Introduction
Chemoradiation therapy (CRT) has become widely
accepted as the standard treatment for locally advanced cervical
cancer, as declared in the Clinical Announcement by the US National
Cancer Institute in 1999 (1,2). However, the benefits of CRT decrease
with increasing International Federation of Gynecology and
Obstetrics (FIGO) stage (3).
Additionally, CRT may only benefit 1 out of 10 patients with
cervical cancer, due to differences between tumors and the high
frequency of side effects (4).
Therefore, a more precise stratification of the patients that
respond to CRT and the identification of prognostic markers for
cervical cancer are required.
Several pretreatment variables have been associated
with the clinical outcome of patients with cervical carcinoma,
including the FIGO stage, tumor size, lymph node status, hemoglobin
level, histological type and patient age (5). Previously, in addition to the
clinicopathological factors, numerous molecular markers in cervical
cancer tissue specimens or serum samples have been investigated to
identify the association between the survival rate and response to
CRT (6,7). According to the studies reviewed by
Noordhuis et al, angiogenesis and hypoxia markers, including
hypoxia-inducible factor-1α and vascular endothelial growth factor,
epidermal growth factor receptor pathway markers and
cyclooxygenase-2 were identified as prognostic markers, mainly by
immunohistochemistry (6). In addition
to protein biomarkers, which are derived from messenger RNAs
(mRNAs), recent prognostic studies have investigated a large group
of non-coding RNAs (ncRNAs).
Although ncRNAs are transcribed from the genomic
region, they are not translated into proteins (8). In higher organisms the
non-protein-coding regions of DNA are increased; in humans, ~98% of
DNA consists of non-protein coding regions (9), and numerous ncRNAs may be transcribed
from these regions. Furthermore, a recent study has suggested that
numerous types of ncRNAs are also transcribed from protein-coding
regions of DNA (10). Therefore,
ncRNAs are hypothesized to be important in determining the
complexity of higher eukaryotes and other physiological phenomena
and are considered to be as critical as mRNA coding proteins.
Although the functions of ncRNAs are not well known, ncRNAs may be
crucial in the development, physiology and pathology of cells,
including cancer. MicroRNAs, which belong to a small group of
ncRNAs, are the most well-studied ncRNAs, including microRNA-200a
and microRNA-9, which have been demonstrated to be predictors of
cervical cancer (11). Long
non-coding RNAs (lncRNAs) are tentatively defined as ncRNAs >200
nucleotides in length (12–14). Numerous lncRNAs are downregulated in
various cancers, which demonstrate oncogenic and tumor suppressive
roles (13). There are >60 lncRNAs
reported to be associated with cancer (13); however, the function of the majority
of lncRNAs, and the association between lncRNAs and the prognosis
of cervical cancer, remains to be determined (12–14).
In general, clinical tissue samples are preserved in
formalin-fixed, paraffin-embedded (FFPE) blocks. Previous studies
demonstrate the feasibility of quantifying gene expression by using
RNA isolated from blocks of FFPE tumor tissue (15–18).
The aim of the present retrospective study was to
examine expression of lncRNAs in pretreatment FFPE tissue samples
of patients with cervical squamous cell carcinoma (CSCC) that
underwent platinum-based CRT, and to analyze the association
between lncRNA expression and the clinical prognosis of the
patients.
Materials and methods
Patients and tissue samples
Between March 2001 and March 2013, consecutive
patients that were treated with definitive CRT for cervical cancer
at the University of Tokyo Hospital (Tokyo, Japan) and met the
following eligibility criteria were selected for the present study:
A diagnosis of pathologically confirmed CSCC; FFPE tumor tissue
biopsies were available prior to treatment; CSCC tumors were FIGO
clinical stage IB1/IIA1, with pelvic lymph node (PLN) metastasis,
or stage IB2 or IIA2-IVA; the patients had undergone complete
definitive CRT without any prior treatment for CSCC; and the
patients had been followed-up at the University of Tokyo Hospital.
Patients with distant metastasis, including para-aortic lymph node
metastasis or uncontrollable other malignancies, were excluded from
the present study.
Sections from the FFPE tumor blocks were stained
with hematoxylin and eosin (Merck Millipore, Darmstadt, Germany),
and reviewed by two experienced pathologists at the University of
Tokyo Hospital to determine the suitability for analysis of the
tumor content. Among the 58 patients that met the eligibility
criteria, 9 were excluded from the present study, generally due to
low tumor content in the FFPE samples. Therefore, 49 patients were
analyzed in the current study.
The median age of the patients was 55 years (range,
29–82 years). The majority of patients were diagnosed with a FIGO
stage of IIIB (n=24). The median maximum tumor diameter was 5.5 cm
(range, 2.0–9.7 cm), as measured by T2 weighted magnetic resonance
imaging (MRI). Patient and tumor characteristics are reported in
Table I.
 | Table I.Patient and tumor
characteristics. |
Table I.
Patient and tumor
characteristics.
| Characteristic | n (%) |
|---|
| Total | 49 (100) |
| Age range, years
(median) | 29–82 (55) |
| FIGO stage |
|
| IB | 2 (4) |
|
IIA | 1 (2) |
|
IIB | 10 (20) |
|
IIIA | 7 (14) |
|
IIIB | 24 (49) |
|
IVA | 5 (10) |
| Pelvic lymph node
metastasis |
|
|
Positive | 17 (35) |
|
Negative | 32 (65) |
| Initial hemoglobin
range, g/dl (median) | 6.9–14.2
(12.0) |
| Maximum tumor
diameter range, cm (median) | 2.0–9.7 (5.5) |
| Concurrent
chemotherapy |
|
|
Tri-weekly CDDP 75
mg/m2 | 18 (37) |
|
Tri-weekly NDP 75–100
mg/m2 | 18 (37) |
| Weekly
CDDP 40 mg/m2 | 12 (24) |
|
Other | 1 (2) |
| RT duration range,
days (median) | 35–89 (45) |
The last patient follow-up took place in March 2014.
At the time of data analysis, 34 patients (69%) were alive and 15
patients (31%) had succumbed to cause-specific (13 patients; 27%)
and other (2 patients; 4%) diseases. The overall median follow-up
period was 44.1 months (range, 5.2–142.1 months).
The present study was approved by the Institutional
Review Board of the University of Tokyo Hospital {approved on May
21, 2013 [no. 10152]; minor revision approved on September 9, 2013
[no. 10152-(1)] and January 30, 2014
[no. 10152-(2)]}. Written informed
consent was obtained from all patients. All patient identifiers
were removed prior to the analysis of the data.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
From each FFPE block, 10–15 tissue cores, measuring
10 µm in thickness, were removed. Total RNA was extracted from the
tissue cores with RecoverAll™ Total Nucleic Acid Isolation kit for
FFPE (Thermo Fisher Scientific, Inc., Waltham, MA, USA), according
to the manufacturer's protocol. Complementary DNA (cDNA) was
obtained by reverse transcribing the total RNA with a PrimeScript™
RT reagent kit with gDNA Eraser (Perfect Real Time) (Takara Bio
Inc., Otsu, Shiga, Japan). RT-qPCR was performed with
SYBR® Premix Ex Taq™ II (Tli RNaseH Plus; Takara Bio
Inc.) and the Eco Real-Time PCR System (Illumina, Inc., San Diego,
CA, USA). lncRNA values were normalized to those of GAPDH, which
was used as an internal control.
lncRNAs examined
The present study focused on lncRNAs that had been
implicated in various types of cancer, according to previous
studies (12–14). Thus, the follow 5 lncRNAs were
examined: X inactive-specific transcript (XIST), Tsix, telomerase
RNA component (TERC), dihydrofolate reductase (DHFR) upstream
transcripts, antisense insulin-like growth-factor type-II receptor
RNA (Air).
Primers
The primer sequences used to amplify GAPDH and the 5
lncRNAs are reported in Table II.
Since FFPE treatment and storage of tissue samples may lead to
damage across the length of RNA, primers were used to generate
amplicons that were as short as possible. These primers were
selected from previous studies (19–22) or
produced using GENETYX®-MAC software, version 13
(GENETYX Co., Tokyo, Japan).
 | Table II.Primer sequences for GAPDH and 5
lncRNAs. |
Table II.
Primer sequences for GAPDH and 5
lncRNAs.
|
Oligonucleotides | Sequence |
|---|
| GAPDH (20) |
|
|
Foward |
GCACCGTCAAGGCTGAGAAC |
|
Reverse |
TGGTGAAGACGCCAGTGGA |
| XIST (21) |
|
|
Foward |
AATGGAACGGGCTGAGTTTTAG |
|
Reverse |
TCATCCGCTTGCGTTCATAG |
| Tsix (21) |
|
|
Foward |
AGTTGTGACCGATTTGGAGGGCTTACG |
|
Reverse |
GTATGGAGTCACCAGGTTCCCAGAGAAAGAC |
| TERCa |
|
|
Foward |
TTCAGGCCGCAGGAAGAGGA |
|
Reverse |
ACGTCCCACAGCTCAGGGAA |
| DHFR upstream
transcripts (19) |
|
|
Foward |
ACCTGGTCGGCTGCACCT |
|
Reverse |
TTGCCCTGCCATGTCTCG |
| Air (22) |
|
|
Foward |
GCAGCAAGAAGCACAGCAC |
|
Reverse |
GATGTCTGCGTGGTAACTGG |
Pretreatment evaluation
The pretreatment state of the patients was
clinically evaluated by physical and pelvic examination without
anesthesia, biopsy of the primary tumor, complete blood cell count
and biochemistry profile, computed tomography (CT) of the chest,
abdomen and pelvis, MRI of the pelvis, drip infusion pyelography,
cystoscopy and rectoscopy. [18F]-fluoro-2-deoxy-D-glucose positron
emission tomography (FDG-PET) was routinely performed from 2008.
PLNs >10 mm in diameter that were observed on CT or MRI, or
revealed by FDG-PET were considered to be metastases. The patients
were assigned to a clinical stage on the basis of the FIGO
classification.
Treatment schedule
CRT consisted of an external beam of radiation
therapy (EBRT) to the whole pelvis followed by a high-dose rate of
intra-cavitary brachytherapy (HDR-ICBT) and platinum-based
chemotherapy (CTx).
EBRT was administered to the whole pelvis with 6 or
10 MV photon-beams. No patients received prophylactic
extended-field radiation therapy (RT). The daily dose was 1.8–2.0
Gy, which was administered to the mid-pelvis once a day, 5 days a
week. Depending on the tumor size, the whole pelvis was irradiated
using the four-field box technique up to a dose of 20–40 Gy, then a
boost up to a dose of 50–50.4 Gy was administered to the
parametrium, with a 4 cm-wide central block performed using the
antero-posterior parallel two-field technique. If necessary, a
boost of 10 Gy in 2 Gy fractions was administered to the PLN.
Subsequent to the exposure of the central block to
EBRT, HDR-ICBT was started, using an Iridium-192 remote
afterloading technique, in 6 Gy fractions, once or twice a week,
for a minimum of 3–4 fractions (microSelectron HDR-V3; Elekta AB,
Stockholm, Sweden). In the majority of patients, a tandem and ovoid
applicator (Elekta AB) were used in combination for treatment at
the primary tumor (point A), while in patients with a lower vaginal
extension, a tandem-cylinder (Elekta) was used for treatment at a
distance of 5 mm from the applicator surface. All HDR-ICBTs were
devised based on CT using PLATO software version 14.2.6 (Nucletron,
Veenendaal, The Netherlands) for each application.
The cumulative doses administered to the tumor from
EBRT and HDR-ICBT, were normalized to the biologically equivalent
doses in 2 Gy fractions (GyEQD2) based on the
linear-quadratic model using an α/β ratio of 10 Gy. The median
total dose to point A was 70.9 GyEQD2 (range,
53.4–85.6).
CTx was administered on the first day of RT. Between
2001 and 2007, cisplatin was administered at a dose of 75
mg/m2 tri-weekly, for 3 cycles. Subsequent to 2007,
cisplatin (40 mg/m2 weekly, for 6 cycles) or nedaplatin
(75–100 mg/m2 tri-weekly, for 3 cycles) became the
principal drugs of the CTx regimen. Patients with an advanced age,
lower performance status or renal dysfunction required a reduction
in the CTx dose.
Follow-up and analysis of response and
survival
Follow-up was performed every month for the first
year, every 2–3 months for the second year and 3–6 months
thereafter. Follow-up procedures consisted of physical and pelvic
examination, cervical Papanicolaou smears and tumor markers. Chest
to pelvic CT was performed with an interval of 3–6 months for the
first 2 years, and 6–12 months thereafter. Pelvic MRI or FDG-PET
was performed if required.
Complete remission was defined as no evidence of
disease 3 months subsequent to the end of treatment, as evaluated
by clinical and radiographic examinations. A diagnosis of tumor
progression or recurrence was based on physical or radiographic
examination or pathological confirmation.
Statistical analysis
Statistical analysis was performed using StatView
Dataset File software version 5.0J for Windows (SAS Institute,
Inc., Cary, NC, USA). The survival period was defined as the time
between the start of RT and cancer progression, mortality from any
cause or the last follow-up date. Survival curves were estimated
using the Kaplan-Meier method, and log-rank tests were used to
compare the survival distributions. Differences in patient or tumor
characteristics were analyzed by the χ2 test or Fisher's
exact test for 2 × 2 columns. Cox proportional hazards model was
used for multivariate analysis. P<0.05 was considered to
indicate a statistically significant difference.
Results
The 4-year overall survival (OS) rates were 71.2±6.8
for all 49 patients and 73.5±7.6% for the 36 patients with FIGO
stage III–IVA tumors. Among the 5 lncRNAs, XIST expression was
significantly associated with prognosis. The 49 patients were
classified into high (n=24) and low (n=25) XIST expression groups,
according to the median value of XIST expression. The Kaplan-Meier
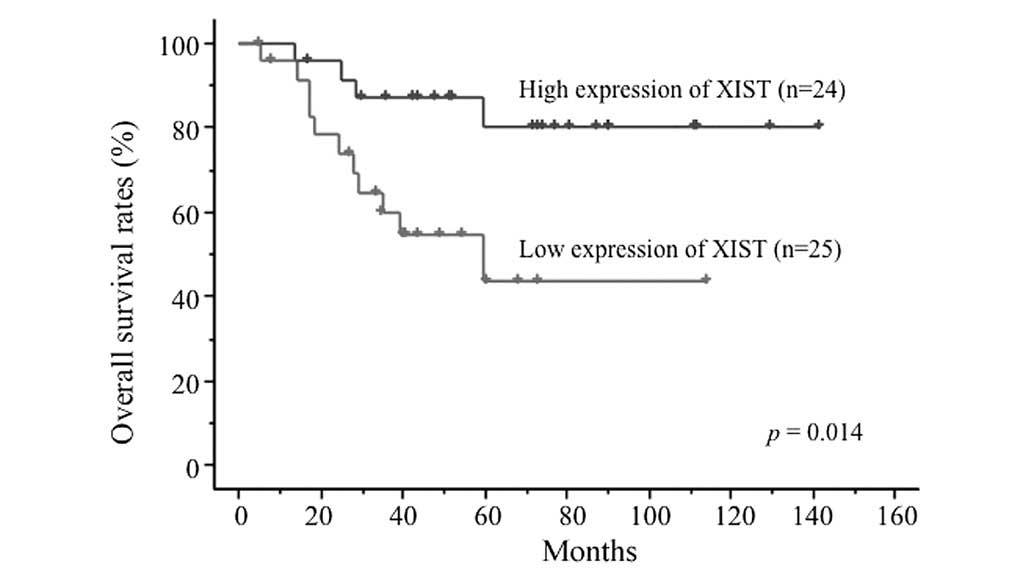
survival curves demonstrated that XIST expression levels were
significantly associated with OS rates. The 4-year OS rates were
87.1±6.9% in the high expression group and 54.4±10.8% in the low
expression group (P=0.014; Fig. 1).
Similarly, the 4-year progression-free survival (PFS) rates were
74.5±9.0 and 49.8±10.3%, in the high and low XIST expression
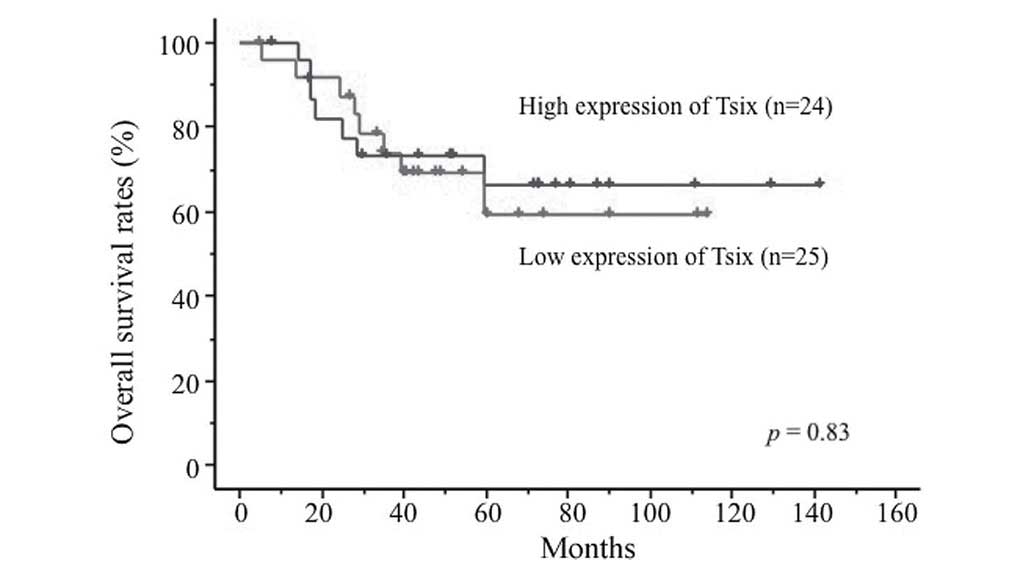
groups, respectively (P=0.065). By contrast, the expression levels
of Tsix, TERC, DHFR upstream transcripts and Air were not
associated with prognosis (Fig. 2;
Table III). Tumor size was also a
variable that was significantly associated with OS rates (P=0.035).
Clinicopathological factors were compared between the high XIST
expression and low XIST expression groups (Table IV). XIST upregulation was not
associated with any of the clinicopathological factors.
Multivariate analysis also demonstrated that the XIST expression
level was significantly associated with OS rates (Table V). These results strongly suggest that
XIST expression levels may be a potential prognostic factor for
cervical cancer OS rates. Therefore, the present study concludes
that overexpression of XIST may be important in CSCC progression
and development.
 | Table III.Univariate analysis for OS rate by
log-rank test. |
Table III.
Univariate analysis for OS rate by
log-rank test.
| Variable | n | 2 year OS rate,
% | 4 year OS rate,
% | P-value |
|---|
| Age, years |
|
|
| 0.240 |
|
<56 | 25 | 91.7±5.6 | 75.0±8.8 |
|
|
≥56 | 24 | 82.4±8.0 |
67.0±10.4 |
|
| FIGO stage |
|
|
| 0.780 |
|
I–II | 13 |
81.8±11.6 |
63.6±14.5 |
|
|
III–IV | 36 | 88.8±5.3 | 73.5±7.6 |
|
| Nodal status |
|
|
| 0.450 |
| N0 | 32 | 89.9±5.5 | 68.0±8.9 |
|
| N1 | 17 | 82.4±9.2 |
76.5±10.3 |
|
| Max. tumor
diameter, cm |
|
|
| 0.035 |
| ≤5 | 21 | 94.7±5.1 | 84.2±8.4 |
|
|
>5 | 28 | 82.0±7.3 | 61.9±9.6 |
|
| RT dose,
GyEQD2 |
|
|
| 0.980 |
|
≤70 | 24 | 82.8±7.8 | 73.5±9.3 |
|
|
>70 | 25 | 91.7±5.6 | 68.8±9.9 |
|
| Initial hemoglobin,
g/dl |
|
|
| 0.500 |
|
≤12 | 25 | 84.0±7.3 | 63.5±9.7 |
|
|
>12 | 24 | 90.9±6.1 | 80.7±8.7 |
|
| XIST |
|
|
| 0.014 |
|
High | 24 | 95.8±4.1 | 87.1±6.9 |
|
|
Low | 25 | 78.4±8.6 |
54.4±10.8 |
|
| Tsix |
|
|
| 0.830 |
|
High | 24 | 82.2±8.1 | 73.0±9.4 |
|
|
Low | 25 | 91.7±5.6 | 69.2±9.8 |
|
| TERC |
|
|
| 0.910 |
|
High | 24 | 87.1±6.9 | 74.1±9.1 |
|
|
Low | 25 | 83.3±7.6 |
65.5±10.0 |
|
| DHFR, upstream
transcripts |
|
|
| 0.910 |
|
High | 24 | 87.1±6.9 | 74.1±9.1 |
|
|
Low | 25 | 87.1±6.9 |
67.6±10.2 |
|
| Air |
|
|
| 0.850 |
|
High | 24 | 82.9±7.8 | 74.2±9.1 |
|
|
Low | 25 | 91.5±5.8 |
67.5±10.3 |
|
 | Table IV.Univariate analysis for XIST
expression by χ2 test. |
Table IV.
Univariate analysis for XIST
expression by χ2 test.
| Variables | High XIST
expression group, n (%) | Low XIST expression
group, n (%) | P-value |
|---|
| Total | 24
(100) | 25
(100) |
|
| Age, years |
|
|
|
|
≤55 | 15 (63) | 10 (40) | 0.12 |
|
≥56 | 9
(38) | 15 (60) |
|
| FIGO stage |
|
|
|
|
I–II | 6
(25) | 7
(28) | 0.81 |
|
III–IV | 18 (75) | 18 (72) |
|
| Nodal status |
|
|
|
| N0 | 13 (54) | 19 (76) | 0.11 |
| N1 | 11 (46) | 6
(24) |
|
| Maximum tumor
diameter, cm |
|
|
|
| ≤5 | 10 (42) | 11 (44) | 0.87 |
|
>5 | 14 (58) | 14 (56) |
|
| RT dose,
GyEQD2 |
|
|
|
|
≤70 | 14 (58) | 10 (40) | 0.20 |
|
>70 | 10 (42) | 15 (60) |
|
| Initial hemoglobin,
g/dl |
|
|
|
|
≤12 | 12 (50) | 13 (52) | 0.89 |
|
>12 | 12 (50) | 12 (48) |
|
 | Table V.Multivariate analysis for XIST
expression OS rates by Cox proportional hazards model. |
Table V.
Multivariate analysis for XIST
expression OS rates by Cox proportional hazards model.
| Variables | HR | 95% CI | P-value |
|---|
| Age |
|
|
|
| XIST,
high vs. low | 0.27 | 0.08–0.86 | 0.027 |
| Age,
<56 vs. ≥56, years | 0.59 | 0.21–1.70 | 0.330 |
| FIGO stage |
|
|
|
| XIST,
high vs. low | 0.26 | 0.08–0.83 | 0.023 |
| FIGO
stage, I–II vs. III–IV | 1.00 | 0.31–3.22 | 1.000 |
| Nodal status |
|
|
|
| XIST,
high vs. low | 0.27 | 0.08–0.87 | 0.028 |
| Nodal
status, N0 vs. N1 | 1.16 | 0.36–3.73 | 0.800 |
| Max. tumor
diameter |
|
|
|
| XIST,
high vs. low | 0.21 | 0.06–0.71 | 0.012 |
| Max.
tumor diameter, ≤5 vs. >5, cm | 0.23 | 0.06–0.84 | 0.027 |
Discussion
The following 5 lncRNAs examined in the present
study had been implicated in cancer, according to previous studies
(12–14): XIST, Tsix, TERC, DHFR upstream
transcripts and Air.
XIST is involved in X chromosome inactivation (XCI)
in the cells of females and allows X chromosome equilibration in
males. XIST expression has been downregulated in various human
cancers. Loss of XCI and downregulation of XIST expression are
commonly observed in basal-like cancer, breast cancer
susceptibility gene 1-null triple negative breast cancer (23–29) and
ovarian cancer cell lines (30,31). In
ovarian cancer cell lines, XIST expression may act as a prognostic
marker associated with a chemotherapeutic response (32).
Tsix is transcribed in the antisense orientation
from XIST and fully overlaps with the XIST gene (33). Tsix inhibits XIST expression in
cis (via interactions between different X chromosome
regions) by several mechanisms. In one mechanism, Tsix binds to
complementary XIST RNA and renders it non-functional. Following
this binding, XIST is made inactive through dicer, which is a type
of endoribonuclease, preferentially cleaves double-stranded RNA
(33). However, other mechanisms are
also currently being studied.
TERC is a component of telomerase that extends
telomeres (34). An increase in TERC
gene expression has been frequently detected in a variety of human
cancers (35). Furthermore,
amplification of TERC and overexpression of telomerase are
associated with cervical tumorigenesis (36). Therefore, testing whether TERC has
been amplified in cervical cancer may be used in addition to
cytology screening and human papilloma virus testing, particularly
in high-risk patients.
DHFR upstream transcripts are transcribed upstream
of the DHFR gene and regulate DHFR expression by forming a triple
helix with the promoter and disassociating from the pre-initiation
complex (19). It has been reported
that this lncRNA may be linked to cancer (13); however, the molecular mechanism
remains unidentified. DHFR is the enzyme that converts
dihydrofolate into tetrahydrofolate. This reaction is essential for
de novo nucleic acid synthesis. DHFR upstream transcripts
may affect de novo nucleic acid synthesis and this dysbolism
may be conducive to tumorigenesis (37).
The mannose 6-phosphate/insulin-like growth-factor
type-II receptor (M6P/IGF-IIR) is considered to act as a suppressor
of tumor growth in various types of cancer (38). Air regulates genomic imprinting of a
cluster of autosomal genes, including IGF-IIR, Slc22a2 and Slc22a3
in cis in mouse chromosome 17 (39). Although full-length transcripts have
yet to be characterized in humans, this lncRNA may be associated
with human cancers (13) and may
affect cancer-associated gene expression at an epigenetic level
(40).
To the best of our knowledge, the present study is
the first to demonstrate an association between the XIST expression
level and the prognosis of CSCC treated with platinum-based CRT.
High expression levels of XIST were clinically associated with
increased OS rates. This observation reinforces the theory that
XIST lncRNA may be used to predict the prognosis of CSCC treated
with CRT.
Since lncRNAs are produced from the majority of the
regions in the genome, they are emerging as key molecules in human
cancer and may be useful as novel biomarkers for the diagnosis,
prognosis and prediction of response to treatment. In particular,
lncRNAs have been hypothesized to possess tumor suppressive and
oncogenic functions in various cancer types. Homeobox transcript
antisense intergenic RNA (HOTAIR) has been reported to be
associated with the progression and prognosis of cancers, including
breast, esophageal, lung, liver and endometrial carcinomas
(41–46). In endometrial carcinoma, an increased
level of HOTAIR was demonstrated to be associated with the depth of
myometrial invasion, lymphovascular space invasion and a poorer OS
rate. Therefore, this lncRNA may be a novel biomarker of prognosis
in cancer patients (47).
The XIST gene is located in the X chromosome
inactivation center and its product is transcribed from the
inactive X chromosome (48,49). XIST then spreads along the X
chromosome from which it was transcribed. XIST lncRNA is important
in the initiation of XCI in female cells, which achieves dosage
equilibration of X-linked genes with males. Since XCI silences
several hundred genes, including oncogenes, misexpression of XIST
may potentially be a mechanism underlying tumorigenesis (49–51).
Recently, Yildirim et al demonstrated a causal link between
XIST expression and cancer in mice (52). Deleting XIST in the mouse blood
compartment was found to lead to a highly aggressive
myeloproliferative neoplasm and myelodysplastic syndrome with 100%
penetrance (52). This result
suggests that upregulation of X-linked genes following the deletion
of XIST leads to genome-wide alterations in key developmental and
homeostatic pathways, which in turn drive cancer formation and
progression (52). Furthermore, in
theory, XIST is particularly important in female-specific cancer.
Certain studies have claimed that XIST is involved in female
cancers (14,23–32). Taken
together, these findings, and the results from the present study,
demonstrate that a low XIST level may lead to the depression of X
chromosome inactivation and the continuous gain of X chromosome
reactivation. This phenomenon may, in turn, upregulate the
expression of numerous cancer-associated genes, including genes
responsible for the aggressiveness of cancer, in the X chromosome.
This may lead to low XIST group patients not being treated
effectively with CRT. In addition, high XIST levels may trigger the
reduction of numerous genes responsible for RT and/or CTx
resistance in the X chromosome, and this may boost the
effectiveness of RT and CTx.
Since XIST is able to control cancer, it may be
reasonable to propose the reactivation of XIST as a therapeutic
strategy in cancer. In tumors, XIST expression may be reactivated
by small molecules, such as XIST expression vector, offering a
novel therapeutic approach that would target epigenetically
functional lncRNAs. If molecules that induce XIST expression are
identified, artificially improving the prognosis of a patient may
become feasible.
It is notable that there was no association between
XIST and Tsix in the present study. In general, Tsix is transcribed
from the antisense lncRNA XIST and controls XIST upregulation, and
XIST and Tsix often work in combination (53). However, the inherent functions of XIST
are considered to be associated with the prognosis of CSCC. The
findings of the present study may lead to an improved understanding
of the emerging roles of XIST in cancer therapy.
Comprehensive analysis of gene expression using RNA
from fresh frozen tumor specimens is important to improve the
understanding of cancer pathogenesis, progression and prognosis.
However, it is challenging for studies to treat frozen tumor
specimens with degradable biomolecules, as frozen tissue samples
are not readily available. By contrast, since all biomolecules are
fixed, FFPE tissues may be treated at room temperature for extended
periods of time, and therefore FFPE tissues are the most widely
available specimens. Although formalin may cause the degradation
and modification of nucleic acids, leading to a poor recovery of
nucleic acid from preserved tissues, presently there are various
commercially available kits for the isolation of nucleic acids from
FFPE tissue sections (15–18).
Various types of protein markers for the prognosis
of cancer have been explored by numerous studies. At present,
lncRNAs, in addition to HOTAIR and XIST, may also be considered as
candidates for prognostic biomarker roles. Experiments to
investigate the role lncRNAs play in cancer may be performed using
FFPE specimens routinely preserved in numerous hospitals or
laboratories. Additionally, as RT-qPCR possesses a high
quantitative capability, precisely-analyzed data may be obtained.
In the present study, only 5 lncRNAs were analyzed; there may be
other lncRNAs that are associated with the prognosis of CSCC
patients. Additional studies are required to investigate other
lncRNAs that may associate with cancer prognosis and the mechanisms
by which they affect cancer.
There were certain limitations to the present study.
First, due to the nature of a retrospective study, there was a lack
of consistency of treatment. Second, the present study investigated
a small sample size. Over a 13-year period, 263 patients were
referred for definitive RT for cervical cancer treatment at the
University of Tokyo Hospital. Of these 263 patients, only 49 were
included in the present study. The following patients were excluded
from the present study: 104 patients that partially received
HDR-ICBT at the University of Tokyo Hospital and received EBRT and
CTx at other institutions; 4 patients with other malignancies; 8
patients that received prior treatment for cervical cancer; 7
patients without detailed information of treatment or without
follow-up; 14 patients with carcinoma in situ or FIGO stage
IA, IB1 or IIA1 without PLN metastasis; 18 patients with
para-aortic lymph node metastasis; 3 patients with other distant
metastasis; 3 patients with distant metastasis and para-aortic
lymph node metastasis; and 1 patient that did not complete the
planned RT. Out of the remaining 101 patients, 76 patients received
CRT and 25 received RT alone. Among the 76 patients treated with
CRT, 58 were diagnosed with squamous cell carcinoma. The FFPE
tissue samples from 9 patients were unsuitable or lost. Therefore,
the remaining 49 patients were included in the present study. To
minimize variation that may have affected prognosis, the
eligibility criteria were specific, and so the data gathered in the
present study were reliable.
In conclusion, the results of the present study
indicate that the expression of XIST associates with the OS rates
of CSCC patients. However, additional studies are required to
investigate the underlying mechanisms of XIST associated with
prognosis, including the regulation of the response to CRT or
progression of CSCC. Additional studies may reveal novel
therapeutic strategies for CSCC treatment.
References
|
1
|
NCI Press Office, . NCI issues clinical
announcement on cervical cancer: Chemotherapy plus radiation
improves survival. http://www.nih.gov/news/pr/feb99/nci-22.htmAccessed.
August 28–2014
|
|
2
|
NCCN Guidelines®, . NCCN clinical practice
guidelines in oncology: Cervical cancer version 1, 2016. National
Comprehensive Cancer Network, Inc.; Fort Washington, PA, USA:
2015
|
|
3
|
Chemoradiotherapy for Cervical Cancer
Meta-Analysis Collaboration, . Reducing uncertainties about the
effects of chemoradiotherapy for cervical cancer: A systematic
review and meta-analysis of individual patient data from 18
randomized trials. J Clin Oncol. 26:5802–5812. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nakano T, Ohno T, Ishikawa H, Suzuki Y and
Takahashi T: Current advancement in radiation therapy for uterine
cervical cancer. J Radiat Res. 51:1–8. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hauspy J, Harley I and Fyles AW: Uterine
cervix cancerPrognostic Factors in Cancer. Gospodarowicz MK,
O'Sullivan B and Sobin LH: 3rd. John Wiley & Sons Inc.;
Hoboken, NJ: pp. 219–222. 2006
|
|
6
|
Noordhuis MG, Eijsink JJ, Roossink F, de
Graeff P, Pras E, Schuuring E, Wisman GB, de Bock GH and van der
Zee AG: Prognostic cell biological markers in cervical cancer
patients primarily treated with (chemo)radiation: A systematic
review. Int J Radiat Oncol Biol Phys. 79:325–334. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Harima Y, Ikeda K, Utsunomiya K, Komemushi
A, Kanno S, Shiga T and Tanigawa N: Apolipoprotein C-II is a
potential serum biomarker as a prognostic factor of locally
advanced cervical cancer after chemoradiation therapy. Int J Radiat
Oncol Biol Phys. 87:1155–1161. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cech TR and Steitz JA: The noncoding RNA
revolution-trashing old rules to forge new ones. Cell. 157:77–94.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Elgar G and Vavouri T: Tuning in to the
signals: Noncoding sequence conservation in vertebrate genomes.
Trends Genet. 24:344–352. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang Y, Yang L and Chen LL: Life without
A tail: New formats of long noncoding RNAs. Int J Biochem Cell
Biol. 54:338–349. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hu X, Schwarz JK, Lewis JS Jr, Huettner
PC, Rader JS, Deasy JO, Grigsby PW and Wang XA: A microRNA
expression signature for cervical cancer prognosis. Cancer Res.
70:1441–1448. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hauptman N and Glavač D: Long non-coding
RNA in cancer. Int J Mol Sci. 14:4655–4669. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cheetham SW, Gruhl F, Mattick JS and
Dinger ME: Long noncoding RNAs and the genetics of cancer. Br J
Cancer. 108:2419–2425. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ismailov S, Rashitov M, Kobayashi M,
Shibata N, Kato Y, Omi Y, Iihara M and Okamoto T: Trefoil factor 3
(TFF3) mRNA expression level in follicular thyroid tumors using
formalin-fixed, paraffin-embedded (FFPE) blocks. O J Pathol.
3:78–84. 2013. View Article : Google Scholar
|
|
16
|
von Ahlfen S, Missel A, Bendrat K and
Schlumpberger M: Determinants of RNA quality from FFPE samples.
PLoS One. 2:e12612007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sánchez-Navarro I, Gámez-Pozo A,
González-Barón M, Pinto-Marín A, Hardisson D, López R, Madero R,
Cejas P, Mendiola M, Espinosa E and Vara JA: Comparison of gene
expression profiling by reverse transcription quantitative PCR
between fresh frozen and formalin-fixed, paraffin-embedded breast
cancer tissues. Biotechniques. 48:389–397. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mittempergher L, de Ronde JJ, Nieuwland M,
Kerkhoven RM, Kerkhoven RM, Simon I, Rutgers EJ, Wessels LF and
Van't Veer LJ: Gene expression profiles from formalin fixed
paraffin embedded breast cancer tissue are largely comparable to
fresh frozen matched tissue. PLoS One. 6:e171632011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Martianov I, Ramadass A, Barros A Serra,
Chow N and Akoulitchev A: Repression of the human dihydrofolate
reductase gene by a non-coding interfering transcript. Nature.
445:666–670. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Miyagawa R, Tano K, Mizuno R, Nakamura Y,
Ijiri K, Rakwal R, Shibato J, Masuo Y, Mayeda A, Hirose T and
Akimitsu N: Identification of cis- and trans-acting factors
involved in the localization of MALAT-1 noncoding RNA to nuclear
speckles. RNA. 18:738–751. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kumamoto T and Oshio S: Effect of fetal
exposure to bisphenol A on brain mediated by X-chromosome
inactivation. J Toxicol Sci. 38:485–494. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Latos PA, Pauler FM, Koerner MV, et al:
Airn transcriptional overlap, but not its lncRNA products, induces
imprinted Igf2r silencing. Science. 338:1469–1472. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ganesan S, Silver DP, Greenberg RA, Avni
D, Drapkin R, Miron A, Mok SC, Randrianarison V, Brodie S, Salstrom
J, et al: BRCA1 supports XIST RNA concentration on the inactive X
chromosome. Cell. 111:393–405. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pageau GJ, Hall LL and Lawrence JB: BRCA1
does not paint the inactive X to localize XIST RNA but may
contribute to broad changes in cancer that impact XIST and Xi
heterochromatin. J Cell Biochem. 100:835–850. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Richardson AL, Wang ZC, De Nicolo A, Lu X,
Brown M, Miron A, Liao X, Iglehart JD, Livingston DM and Ganesan S:
X chromosomal abnormalities in basal-like human breast cancer.
Cancer Cell. 9:121–132. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Silver DP, Dimitrov SD, Feunteun J, Gelman
R, Drapkin R, Lu SD, Shestakova E, Velmurugan S, Denunzio N,
Dragomir S, et al: Further evidence for BRCA1 communication with
the inactive X chromosome. Cell. 128:991–1002. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sirchia SM, Ramoscelli L, Grati FR,
Barbera F, Coradini D, Rossella F, Porta G, Lesma E, Ruggeri A,
Radice P, et al: Loss of the inactive X chromosome and replication
of the active X in BRCA1-defective and wild-type breast cancer
cells. Cancer Res. 65:2139–2146. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sirchia SM, Tabano S, Monti L, Recalcati
MP, Gariboldi M, Grati FR, Porta G, Finelli P, Radice P and Miozzo
M: Misbehaviour of XIST RNA in breast cancer cells. PLoS One.
4:e55592009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vincent-Salomon A, Ganem-Elbaz C, Manié E,
Raynal V, Sastre-Garau X, Stoppa-Lyonnet D, Stern MH and Heard E: X
inactive-specific transcript RNA coating and genetic instability of
the X chromosome in BRCA1 breast tumors. Cancer Res. 67:5134–5140.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Benoît MH, Hudson TJ, Maire G, Squire JA,
Arcand SL, Provencher D, Mes-Masson AM and Tonin PN: Global
analysis of chromosome X gene expression in primary cultures of
normal ovarian surface epithelial cells and epithelial ovarian
cancer cell lines. Int J Oncol. 30:5–17. 2007.PubMed/NCBI
|
|
31
|
Kawakami T, Zhang C, Taniguchi T, Kim CJ,
Okada Y, Sugihara H, Hattori T, Reeve AE, Ogawa O and Okamoto K:
Characterization of loss-of-inactive X in Klinefelter syndrome and
female-derived cancer cells. Oncogene. 23:6163–6169. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huang KC, Rao PH, Lau CC, Heard E, Ng SK,
Brown C, Mok SC, Berkowitz RS and Ng SW: Relationship of XIST
expression and responses of ovarian cancer to chemotherapy. Mol
Cancer Ther. 1:769–776. 2002.PubMed/NCBI
|
|
33
|
Lee JT, Davidow LS and Warshawsky D: Tsix,
a gene antisense to Xist at the X-inactivation centre. Nat Genet.
21:400–404. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Feng J, Funk WD, Wang SS, Weinrich SL,
Avilion AA, Chiu CP, Adams RR, Chang E, Allsopp RC, Yu J, et al:
The RNA component of human telomerase. Science. 269:1236–1241.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cao Y, Bryan TM and Reddel RR: Increased
copy number of the TERT and TERC telomerase subunit genes in cancer
cells. Cancer Sci. 99:1092–1099. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tu Z, Zhang A, Wu R, Jiang J, Li Y, Wulan
N, Li J, Zhang Y, Li Y, Chen Z and Wei L: Genomic amplification of
the human telomerase RNA gene for differential diagnosis of
cervical disorders. Cancer Genet Cytogenet. 191:10–16. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hitchings GH Jr: Nobel lecture in
physiology or medicine - 1988. Selective inhibitors of
dihydrofolate reductase. In Vitro Cell Dev Biol. 25:303–310. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
El-Shewy HM and Luttrell LM: Insulin-like
growth factor-2/mannose-6 phosphate receptors. Vitam Horm.
80:667–697. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nagano T, Mitchell JA, Sanz LA, Pauler FM,
Ferguson-Smith AC, Feil R and Fraser P: The Air noncoding RNA
epigenetically silences transcription by targeting G9a to
chromatin. Science. 322:1717–1720. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mohammad F, Mondal T and Kanduri C:
Epigenetics of imprinted long noncoding RNAs. Epigenetics.
4:277–286. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chisholm KM, Wan Y, Li R, Montgomery KD,
Chang HY and West RB: Detection of long non-coding RNA in archival
tissue: Correlation with polycomb protein expression in primary and
metastatic breast carcinoma. PLoS One. 7:e479982012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen FJ, Sun M, Li SQ, Wu QQ, Ji L, Liu
ZL, Zhou GZ, Cao G, Jin L, Xie HW, et al: Upregulation of the long
non-coding RNA HOTAIR promotes esophageal squamous cell carcinoma
metastasis and poor prognosis. Mol Carcinog. 52:908–915. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liu XH, Liu ZL, Sun M, Liu J, Wang ZX and
De W: The long non-coding RNA HOTAIR indicates a poor prognosis and
promotes metastasis in non-small cell lung cancer. BMC Cancer.
13:4642013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yang Z, Zhou L, Wu LM, Lai MC, Xie HY,
Zhang F and Zheng SS: Overexpression of long non-coding RNA HOTAIR
predicts tumor recurrence in hepatocellular carcinoma patients
following liver transplantation. Ann Surg Oncol. 18:1243–1250.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Geng YJ, Xie SL, Li Q, Ma J and Wang GY:
Large intervening non-coding RNA HOTAIR is associated with
hepatocellular carcinoma progression. J Int Med Res. 39:2119–2128.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
He X, Bao W, Li X, Chen Z, Che Q, Wang H
and Wan XP: The long non-coding RNA HOTAIR is upregulated in
endometrial carcinoma and correlates with poor prognosis. Int J Mol
Med. 33:325–332. 2014.PubMed/NCBI
|
|
48
|
Brown CJ, Ballabio A, Rupert JL,
Lafreniere RG, Grompe M, Tonlorenzi R and Willard HF: A gene from
the region of the human X inactivation centre is expressed
exclusively from the inactive X chromosome. Nature. 349:38–44.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Weakley SM, Wang H, Yao Q and Chen C:
Expression and function of a large non-coding RNA gene XIST in
human cancer. World J Surg. 35:1751–1756. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu Y, Wang L and Zheng P: X-linked tumor
suppressors: Perplexing inheritance, a unique therapeutic
opportunity. Trends Genet. 26:260–265. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Spatz A, Borg C and Feunteun J:
X-chromosome genetics and human cancer. Nat Rev Cancer. 4:617–629.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yildirim E, Kirby JE, Brown DE, Mercier
FE, Sadreyev RI, Scadden DT and Lee JT: Xist RNA is a potent
suppressor of hematologic cancer in mice. Cell. 152:727–742. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Froberg JE, Yang L and Lee JT: Guided by
RNAs: X-inactivation as a model for lncRNA function. J Mol Biol.
425:3698–3706. 2013. View Article : Google Scholar : PubMed/NCBI
|