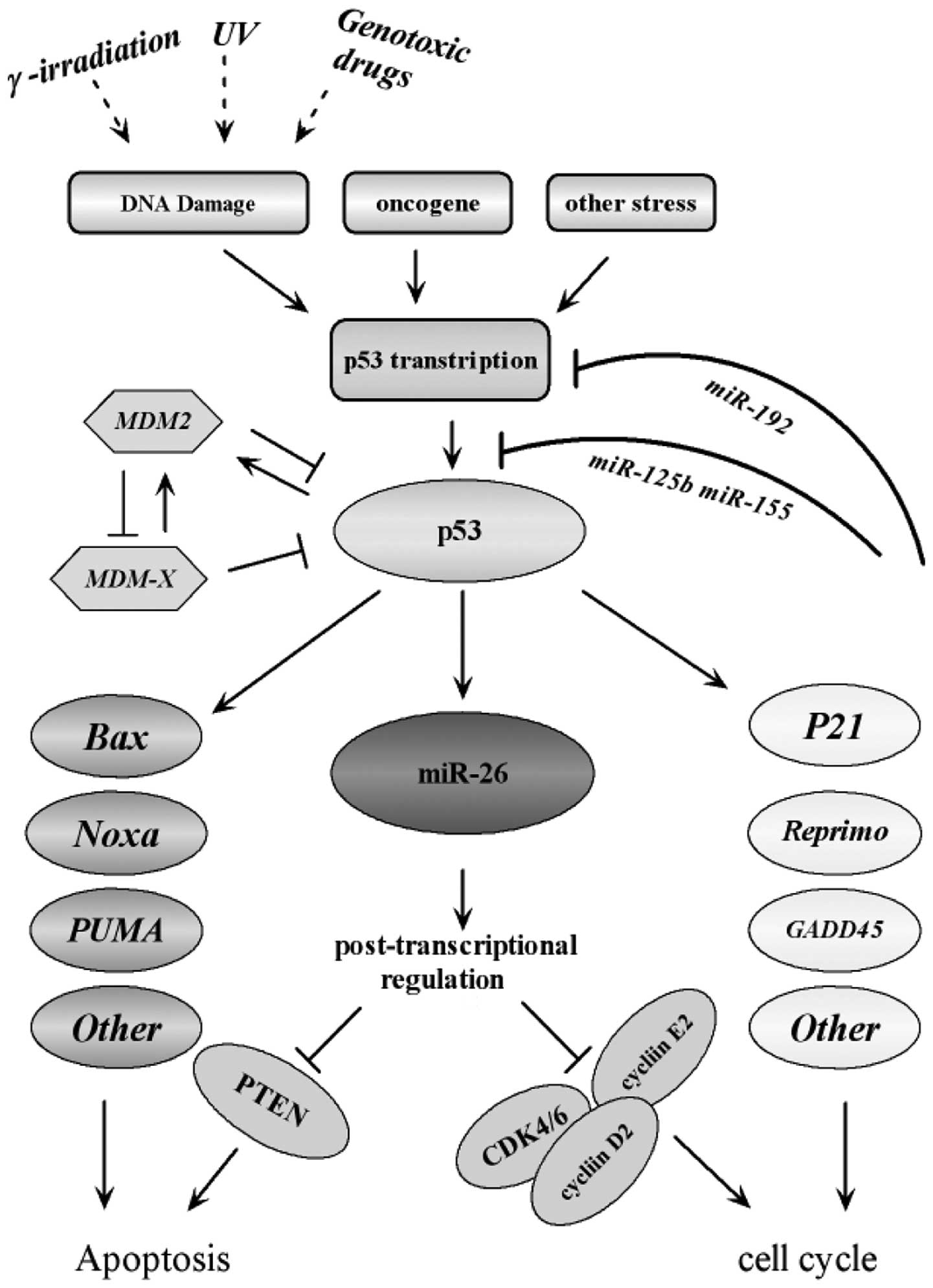 | Figure 4Interaction of miR-26a with the p53
signaling pathway. The diagram illustrates a simple scheme
highlighting the modes of interaction and regulatory loops that
exist between miR-26a and the p53 signaling pathway. It illustrates
that miR-26a is a direct transcriptional target of p53, which in
turn downregulates genes required for proliferation and survival.
Along with other p53 targets, including p21 and BAX, miR-26a
promotes cell cycle arrest and apoptosis in response to cancer
related stress. The figure was adapted according to the KEGG
database (http://www.genome.jp/kegg-bin/show_pathway). ATM,
ataxia telangiectasia mutated; ATR, ataxia telangiectasia and
RAD3-related; CDK, cyclin-dependent kinase; CHK, checkpoint kinase;
ROS, reactive oxygen species; miR, microRNA; PTEN, phosphatase and
tensin homolog; UV, ultraviolet; MDM2, mouse double minute 2
homolog; PUMA, p53 upregulated modulator of apoptosis; GADD45,
growth arrest and DNA damage-inducible 45. |
Currently, more than half of cancer patients receive
radiation treatment. Although radiation treatment is very
effective, it requires the development of radiation-sensitive
agents to achieve tumor specific treatment. It is now apparent that
numerous types of tumor are addicted to a loss of wild-type p53
function, providing a rationale for therapeutic reactivation of
p53. Therefore, using specific miRNAs that may function downstream
of p53 in several types of cancer possessing deleted or mutated p53
may represent a novel approach for treatment, by resensitizing
these tumors to DNA damaging chemotherapy drugs.
The focus of the present study was to investigate
the differential expression of selected miRNAs in HepG2 cells in
response to DNA damage induced by UV irradiation. Our results
provide further evidence linking the altered expression of miRNAs
with cellular stress. In the present study, several miRNAs, which
may be involved in radiation sensitivity, were identified. One of
particular interest is miR-26a, which is involved in the p53
signaling pathway and demonstrated to specifically sensitize liver
cancer cells against radiation treatment. Therefore, miR-26a may be
a potential target to enhance the efficacy of current cancer
therapies, particularly for radiotherapy alone or in combination
with drugs.
Acknowledgements
This study was financially supported by the National
Basic Research Program of China (973 Program: 2013CB932902) and the
NSFC (no. 61071047,81071230).
References
|
1
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar
|
|
2
|
O’Donnell KA, Wentzel EA, Zeller KI, Dang
CV and Mendell JT: c-Myc-regulated microRNAs modulate E2F1
expression. Nature. 435:839–843. 2005.PubMed/NCBI
|
|
3
|
Si ML, Zhu S, Wu H, Lu Z, Wu F and Mo YY:
miR-21-mediated tumor growth. Oncogene. 26:2799–2803. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Qi LQ, Bart J, Tan LP, et al: Expression
of miR-21 and its targets (PTEN, PDCD4, TM1) in flat epithelial
atypia of the breast in relation to ductal carcinoma in situ and
invasive carcinoma. BMC Cancer. 9:1632009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pineau P, Volinia S, McJunkin K, et al:
miR-221 overexpression contributes to liver tumorigenesis. Proc
Natl Acad Sci USA. 107:264–269. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Voorhoeve PM, Le Sage C, Schrier M, et al:
A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in
testicular germ cell tumors. Cell. 124:1169–1181. 2006. View Article : Google Scholar
|
|
7
|
Liu AM, Poon RT and Luk JM: MicroRNA-375
targets Hippo-signaling effector YAP in liver cancer and inhibits
tumor properties. Biochem Biophys Res Commun. 394:623–627. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang Y, Chaerkady R, Beer MA, Mendell JT
and Pandey A: Identification of miR-21 targets in breast cancer
cells using a quantitative proteomic approach. Proteomics.
9:1374–1384. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yamamichi N, Shimomura R, Inada KI, et al:
Locked nucleic acid in situ hybridization analysis of miR-21
expression during colorectal cancer development. Clin Cancer Res.
15:4009–4016. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lu J, Getz G, Miska EA, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gaur A, Jewell DA, Liang Y, et al:
Characterization of microRNA expression levels and their biological
correlates in human cancer cell lines. Cancer Res. 67:2456–2468.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lau NC, Lim LP, Weinstein EG and Bartel
DP: An abundant class of tiny RNAs with probable regulatory roles
in Caenorhabditis elegans. Science. 294:858–862. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ji JF, Shi J, Budhu A, et al: MicroRNA
expression, survival, and response to interferon in liver cancer. N
Engl J Med. 361:1437–1447. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Szumiel I: Intrinsic radiation
sensitivity: cellular signaling is the key. Radiat Res.
169:249–258. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
An IS, An S, Kang SM, et al: Titrated
extract of Centella asiatica provides a UVB protective effect by
altering microRNA expression profiles in human dermal fibroblasts.
Intl J Mol Med. 30:1194–1202. 2012.
|
|
16
|
Chen CF, Ridzon DA, Broomer AJ, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chaudhry MA: Real-time PCR analysis of
micro-RNA expression in ionizing radiation-treated cells. Cancer
Biother Radiopharm. 24:49–55. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guo L, Huang ZX, Chen XW, et al:
Differential expression profiles of microRNAs in NIH3T3 cells in
response to UVB irradiation. Photochem Photobiol. 85:765–773. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(T)(−Delta Delta C) method. Methods. 25:402–408.
2001.PubMed/NCBI
|
|
20
|
Alexiou P, Maragkakis M, Papadopoulos GL,
Simmosis VA, Zhang L and Hatzigeorgiou AG: The DIANA-mirExTra web
server: from gene expression data to microRNA function. PLoS One.
5:e91712010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kota J, Chivukula RR, O’Donnell KA, et al:
Therapeutic microRNA delivery suppresses tumorigenesis in a murine
liver cancer model. Cell. 137:1005–1017. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Murakami Y, Yasuda T, Saigo K, et al:
Comprehensive analysis of microRNA expression patterns in
hepatocellular carcinoma and non-tumorous tissues. Oncogene.
25:2537–2545. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Löbrich M and Jeggo PA: The impact of a
negligent G2/M checkpoint on genomic instability and cancer
induction. Nat Rev Cancer. 7:861–869. 2007.PubMed/NCBI
|
|
24
|
Spitz DR, Azzam EI, Li JJ and Gius D:
Metabolic oxidation/reduction reactions and cellular responses to
ionizing radiation: a unifying concept in stress response biology.
Cancer Metastasis Rev. 23:311–322. 2004. View Article : Google Scholar
|
|
25
|
Luo H, Zou J, Dong Z, Zeng Q, Wu D and Liu
L: Up-regulated miR-17 promotes cell proliferation, tumour growth
and cell cycle progression by targeting the RND3 tumour suppressor
gene in colorectal carcinoma. Biochem J. 442:311–321. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Creevey L, Ryan J, Harvey H, Bray IM,
Meehan M, Khan AR and Stallings RL: MicroRNA-497 increases
apoptosis in MYCN amplified neuroblastoma cells by targeting the
key cell cycle regulator WEE1. Mol Cancer. 12:232013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Meng F, Henson R, Lang M, et al:
Involvement of human micro-RNA in growth and response to
chemotherapy in human cholangiocarcinoma cell lines.
Gastroenterology. 130:2113–2129. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Varnholt H: The role of microRNAs in
primary liver cancer. Ann Hepat. 7:104–113. 2008.
|
|
29
|
He L, He XY, Lowe SW and Hannon GJ:
microRNAs join the p53 network--another piece in the
tumour-suppression puzzle. Nat Rev Cancer. 7:819–822. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Suzuki HI, Yamagata K, Sugimoto K, Iwamoto
T, Kato S and Miyazono K: Modulation of microRNA processing by p53.
Nature. 460:529–533. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rossi JJ: New hope for a microRNA therapy
for liver cancer. Cell. 137:990–992. 2009. View Article : Google Scholar : PubMed/NCBI
|