Introduction
Chronic myeloid leukemia (CML) is a
myeloproliferative disorder characterized by the expansion of a
clone of pluripotent hematopoietic stem cells carrying the chimeric
BCR-ABL fusion gene. The Philadelphia (Ph) chromosome created as a
result of t(9;22) (q34;q11) is observed in more than 90% of CML
patients. The BCR-ABL fusion gene is formed by transposing the 3′
portion of the ABL oncogene from 9q34 to the 5′ portion of the BCR
gene on chromosome 22, and this fusion gene encodes a constitutive
active tyrosine kinase (1). BCR
breakpoints localize on 1 of 3 breakpoint cluster regions (bcr):
major bcr spanning BCR exons 12–16, minor bcr spanning BCR
alternative exons 2′ and 2 and μbcr spanning downstream of exon 19.
The majority of the BCR-ABL fusion transcripts are e13a2, e14a2,
with e1a2 and e19a2 being less common (2). Extremely rare CML cases with fusion
transcripts, such as e13a3, in which ABL exon 3 rather than exon 2
is fused to BCR, were previously reported (3–6). Cases
reported to have the e13a3 transcript showed the Ph chromosome on
karyotype analysis. A cryptic or variant rearrangement in which the
typical Ph chromosome is not visible at the karyotype level is
noted in 5–10% of CML patients (7).
This rearrangement can be found in cases of normal or complex
karyotypes. The mechanisms for these rearrangements are difficult
to determine. This study reported a rare Ph chromosome-positive CML
case involving the e13a3 BCR-ABL transcript and new complex
aberrations, including four chromosomal breakpoints.
Materials and methods
Case report
A 25-year-old female was diagnosed as suffering from
CML in the chronic phase (CP) after a blood cell count was
initiated in November 2009 due to hepatosplenomegaly and loss of
weight. The hematologic parameters were: white blood cells (WBC)
122×109/l (96.6% neutrophils, 1.55% lymphocytes, 0.170%
monocytes, 1.68% eosinophiles and 0.04% basophiles). The platelet
count was 156×109/l and the hemoglobin level was 10.3
g/dl. No treatment had been administered prior to the test. The
patient was referred to our laboratory one month later, after
treatment with hydroxyurea (2,000 mg/day). Thus, the previous
relevant symptoms disappeared. The more recent hematological
parameters were: WBC 4.5×109/l (42.9% neutrophils, 44.1%
lymphocytes and 13% monocytes). The platelet count was
375×109/l and the hemoglobin level was 9.9 g/dl.
Banding cytogenetics
Chromosome analysis was performed using the
GTG-banding technique according to standard procedures (8). A total of 20 metaphases, obtained from
the unstimulated bone marrow of the patient, were analyzed.
Karyotypes were described according to the International System for
Human Cytogenetic Nomenclature (9).
Molecular cytogenetics
Fluorescence in situ hybridization (FISH)
using a LSI BCR/ABL dual color dual fusion translocation probe
(Abbott molecular/Vysis, Des Plaines, IL, USA) was applied
according to the manufacturer’s instructions. Array-proven
multicolor banding (aMCB) probe sets based on
microdissection-derived region-specific libraries for chromosomes
9, 12, 19 and 22 were applied as previously described (10,11). A
total of 20 metaphase spreads were analyzed, using a fluorescence
microscope (AxioImager.Z1 mot, Zeiss) equipped with appropriate
filter sets to discriminate between a maximum of five fluorochromes
and the counterstain DAPI (4′,6-diamino-2-phenylindole). Image
capturing and processing were carried out using an Isis imaging
system (MetaSystems, Altlussheim, Germany) for the MCB
evaluation.
RNA isolation and reverse
transcriptase-polymerase chain reaction
Total RNA was extracted from 1.5 ml of fresh
peripheral blood immediately after collection using the InviTrap
RNA kit (Invitek, Germany). A negative control was used to monitor
RNA isolation. RNA concentration was estimated by absorbance at 260
nm and its purification was determined by a 1.8:2.0 ratio of
absorbance values at 260:280 nm. The RNA solutions were stored at
−80°C. Reverse transcription (RT) for the complementary DNA (cDNA)
synthesis, polymerase chain reaction (PCR) and nested PCR
amplification of the BCR-ABL gene were performed using a
ready-to-use Genequality BCR-ABL kit (AB Analitica, Italy) and a
GeneAmp® PCR System 9700 thermocycler. Two positive
controls were used to monitor amplification, one with
β2-microglobuline and the second b3a2 with the patient sample. The
negative controls used serial water. PCR products were
electrophoresed on an ethidium bromide-stained 2.5% agarose-TAE-gel
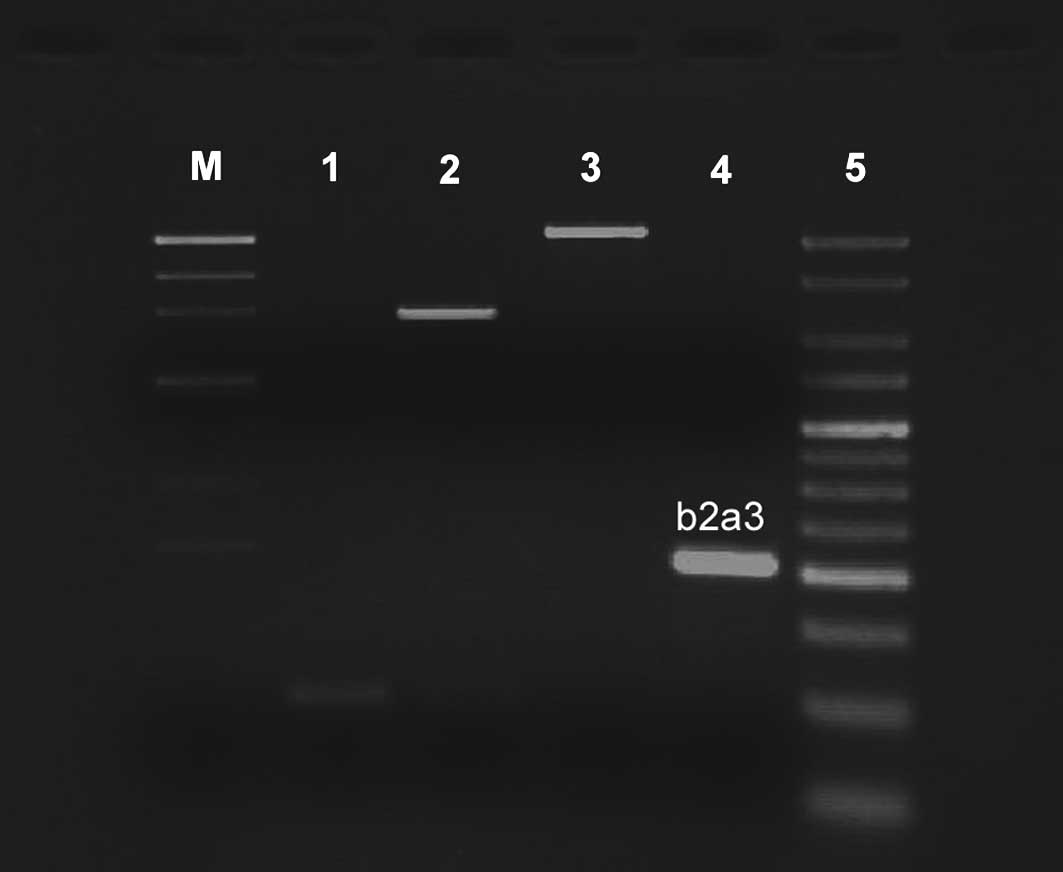
and observed under UV light. The type of amplified BCR/ABL cDNA was
established on the basis of the size compared with the molecular
markers and the primers used (b3a2, 353 bp; b2a2, 278 bp; b3a3, 179
bp and b2a3 (e13a3), 104 bp).
Results
Karyotyping was performed before and after the
initiation of chemotherapy treatment, showing the following
karyotypic changes: a complex karyotype 48,XX,+8,+der(12)t(12;19),t(9;12;19;22)/47,XX,+8,t(9;12;19;22)/47,XX,+der(12)t(12;19),t(9;12;19;22)/46,XX,t(9;12;19;22)
was determined using GTG-banding (Fig.
1) and was further specified by molecular cytogenetic studies
(Fig. 2). A dual-color-FISH using a
probe specific for BCR and ABL showed a typical Ph chromosome with
a BCR/ABL fusion gene. However, sections of chromosome 22 were
present on a der(19) (Fig. 2F). A multi-color (M)-FISH, was
applied to exclude further cryptic rearrangements (Fig. 2A). Thus, the four chromosomes 9, 12,
19 and 22 were found to be involved. aMCB using probes for the
corresponding chromosomes were performed as previously reported
(11). A complex translocation
among the four chromosomes was detected (Figs. 2B-E), and the following final
karyotypes were obtained: 48,XX,+8,+der(12)t(12;19)(p11.2;q13.3),t(9;12;19;22)(q34.1;p11.2;q13.3;q11.2)[8]/47,XX,+8,t(9;12;19;22)(q34.1;p11.2;q13.3;q11.2)[2]/47,XX,+der(12)t(12;19)(p11.2;q13.3),t(9;12;19;22)(q34.1;p11.2;q13.3;q11.2)[2]/46,XX,t(9;12;19;22)(q34.1;p11.2;q13.3;q11.2)[8].
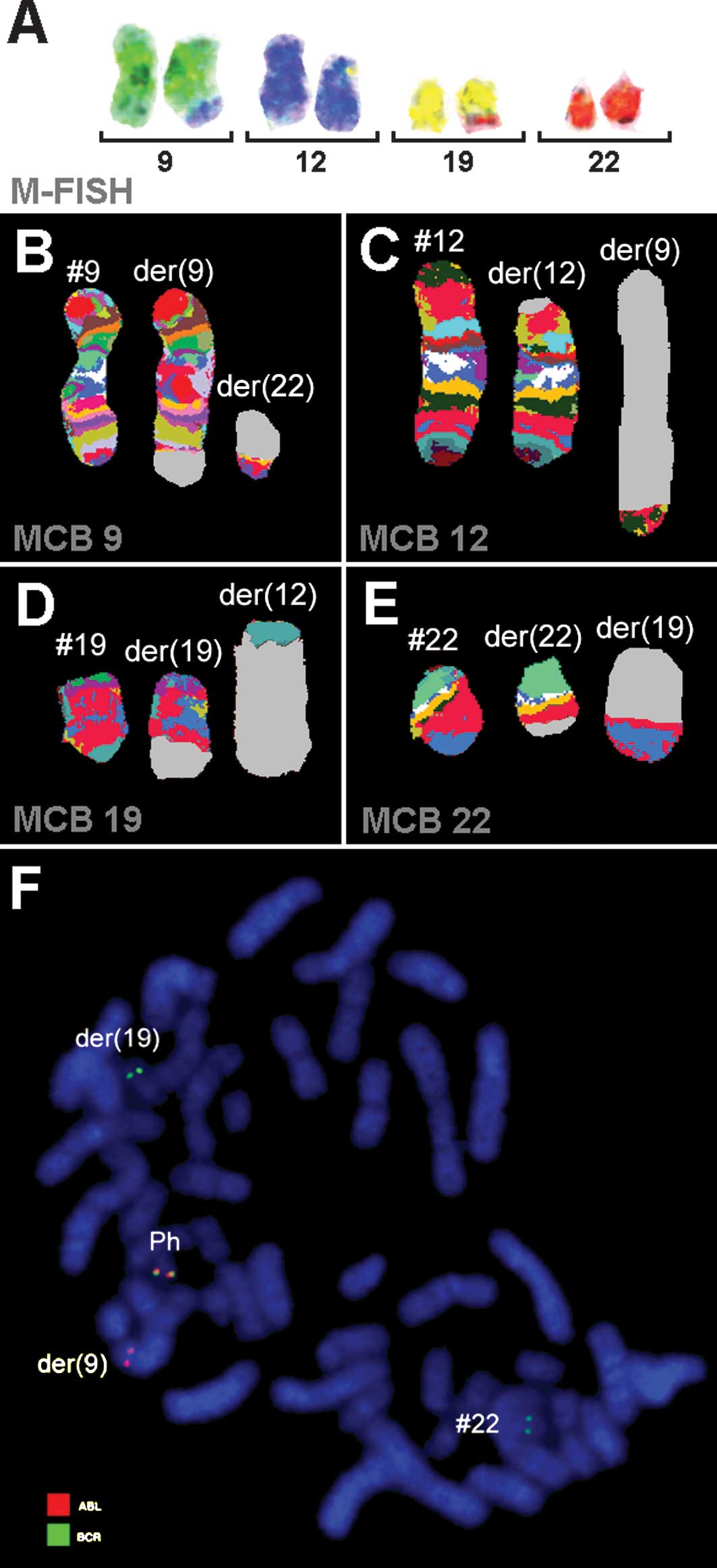 | Figure 2Karyotype and chromosomal aberrations
were confirmed using molecular cytogenetic approaches. (A) M-FISH
confirmed the complexity of the karyotype: 48,XX,+8,+der(12),t(9;12),t(12;19),t(19;22). (B-E) The
application of aMCB analysis using probe sets for chromosomes 9,
12, 19 and 22 is shown. The normal chromosomes are shown to the
left of the image, and the derivative of the four chromosomes to
the middle and right. Using aMCB probes, the light gray areas show
unstained regions on the derivative chromosomes. (F) Fluorescence
in situ hybridization (FISH) using probes for BCR (green)
and ABL (red) confirmed the involvement of chromosome 19 in the
rearrangement present in this case. #, chromosome; der, derivative
chromosome; Ph, Philadelphia chromosome. |
It is likely that due to low chromosomal resolution,
the complexity of the karyotype was initially missed. RT-PCR
analysis of the fusion transcript showed a band corresponding to
the e13a3 (b2-a3) transcript. However, the band was ~104 bp
(Fig. 3).
Discussion
We described a rare Ph chromosome-positive CML case
with the e13a3 BCR-ABL transcript and a new complex variant
translocation t(9;12;19;22)(q34.1;p11.2;q13.3;q11.2) that was
detected. Apart from a trisomy 8 and an additional derivative
chromosome 12, two additional chromosomal alterations were present.
To the best of our knowledge, this translocation has yet to be
elucidated in CML (12).
In 5–10% of Ph chromosome CML cases complex
translocations in addition to those and/or besides chromosomes 9
and 22 (13) are noted. At present
it appears that in such rearrangements any other chromosome may be
involved. However, it has been suggested that the distribution of
chromosomes and breakpoints is non-random with the chromosomal
bands most susceptible to breakage being 1p36, 3p21, 5q31, 6p21,
9q22, 10q22, 11q13, 12p13, 17p13, 17q21, 17q25, 19q13, 21q22, 22q12
and 22q13 (7), showing one match
with the present case, i.e., 19q13.
The progression of CML from CP to blast crisis is
frequently associated with non-random secondary chromosomal
aberrations such as +8, i(17q), +19 and an extra Ph chromosome
(14).
Trisomy 12 is the most frequently reported
chromosome abnormality in B-cell chronic lymphocytic leukemia
(B-CLL). It is found in one third of cytogenetically abnormal CLL
by conventional karyotype (15) and
in approximately 11–46% of cases when interphase FISH is used
(16).
The majority of CML patients express e13a2 (b2-a2)
or e14a2 (b3-a2) of BCR-ABL mRNA encoding for p210 Bcr-Abl tyrosine
kinase. These two types are detected by RT-PCR (17–19).
CML with the e13a3 transcript is extremely rare, and RT-PCR for
e13a2 and e14a2 is usually negative because ABL exon 2 is deleted
(20). Our case was diagnosed using
conventional G-band, FISH, M-FISH, MCB and RT-PCR.
In conclusion, we reported a rare Ph
chromosome-positive CML case in CP with the rare e13a3 BCR-ABL
transcript and new complex variant translocation involving the
chromosomes 9, 12, 19 and 22.
Acknowledgements
We thank Professor I. Othman, the Director General
of the Atomic Energy Commission of SYRIA (AECS) and Dr N. MirAli,
Head of Molecular Biology and Biotechnology Department for their
support. This work was supported by the AECS, and in part by the
Stefan-Morsch-Stiftung, Monika-Kutzner-Stiftung and the DAAD
(D/07/09624).
References
|
1
|
Shtivelman E, Lifshitz B, Gale RP and
Canaani E: Fused transcript of abl and bcr genes in chronic
myelogenous leukemia. Nature. 315:550–554. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Deiningger MW, Goldman JM and Melo JV: The
molecular biology of chronic myeloid leukemia. Blood. 96:3343–3356.
2000.PubMed/NCBI
|
|
3
|
Al-Ali HK, Leiblein S, Kovacs I, Hennig E,
Niederwieser D and Deininger MW: CML with an e1a3 BCR-ABL fusion:
rare, benign, and a potential diagnostic pitfall. Blood.
100:1092–1093. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu LG, Tanaka H, Ito K, Kyo T, Ito T and
Kimura A: Chronic myelogenous leukemia with e13a3 (b2a3) type of
BCR-ABL transcript having a DNA breakpoint between ABL exons a2 and
a3. Am J Hematol. 74:268–272. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Snyder DS, McMahon R, Cohen SR and Slovak
ML: Chronic myeloid leukemia with e13a3 BCR-ABL fusion: benign
course responsive to imatinib with an RT-PCR advisory. Am J
Hematol. 75:92–95. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pienkowska-Grela B, Woroniecka R, Solarska
I, Kos K, Pastwińska A, Konopka L and Majewski M: Complete
cytogenetic and molecular response after imatinib treatment for
chronic myeloid leukemia in a patient with atypical karyotype and
BCR-ABL b2a3 transcript. Cancer Genet Cytogenet. 174:111–115. 2007.
View Article : Google Scholar
|
|
7
|
Johansson B, Fioretos T and Mitelman F:
Cytogenetic and molecular genetic evolution of chronic myeloid
leukemia. Acta Haematol. 107:76–94. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Al Achkar W, Wafa A and Almedani S: BCR
translocation to derivative chromosome 2: A new case of chronic
myeloid leukemia with a complex variant translocation and
Philadelphia chromosome. Oncol Lett. 1:445–447. 2010.PubMed/NCBI
|
|
9
|
Shaffer L, Slovak M and Cambell L: ISCN:
An International System for Human Cytogenetic Nomenclature. S
Karger; Basel: 2009
|
|
10
|
Weise A, Mrasek K, Fickelscher I, Claussen
U, Cheung SW, Cai WW, Liehr T and Kosyakova N: Molecular definition
of high-resolution multicolor banding probes: first within the
human DNA sequence anchored FISH banding probe set. J Histochem
Cytochem. 56:487–493. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liehr T, Heller A, Starke H, Rubtsov N,
Trifonov V, Mrasek K, Weise A, Kuechler A and Claussen U:
Microdissection based high resolution multicolor banding for all 24
human chromosomes. Int J Mol Med. 9:335–339. 2002.PubMed/NCBI
|
|
12
|
Mitelman F, Johansson B and Mertens F:
Mitelman Database of Chromosome Aberrations in Cancer. http://cgap.nci.nih.gov/Chromosomes/Mitelman.
2009
|
|
13
|
La Starza R, Testoni N, Lafage-Pochitaloff
M, Ruggeri D, Ottaviani E, Perla G, Martelli MF, Marynen P and
Mecucci CL: Complex variant Philadelphia translocations involving
the short arm of chromosome 6 in chronic myeloid leukemia.
Haematologica. 87:143–147. 2002.
|
|
14
|
Sandberg AA: The Chromosomes In Human
Cancer And Leukemia. 2nd edition. Elsevier Science; New York: pp.
151–172. 1990
|
|
15
|
Juliusson G and Merup M: Cytogenetics in
chronic lymphocytic leukemia. Sem Oncol. 25:19–26. 1998.
|
|
16
|
Que TH, Garcia Marco J, Ellis J, Matutes
E, Brito Babapulle V, Boyle S and Catovsky D: Trisomy 12 in chronic
lymphocytic leukemia detected by fluorescence in situ
hybridization: analysis by stage, immunophenotype, and morphology.
Blood. 82:571–575. 1993.PubMed/NCBI
|
|
17
|
Rowley JD: A new consistent chromosomal
abnormality in chronic myelogenous leukemia identified by
quinacrine fluorescence and Giemsa staining. Nature. 24:290–295.
1973. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lifshitz B, Fainstein E, Marcelle C,
Shtivelman E, Amson R, Gale RP and Canaani E: Bcr genes and
transcripts. Oncogene. 2:113–117. 1988.PubMed/NCBI
|
|
19
|
Hughes TP, Morgan GJ, Martiat P and
Goldman JM: Detection of residual leukemia after bone marrow
transplant for chronic myeloid leukemia: role of polymerase chain
reaction in predicting relapse. Blood. 77:874–878. 1991.PubMed/NCBI
|
|
20
|
Masuko M, Furukawa T, Abe T, Wada R,
Maruyama S, Kitajima T, Shibasaki Y, Toba K, Okada M and Aizawa Y:
A chronic myeloid leukemia patient with atypical karyotype and
BCR-ABL e13a3 transcript caused by complex chromosome
rearrangement. Int J Hematol. 90:230–234. 2009. View Article : Google Scholar : PubMed/NCBI
|