Introduction
Gestational diabetes mellitus (GDM), characterized
by glucose intolerance diagnosed during pregnancy, is one of the
most common complications in pregnancy and affects 3–8% of all
pregnancies (1,2). The prevalence of GDM has increased in
recent decades due to increased average age of pregnant females and
increased risk of obesity (3).
However, GDM is associated with numerous complications including
macrosomia, neonatal metabolic disorders, respiratory distress
syndrome and neonatal death as well as a predisposition for the
development of metabolic syndromes and type 2 diabetes (4,5).
The mammalian placenta serves as an auxiliary fetal
organ at the interface between the mother and the fetus. It is a
vitally important endocrine organ during pregnancy. In addition to
the production of a wide variety of steroids, peptides and further
regulatory factors, the placenta is an endocrine target tissue,
expressing a broad spectrum of hormone receptors and growth factor
receptors (6). Growth and
differentiation of the placenta are fundamental to mammalian
reproduction, and functional impairment of this organ occasionally
leads to severely abnormal pregnancies (7). The aberrant development and function
of the placenta have been suggested as important contributory
factors to GDM-associated complications (8) and, as the condition is resolved
following delivery, it is likely that the placenta has a critical
role in the pathogenesis of GDM (10).
Studies have revealed that microRNA (miRNA)
expression is tissue-specific and it has been reported that some
miRNAs are specific to, and highly expressed in, the placenta
(placental-specific miRNAs) (8,9). In
a recent study, it was shown that certain abnormal pregnancies are
associated with alterations in miRNA expression in the placenta
(9). Two large miRNA clusters
expressed in placenta have been described: C19 MC (located at
chromosome 19q13.41), which comprises 54 predicted miRNAs, and C14
MC (located at chromosome 14q32), which contains 46 miRNAs
(8). It has been reported that
these placenta-specific miRNAs may contribute to the pathology of
abnormal pregnancies, including pre-eclampsia and intrauterine
growth restriction (9). However,
few reports have described the involvement of miRNAs in the
regulation of the development of GDM.
miR-518d is one of the miRNAs from the C19 MC
cluster and is highly expressed in the placenta. In a previous
study, our group showed that the miR-518d microRNA was
differentially expressed in placentas from patients with GDM.
Bioinformatic analysis has predicted the peroxisome
proliferator-activated receptor-α (PPARα) to be a target for
miR-518d. The peroxisome proliferator-activated receptors (PPARs)
are a family of fatty acid receptors that transduce stimuli from
fatty acids into alterations in gene expression. PPARs have a
critical role in lipid homeostasis and inflammation, and have long
been linked to the diabetic phenotype (11). During pregnancy, dynamic
physiological, metabolical and immunological adaptations are
required to ensure fetal development and maternal well-being. A
large number of the metabolic adaptations are mediated by PPARs
(12). Dysregulation of PPARs may
result in GDM, as it has been reported that the expression levels
of the PPARα protein and mRNA are lower in placentas from females
with GDM compared with controls without GDM (13). The present study investigated
whether miR-518d is involved in the development of GDM and whether
PPARα expression is suppressed by miR-518d in placentas from
females with GDM.
Materials and methods
Patients and tissue samples
Human placentas were obtained according to protocols
approved by the Ethics Committee of Nanjing Maternity and Child
Healthcare Hospital, affiliated to Nanjing Medical University. All
the patients provided written informed consent before taking part
in the study. Full-term placental tissue samples were obtained from
elective terminations of pregnancy and uncomplicated cesarean
deliveries, respectively. Placentas were obtained from females who
delivered from 37–40 weeks of gestation. Gestational age, recorded
as completed weeks of gestation, was calculated from the date of
the last menstrual period and/or from ultrasound. Females were
diagnosed with GDM when a 75 g oral glucose tolerance test (OGTT)
revealed either a fasting venous plasma glucose level >5.6
mmol/l glucose and/or a 2 h post-test plasma glucose level >8.6
mmol/l glucose. None of the patients had a previous history of
diabetes mellitus or any known endocrinopathy. Placental tissue was
collected at delivery and immediately transferred on ice for
transport to the laboratory for RNA isolation, protein lysate
preparation and tissue fixation.
Quantitative polymerase chain reaction
(qPCR)
Total RNA was extracted from placentas with TRIzol
reagent (Invitrogen Life Technologies, Carlsbad, CA, USA). miR-518d
was reverse transcribed into complementary DNA (cDNA) using a
TaqMan microRNA assay (Applied Biosystems, Branchburg, NJ, USA)
containing microRNA-specific stem-loop RT primers and a TaqMan
MicroRNA Reverse Transcription kit in a total reaction volume of 50
μl, performed according to the manufacturer’s instructions.
Reverse transcriptase reactions were performed using
a 7300 real-time PCR system (Applied Biosystems, Beijing, China)
and the following thermal cycling parameters: 30 min at 16°C, 30
min at 42°C, 5 min at 85°C and then held at 4°C. miRNA expression
was normalized to the expression level of small nucleolar RNA U6.
The primer of miR-518d was F: 5′-ACACTCCAGCTGGGCAAAGCGCTTCCCTT-3′
and R: 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCTCCAAA-3′.
Luciferase reporter expression assays
using luciferase genes with mutated or wild-type
PPARα-3′-untranslated region (UTR) in human embryonic kidney (HEK)
cells co-transfected with miR-518d or microRNA-scrambled control
(miR-SCR)
Wild-type PPARα-3′-UTR was amplified by PCR from
human cDNA using the primers: F:
5′-CCAAGCTTCGTCCAGTCAACCTGAACCCA-3′ and R:
5′-CGAGCTCCTCCAGGTGCCCAGCGACT-3′. The mutated PPARα-3′-UTR was
amplified using the primers: F: 5′-CCAAGCTTCGTCCAGTCAACCTGAACCCA-3′
and R: 5′-CGAGCTCCACGACGTGCCCAGCGACT-3′. The DNA segments produced
from these amplifications were inserted into the pMIR-REPORT miRNA
Expression Reporter Vector (Ambion, Carlsbad, CA, USA) using the
SacI and HindIII sites. HEK-293 cells cultured in
24-well plates were co-transfected with pMIR-REPORT vectors
containing either the wild-type or mutated PPARα-3′-UTR segments
along with the control vector, renilla luciferase reporter vector
(pRL-TK). These HEK-293 cells were co-transfected with the
precursor microRNAs for miR-518d (pre-miR-518d) and miR-SCR. Assays
were performed to determine the level of gene expression 48 h
post-transfection using the Dual Luciferase Reporter Assay kit
(Promega, Madison, WI, USA). Renilla luciferase activity was
used to normalize the luciferase activity. Three independent
experiments were performed in triplicate.
Immunohistochemistry
Formalin-fixed placental tissue samples were
embedded in paraffin, sectioned at 5 μm, and mounted on
silane-coated slides. The sections were dewaxed and rehydrated
through descending grades of alcohol to distilled water, followed
by blocking of endogenous peroxidase activity using 3% (v/v)
hydrogen peroxidase in phosphate-buffered saline (PBS). The
sections were subjected to microwave antigen retrieval in 0.02 M
ethylenediaminetetraacetic acid (EDTA), washed in PBS and blocked
with goat serum (Beijing ZhongShan Biotechnology, Beijing, China)
for 2 h, then incubated overnight at 4°C with polyclonal anti-PPARα
(1:200, Abcam, Cambridge, MA, USA). Following three washes in PBS,
the sections were incubated with horseradish peroxidase
(HRP)-conjugated secondary antibody (1:1,000; Beijing ZhongShan
Biotechnology, Beijing, China) for 1 h at room temperature.
Immunoreactivity was demonstrated using diaminobenzidine (Sigma,
St. Louis, MO, USA) for increased sensitivity, which produces a
brown precipitate at immunopositive sites. Sections were
counterstained with hematoxylin and mounted with a coverglass. The
negative controls were incubated with immunoglobulin G (IgG)
controls. All the immunostained sections were evaluated in a
blinded manner by two observers.
Protein extraction and western blot
analysis
For western blot analysis, nuclear proteins from 200
mg of frozen placenta were extracted. Protein lysates were prepared
in the presence of protease inhibitors [10 μg/ml aprotinin, 5 μg/ml
leupeptin, 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF), 1
mM Na3VO4 and 1 mM NaF]. Protein
concentrations were determined using the Bradford protein assay.
Samples containing 50 μg of protein were subjected to
electrophoresis on a 12% SDS-PAGE and transferred to a
nitrocellulose membrane (GE Healthcare, San Francisco, CA, USA).
The membranes were blocked in Tris-buffered saline (TBS) containing
5% non-fat milk powder for 1 h and then incubated overnight with
the polyclonal antibodies anti-PPARα (1:500, Abcam, Cambridge, MA,
USA) and anti-GAPDH (1:1,000, Kangcheng, Shanghai, China) diluted
in a solution of 5% non-fat milk powder in TBS. GAPDH was used as a
control. Membranes were washed three times with TBS (10 min each)
and then incubated for 1 h with HRP-conjugated goat anti-rabbit IgG
(1:1,000; Beijing ZhongShan Biotechnology). Specific proteins were
detected using a chemiluminescence ECL kit and AlphaImager
(FluorChem5500, Alpha Innotech, San Leandro, CA, USA). Protein
expression levels were analyzed using AlphaEaseFC software (Alpha
Innotech, San Leandro, CA, USA).
Statistical analysis
Data from at least three independent experiments
were expressed as the mean ± standard deviation (SD). The
differences between groups were analyzed using the Student’s
t-test. The correlation between the relative expression levels of
miR-518d and the expression levels of PPARα was analyzed using a
two-sided Spearman’s ρ-test. Differences between data were
considered significant if P<0.05.
Results
Clinical characteristics of the
patients
Table I shows the
clinical characteristics of the patients included in the present
study and compares these characteristics in females with GDM (n=40)
with those of females in the control group (n=40). No significant
differences were identified between the two groups with respect to
maternal age, gravida, parity or number of weeks of gestation at
delivery. Females with GDM had a significantly greater mean body
mass index (BMI) and the neonates had a significantly higher birth
weight than those whose mothers were in the control group.
 | Table IClinical characteristics of females
with GDM and controls. |
Table I
Clinical characteristics of females
with GDM and controls.
| Variables | Control (n=40) | GDM (n=40) | P-value |
|---|
| Maternal age
(years) | 30.23±3.02 | 30.55±3.30 | NS |
| Gestational age at
delivery (week) | 39.64±0.92 | 39.61±0.76 | NS |
| Body mass index
(kg/m2) | 21.87±2.56 | 23.54±3.46 | 0.023 |
| Neonatal birth weight
(kg) | 3.572±0.306 | 4.007±0.489 | 0.004 |
| Smokers during
pregnancy | 0 | 0 | NS |
| Placental weight
(kg) | 551±121 | 572±132 | NS |
Aberrant expression of miR-518d in GDM
placenta
To investigate whether miR-518d is involved in the
development of GDM, PCR analysis was used to study the expression
levels of miR-518d in placental tissue from patients with GDM and
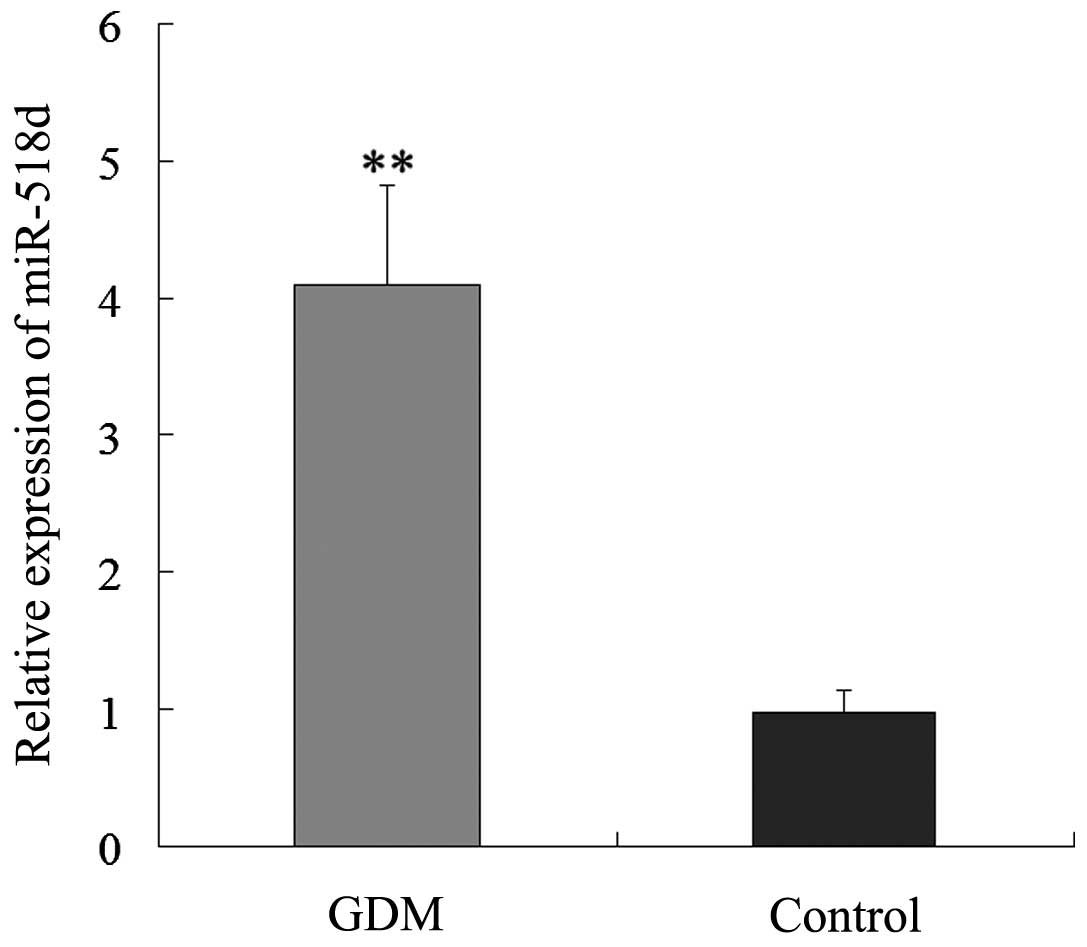
normal controls. The expression levels of miR-518d were
significantly higher in the placenta of patients with GDM than
those in the placentas of the normal controls (Fig. 1). The results suggest that miR-518d
may be associated with the development of GDM.
PPARα is a direct target of miR-518d
To identify the potential mechanism by which
miR-518d is associated with GDM, three bioinformatic algorithms
(TargetScan, PicTar and miRanda) were applied to identify potential
target genes for miR-518d. Among the potential candidates, the
study focused on the PPARα gene as PPARs are connected with the
diabetic phenotype (11). One
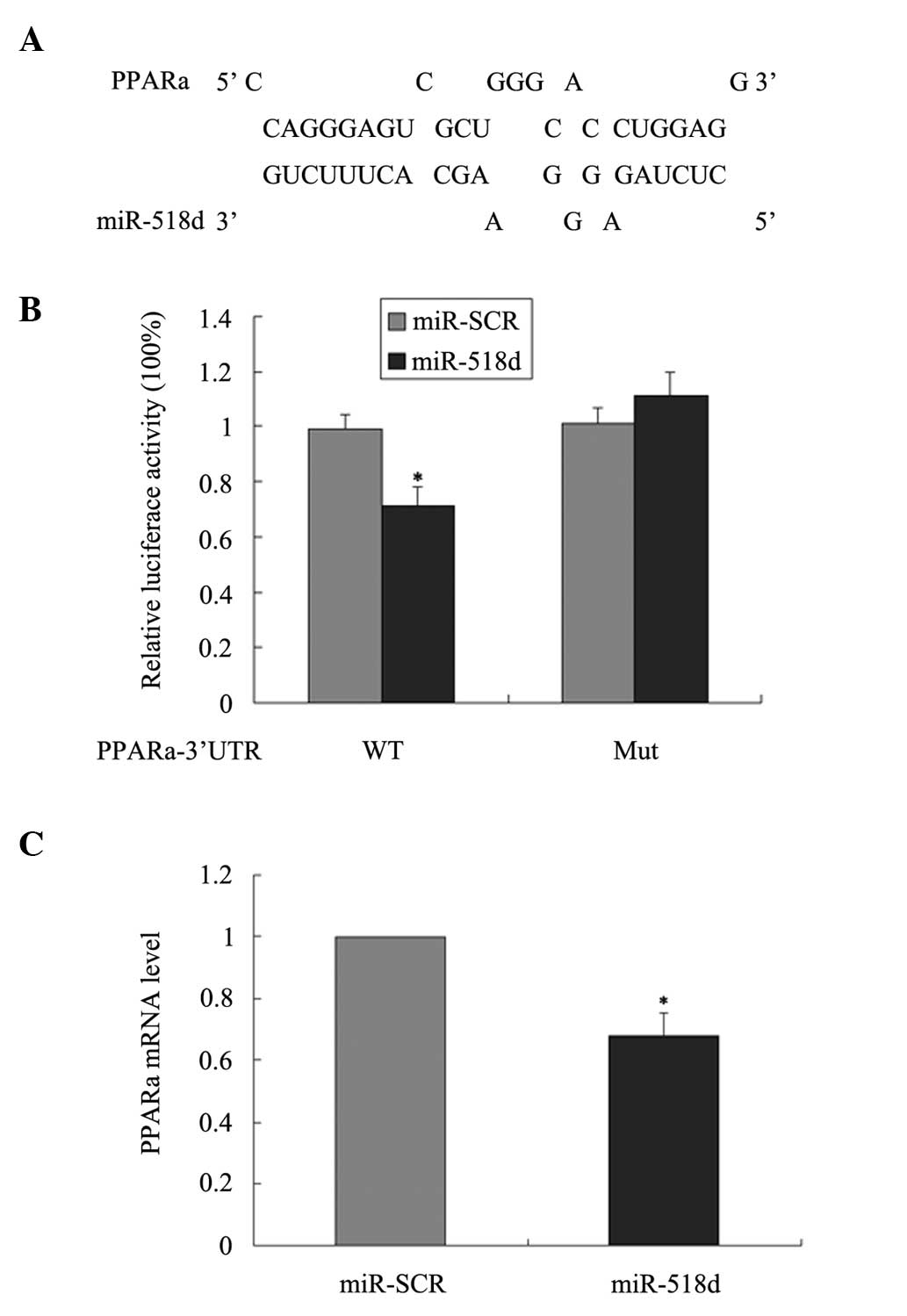
miR-518d-binding site was found in the 3′-UTR of PPARα mRNA
(Fig. 2A). However, there are no
reports as yet describing PPARα regulation by miRNA-518d.
Expression of luciferase gene with
mutated or wild-type PPARα-3′-UTR in HEK cells co-transfected with
miR-518d or miR-SCR
Expression assays were performed with the luciferase
reporter gene system using the wild-type PPARα-3′-UTR or a mutated
version to validate the miR-518d target prediction. The vector was
constructed by inserting the wild-type sequence of the PPARα 3′-UTR
PPARα mRNA (PPARα-3′-UTR) or a mutated seed sequence of the
miR-518d-binding site (PPARα-3′-UTR-mut) into the 3′-UTR of the
pMIR-REPORT luciferase reporter. Co-transfection of the vector with
the wild-type PPARα-3′-UTR and the miR-518d precursor,
pre-miR-518d, inhibited luciferase activity, whereas
co-transfection of the vector with PPARα-3′-UTR-mut and
pre-miR-518d caused no inhibition of luciferase activity (Fig. 2B). These results validated the
hypothesis that miR-518d is able to bind to the 3′-UTR of PPARα
mRNA. The impact of miR-518d on the expression of PPARα was also
investigated. qPCR revealed that PPARα mRNA levels decreased
significantly 48 h following transfection of HEK-293 cells with
pre-miR-518d (Fig. 2C).
Expression and subcellular location of
PPARα in the placenta of patients with GDM
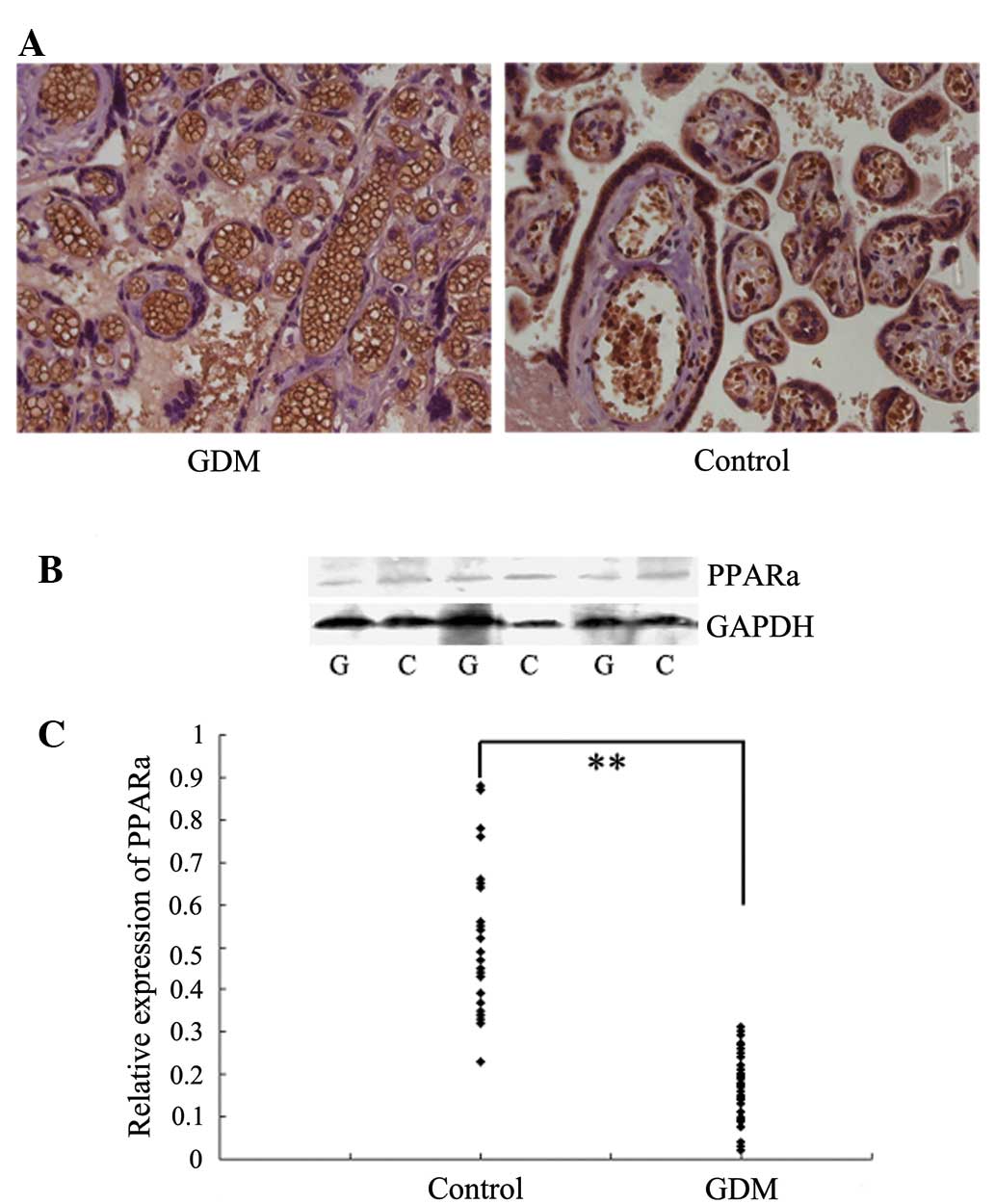
Immunohistochemical staining showed that PPARα was
located in the nuclei of the syncytiotrophoblasts, and its
expression was reduced in placentas of patients with GDM than in
the control placentas (Fig. 3A).
Western blot analysis was performed to confirm the differential
expression of PPARα in placentas of patients with GDM and the
control placentas. The expression levels of PPARα were
significantly reduced in placentas from patients with GDM than
those in the controls (Fig. 3B and
C).
The correlation between PPARα and
miR-518d in females with GDM
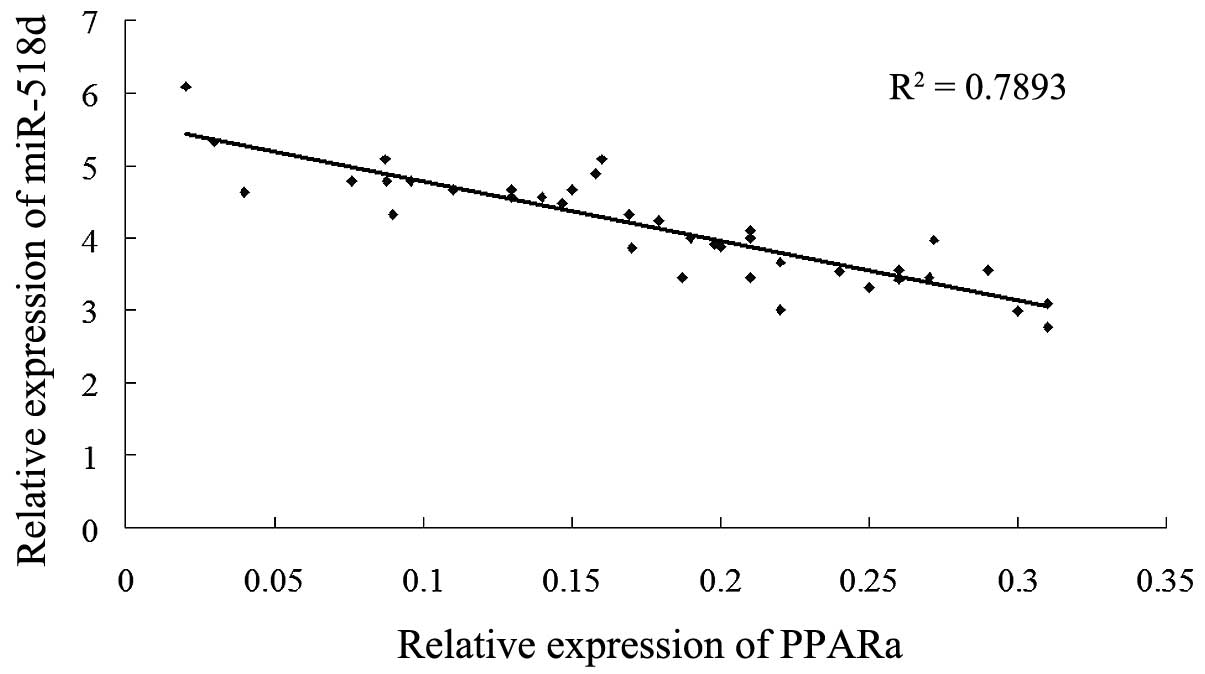
The potential correlation between the levels of
miRNA-518d expression and PPARα protein levels in the placentas of
females with GDM was assessed using Spearman’s correlation
analysis. The levels of miR-518d were negatively correlated with
the protein levels of PPARα in females with GDM (Fig. 4; R2=0.7893,
P<0.01).
Discussion
Recent data indicate that microRNAs have a
fundamental role in a variety of physiological and pathological
processes. miRNA analyses indicate that diverse affected tissue
types have miRNA expression profiles that are significantly
different from normal tissue (9).
Studies of microRNA expression revealed that some microRNAs are
abundantly expressed in the placenta (9). However, the signature of miRNAs in
the placenta has yet to be elucidated. Recently, analyses of the
expression of small RNAs in the placenta by small RNA library
sequencing confirmed that most placenta-specific miRNAs were linked
to the C19 MC cluster and some of them have been reported to be
associated with pre-eclampsia or premature labor (14). However, the possible involvement of
the C19 MC cluster in GDM remains to be elucidated.
MiR-518d is a member of the C19 MC cluster. In a
microarray analysis previously performed by our group, the microRNA
expression profiles in placentas of patients with GDM were compared
with those of normal placentas, which revealed that miR-518d was
differentially expressed in placentas of patients with GDM.
miR-518d has been reported to be upregulated >10-fold in third
trimester trophoblast cells compared with first trimester
trophoblast cells, indicating that miR-518d may be involved in the
regulation of trophoblast proliferation and invasion (14). However, the involvement of
placental miR-518d in the molecular mechanisms of GDM requires to
be elucidated.
In the present study, expression levels of miR-518d
in the placentas of females with GDM were compared with those of
normal pregnant females. miR-518d was aberrantly upregulated in
placentas of females with GDM compared with controls. This
indicates that miR-518d may be associated with the pathogenesis of
GDM. The PPARα gene was selected as a potential target for miR-518d
as PPARα is highly expressed in tissues that catabolize fatty
acids. It is a transcription factor that controls fatty acid uptake
and metabolism, and it upregulates fatty acid β oxidation in these
tissues (16,17). It has been reported that regulation
of PPARα expression and activity contributes to maintaining a
homeostatic balance between cellular fatty acid and glucose
utilization via activation of its target genes (11). Accordingly, activation of PPARα
increases sensitivity to insulin as well as thrombosis and vascular
inflammation (18–20). By contrast, it appears that
inhibition of PPARα suppresses sensitivity to insulin and increases
hepatic glucose production. GDM is a common complication of
pregnancy. GDM is able to lead to dyslipidemia, and is aggravated
by obesity. Placental cholesterol and fatty acid transfer have
critical roles in the development of GDM (21), and it is possible that the
regulation of placental PPARα may be involved in this process.
Consistent with previous studies, the present study shows that the
expression levels of PPARα protein were significantly reduced in
the placentas of females with GDM (15). As the level of miR-518d is
upregulated and the level of PPARα is downregulated in placentas of
females with GDM, it is hypothesized that miR-518d is involved in
the pathophysiology of GDM via its effect on its target gene PPARα.
Using the luciferase reporter gene system and mutation assays, the
present study confirmed that miR-518d is able to target PPARα
directly by binding to the 3′-UTR of PPARα mRNA. In addition, it
was revealed that miR-518d levels were negatively correlated with
the levels of PPARα protein in placentas from females with GDM.
In conclusion, the present study has provided
evidence that miR-518d has an important role in the pathophysiology
of GDM via an inhibitory effect on the expression of PPARα, which
may disrupt the balance of fatty acid uptake and metabolism and
result in an increased resistance to insulin. Elucidation of this
mechanism may offer opportunities for application of miR-518d in
future clinical management of females with GDM.
Acknowledgements
The present study was financially supported by the
National Natural Science Foundation of China (81000258, 81100436),
the Natural Science Foundation of Jiangsu Province (BK2010586), the
Bureau of Nanjing City Science and Technology Development Fund
(201104014), the Open topic of State Key Laboratory of Reproductive
Medicine (SKLRM-KF-201109) and the Nanjing Medical Science and
Technique Development Foundation (QRX11210, QRX11211).
Abbreviations:
|
GDM
|
gestational diabetes mellitus
|
|
PPARα
|
peroxisome proliferator-activated
receptor-alpha
|
|
miR-SCR
|
miR-scrambled control
|
|
OGTT
|
oral glucose tolerance test
|
|
pRL-TK
|
renilla luciferase reporter vector
|
References
|
1
|
Barnes-Powell LL: Infants of diabetic
mothers: the effects of hyperglycemia on the fetus and neonate.
Neonatal Netw. 26:283–290. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Uzelac PS, Li X, Lin J, Neese LD, Lin L,
Nakajima ST, Bohler H and Lei Z: Dysregulation of leptin and
testosterone production and their receptor expression in the human
placenta with gestational diabetes mellitus. Placenta. 31:581–588.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferrara A: Increasing prevalence of
gestational diabetes mellitus: a public health perspective.
Diabetes Care. 30(Suppl 2): S141–S146. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee AJ, Hiscock RJ, Wein P, et al:
Gestational diabetes mellitus: clinical predictors and long-term
risk of developing type 2 diabetes: a retrospective cohort study
using survival analysis. Diabetes Care. 30:878–883. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thadhani R, Powe CE, Tjoa ML, et al:
First-trimester follistatin-like-3 levels in pregnancies
complicated by subsequent gestational diabetes mellitus. Diabetes
Care. 33:664–669. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Murphy VE, Smith R, Giles WB and Clifton
VL: Endocrine regulation of human fetal growth: The role of the
mother, placenta, and fetus. Endocr Rev. 27:141–169. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Luo SS, Ishibashi O, Ishikawa G, et al:
Human villous trophoblasts express and secrete placental-specific
microRNAs into maternal circulation via exosomes. Biol Reprod.
81:717–729. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fu GD, Brkić J, Hayder H and Peng C:
MicroRNAs in human placental development and pregnancy
complications. Int J Mol Sci. 14:5519–5544. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kotlabova K, Doucha J and Hromadnikova I:
Placental-specific microRNA in maternal circulation -
identification of appropriate pregnancy-associated microRNAs with
diagnostic potential. J Reprod Immuno. 89:185–191. 2011. View Article : Google Scholar
|
|
10
|
Gauster M, Desoye G, Tötsch M and Hiden U:
The placenta and gestational diabetes mellitus. Cure Diab Rep.
12:16–23. 2012. View Article : Google Scholar
|
|
11
|
Yilmaz-Aydogan H, Kurnaz O, Kucukhuseyin
O, et al: Different effects of PPARα, PPARG and apoE SNPs on serum
lipids in patients with coronary heart disease based on the
presence of diabetes. Gene. 523:20–26. 2013.
|
|
12
|
Arck P, Toth B, Pestka A and Jeschke U:
Nuclear receptors of the perocisome proliferator-activated receptor
(PPAR) family in gestational diabetes: from animal models to
clinical trials. Biol Reprod. 83:168–176. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shi ZM, Wang J, Yan Z, et al: MiR-128
inhibits tumor growth and angiogenesis by targeting p70S6K1. PLoS
One. 7:e327092012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Morales-Prieto DM, Chaiwangyen W,
Ospina-Prieto S, Schneider U, Herrmann J, Gruhn B and Markert UR:
MicroRNA expression profiles of trophoblastic cells. Placenta.
33:725–734. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Holdsworth-Carson SJ, Lim R, Mitton A,
Whitehead C, Rice GE, Permezel M and Lappas M: Peroxisome
proliferator-activated receptors are altered in pathologies of the
human placenta: gestational diabetes mellitus, intrauterine growth
restriction and preeclampsia. Placenta. 31:222–229. 2010.
View Article : Google Scholar
|
|
16
|
Cresci S, Huss JM, Beitelshees AL, et al:
A PPARα promoter variant impairs ERR-dependent transactivation and
decreases mortality after acute coronary ischemia in patients with
diabetes. PLoS One. 5:e125842010.
|
|
17
|
Beaven SW and Tontonoz P: Nuclear
receptors in lipid metabolism: targeting the heart of dyslipidemia.
Annu Rev Med. 57:313–329. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Flavell DM, Pineda Torra I, Jamshidi Y, et
al: variation in the PPARα gene is associated with altered function
in vitro and plasma lipid concentrations in Type II diabetic
subjects. Diabetologia. 43:673–680. 2010.
|
|
19
|
Matsuda S, Kobayashi M and Kitagishi Y:
Expression and function of PPARs in placenta. PPAR Res.
2013:2565082013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Arck P, Toth B, Pestka A and Jeschke U:
Nuclear receptors of the peroxisome proliferator-activated receptor
(PPAR) family in gestational diabetes: from animal models to
clinical trials. Biol Reprod. 83:168–176. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dubé E, Ethier-Chiasson M and Lafond J:
Modulation of cholesterol transport by insulin-treated gestational
diabetes mellitus in human full-term placenta. Biol Reprod.
88:162013.PubMed/NCBI
|