Introduction
Breast cancer is the most common type of cancer
among women and the fifth most common cause of mortality from
cancer in China (1). Triple-negative
breast cancer (TNBC) accounts for between 10 and 17% of cases of
breast cancer (2,3). TNBC is an immunohistochemical
description of breast cancer characterized by negative staining for
estrogen receptor (ER), progesterone receptor (PR) and human
epidermal growth factor receptor 2 (HER2) (4). TNBC is associated with younger women and
with an increased risk of visceral and/or central nervous system
metastases during the first 1–3 years of follow-up following
diagnosis (5). Owing to the
underlying biological heterogeneity within TNBC and lack of special
target therapy, the concept of a standard approach to TNBC
treatment is inappropriate (6). TNBC
is always more shared in individuals harboring mutations of breast
cancer type 1/2 susceptibility proteins (BRCA1/2), and aberrations
in BRCA1/2 may sensitize breast cancer cells to cisplatin (7). The addition of cisplatin significantly
improved survival times in unselected patients in 1988, with other
studies confirming activity for cisplatin when used as first-line
chemotherapy in TNBC (8–10). Results indicated that the combination
of gemcitabine and cisplatin gave a favorable clinical response and
managed toxicity as a first-line chemotherapeutic agent in patients
with metastatic TNBC, in particular patients with the basal-like
disease subtype (8). However, the
molecular mechanisms underlying cisplatin sensitivity and cisplatin
resistance in TNBC are unclear. Previous studies have demonstrated
that platinum-based combination chemotherapy may represent an
optimum treatment in TNBC, particularly when patients received
anthracycline and/or taxanes, or exhibit homogeneous DNA damage
repair defections (9,11). Although promising, treatment with
cisplatin-based combination therapies faces certain hurdles,
including the mechanism of platinum resistance and lack of
biomarkers to predict responses to cisplatin (12,13).
Methylation of the ERα promoter has previously been
demonstrated to be associated with sporadic TNBC, ranging between
30 and 40% in breast cancer samples. It has been demonstrated that
methylation of the ERα1, ERα3, ERα4 or ERα5 promoters is associated
with basal-like breast cancer (14).
ERα and BRCA1 promoter methylation may contribute to poor
disease-free survival (DFS) and overall survival (OS) times.
Fanconi anemia complementation group F (FANCF) promoter methylation
in its CpG islands caused the inactivation of FANCF and acquired
cisplatin resistance during tumor progression in ovarian tumors
(15). It is not clear whether the
ERα methylation would have a similar effect to that in FANCF,
causing primary cisplatin resistance in TNBC. To investigate this
hypothesis, ERα methylation in the same panel of primary or
recurrent breast tumor samples from patients with breast cancer was
measured; subsequently, a cisplatin sensitivity test was conducted
and the association between ERα methylation and cisplatin
resistance was evaluated. To the best of our knowledge, the present
study is the first to be designed to identify ERα methylation
markers of resistance in patients with TNBC.
Materials and methods
Patient materials
Between March 2013 and July 2015, 35 women (median
age, 47 years; range, 27–69 years) with TNBC were enrolled in the
present study. Primary breast tumor tissues or recurrent breast
tumor tissues were obtained by surgical resection at the Department
of Breast Surgery or Department of Medical Oncology, Liaoning
Cancer Hospital and Institute (Shenyang, China). Tissue sections
containing >30% tumor cells were selected to detect drug
sensitivity including more than eight protocols, based on the
National Comprehensive Cancer Network guidelines (16), including cisplatin-based chemotherapy
with MTT methods. The association between cisplatin resistance
in vitro and clinicopathological characteristics was
analyzed in patients with TNBC. Tumor tissues immunohistochemically
identified as TNBC were identified as ER-negative (threshold value,
1%) (sc-542, Santa Cruz, 1:200), PR-negative (sc-539, Santa Cruz,
1:250) and HER2-negative [immunohistochemistry (IHC, sc-08, Santa
Cruz, 1:500) 0/1+, or IHC 2+/fluorescence in situ
hybridization (Hercep TestTM, DAKP A/S, Glostrup, Denmark)
non-amplified from the archived pathological reports in the
Liaoning Cancer Hospital and Institute]. The present study used
research protocols approved by the Liaoning Cancer Hospital and
Institute. All samples were obtained with the patient's informed
consent. Diagnoses were confirmed by review of clinicopathological
features; the clinical data collected included age, family
histology, tumor grade, hormone receptor status, lymph node status
and tumor size.
Methylation-specific polymerase chain
reaction (PCR) (MSP)
DNA extract of ERα1, ERα3, ERα4 and ERα5 was
isolated from tumor tissues using phenol/chloroform extraction and
ethanol precipitation in high-solubility SDS/proteinase K solution.
DNA concentration was qualified by determination of optical density
(OD)260/280 and amplified with specific unmethylated and
methylated sequences primers using MSP. Sodium bisulfite-treated
DNA was amplified using methylation- and unmethylation-specific
primers (presented in Table I) and
designated M label and U label, respectively. A total of 2 µg DNA
was denatured using NaOH (final concentration, 0.2 M) for 10 min at
37°C. For samples with 2 µg DNA, salmon sperm DNA (Sigma Aldrich;
Merck KgaA, Darmstadt, Germany) was added as carrier prior to
modification. A total of 30 µl of 10 mM hydroquinone (Sigma
Aldrich; Merck KGaA) and 3 M sodium bisulfite (Sigma Aldrich; Merck
KGaA) at pH 5 were added and mixed, and samples were incubated
under mineral oil at 50°C for 16 h. ERα1, ERα3, ERα4 and ERα5 for
MSP using the six primer pairs as described previously (14) and purified using the Promega Wizard
Genomic DNA Purification kit (A1120, Promega Corporation, Madison,
WI, USA). The positive control consisted of alleles from healthy
volunteers methylated with SssI methyltransferase (New England
Biolabs, Inc., Ipswich, MA, USA), and the negative control was
modified using RNA-free water. PCR amplification was performed with
the reaction mixtures including 12.5 µl Premix Taq with 1 µl (20
µM) of each primer and 100 ng bisulfite-modified DNA template, with
a final volume of 25 µl. The thermocycling conditions were:
Denaturation by heating to 95°C for 10 min, followed by 14
amplification cycles of 94°C for 30 sec, 62°C (ERα1) or 59°C (ERα3,
ERα4 and ERα5) for 45 sec (−0.5°C decreased/cycle) and 72°C for 45
sec, ending with a final extension of 72°C for 10 min. The PCR
products were separated on a 1% agarose gel stained with
GeneFinder™ and images captured by Fluorchem 5500 (ProteinSimple,
San Jose, CA, USA). Methylation was considered to be present if the
methylated label was detected.
 | Table I.Primer pair sequences of ERα1, ERα3,
ERα4 and ERα5. |
Table I.
Primer pair sequences of ERα1, ERα3,
ERα4 and ERα5.
| Name | Primer pair
sequences | Size, bp | Sitea, bp |
|---|
| ERα1 U |
5′-TTTTGGGATTGTATTTGTTTTTGTTG-3′ | 192 | +44 |
|
|
5′-AAACAAAATACAAACCATATCCCCA-3′ |
|
|
| ERα1 M |
5′-TTTTGGGATTGTATTTGTTTTCGTC-3′ | 192 | +236 |
|
|
5′-AACAAAATACAAACCGTATCCCCG-3′ |
|
|
| ERα3 U |
5′-GGATATGGTTTGTATTTTGTTTGT-3′ | 120 | +225 |
|
|
5′-ACAAACAATTCAAAAACTCCAACT-3′ |
|
|
| ERα3 M |
5′-GATACGGTTTGTATTTTGTTCGC-3′ | 130 | +345 |
|
|
5′-CGAACGATTCAAAAACTCCAACT-3′ |
|
|
| ERα4 U |
5′-ATGAGTTGGAGTTTTTGAATTGTTT-3′ | 158 | +310 |
|
|
5′-ATAAACCTACACATTAACAACAACCA-3′ |
|
|
| ERα4 M |
5′-CGAGTTGGAGTTTTTGAATCGTTC-3′ | 151 | +468 |
|
|
5′-CTACGCGTTAACGACGACCG-3′ |
|
|
| ERα5 U |
5′-GGTGTATTTGGATAGTAGTAAGTTTGT-3′ | 120 | +375 |
|
|
5′-CCATAAAAAAAACCAATCTAACCA-3′ |
|
|
| ERα5 M |
5′-GTGTATTTGGATAGTAGTAAGTTCGTC-3′ | 118 | +495 |
|
|
5′-CGTAAAAAAAACCGATCTAACCG-3′ |
|
|
Drug-sensitivity test in vitro
The drug sensitivity of surgical biopsy or surgical
excision tissues was assessed using an MTT assay. The primary or
recurrent breast cancer tissues were collected from each patient
immediately following surgical removal. The cancer cells were
digested from tissues and cultured in Dulbecco's modified Eagle's
medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
with 10% fetal bovine serum (SH30084.03, Hyclone, Thermo Fisher
Scientific, Inc.) at 37°C with 5% CO2, and were used in
the exponential phase of growth. Cultured cells were treated with
cisplatin (A14200156601, QILU Pharmaceutical) at a final
concentration of 0.2 mg/ml, for 24 or 48 h. Drug sensitivity was
evaluated using an MTT assay. A total of 150 µl MTT solution (Sigma
Aldrich; Merck KGaA) at concentration of 5 mg/ml was added for 2 h
at 37°C. After removing the supernatant, 150 ml DMSO (Sigma
Aldrich; Merck KGaA) was added to each well for absorbance reading
at a wavelength of 490 nm using a plate reader (Tecan
Sunrise™, Tecan Group Ltd., Männedorf, Switzerland).
Western blotting
The proteins from breast cancer tissues were
extracted using a Tissue or Cell Total Protein Extraction Kit
(C510003; Sangon Biotech Co., Ltd., Shanghai, China). Protein
content was determined by the Lowry method using bovine serum
albumin as the standard. The samples containing 100 µg proteins
were separated via SDS-PAGE (10% gel). Following transfer to
polyvinylidene fluoride membranes, the samples were blocked using
5% skim milk powder in TBS-T (30 mM Tris-HCl, 125 mM NaCl, 0.1%
Tween 20) for 1 h at room temperature. The PVDF membranes were
incubated with the primary antibody, specific to either
P-glycoprotein (P-gp; cat no. sc-55510; 1:1,500; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) or BRCA1 (cat no. ab9141;
1:500; Abcam, Cambridge, UK) overnight at 4°C. After washing, the
blots were incubated with peroxidase-conjugated affinity-purified
goat anti-mouse or goat anti-rabbit secondary antibody (1:1,500;
cat nos. sc-395758 and sc-45101, respectively; Santa Cruz
Biotechnology, Inc.), as appropriate. Band density of P-gp or BRCA1
was scanned and measured using Fluorchem 5500 software (Alpha
Innotech Co., San Leandro, CA, USA). The column ratios were
determined through scanned β-actin as calibration.
Statistical analysis
All statistical data are presented as the mean ±
standard deviation and the data analyses were performed using SPSS
for Windows (version 13.0; SPSS, Inc., Chicago, IL, USA). A
χ2 test and Fisher's exact test were used for binary
variable comparisons. PFS and OS curves were constructed using the
Kaplan-Meier method and a log-rank test was used to perform
comparisons between patients with or without ER-α promoter
methylation and cisplatin resistance. Factors influencing cell
inhibition by cisplatin, including ER-α promoter methylation and
other prognostic factors, were investigated through univariate
logistic regression and Cox's proportional hazards regression model
with hazards ratio (HR) calculation with 95% confidence interval
(95% CI). P<0.05 was considered to indicate a statistically
significant difference. OS time was calculated from initiation of
treatment to mortality, and individuals alive were censored at the
time of last follow-up.
Results
Cisplatin resistance in
drug-sensitivity test and association with clinicopathological
characteristics
Between 1 March 2013 and 30 July 2015, 35 women with
TNBC, with a median age of 47 years (range, 27–69 years), were
enrolled. Baseline characteristics of the patients were
well-balanced between the drug-sensitivity groups. The
drug-sensitivity test including assessment of sensitivity to
cisplatin alone and in combination with gemcitabine or capecitabine
in all 35 patients with TNBC. Primary cancer cells from patients
with TNBC were exposed to various concentrations of cisplatin
(final concentration, 0.2 µg/ml) for 48 h. On the basis of the
threshold of the majority of drugs in the antitumor
drug-sensitivity test in vitro, cells whose proliferation
was decreased by ≤30% were defined as cisplatin-resistant. Using
this definition, the results in vitro indicated that 23
individuals exhibited cisplatin sensitivity (inhibition ratio of
breast cancer cell, >30%) and 12 patients exhibited cisplatin
resistance. The inhibition rate of primary breast cancer cells from
premenopausal patients was 47.12% and that of primary breast cancer
cells from postmenopausal patients was 44.79%. Cisplatin resistance
in vitro occurred more often in cells from postmenopausal
patients, patients negative for lymph node metastasis (cell
inhibition ratio, 51.24 vs. 40.78% for lymph node
metastasis-positive and -negative tissues, respectively) and larger
tumor size group (cell inhibition ratio, 49.15 vs. 43.91% for tumor
sizes <2 cm and >2 cm, respectively). However, the
differences in tumor size, lymph node metastasis status, age and
menopausal status were not significant (Table II). All of the patients with ERα
methylation (n=8) exhibited cisplatin insensitivity in
vitro. Cell inhibition ratios were 20.25% in the ERα-methylated
group and 53.44% in the ERα-unmethylated group. The multivariate
Cox's model yielded results for the following: Arm 1, age <40
years; age >40 years [hazard ratio (HR), 0.715; 95% confidence
interval (CI), 0.30–1.70; P=0.715]; arm 2, premenopausal vs.
postmenopausal (HR, 0.850; 95% CI, 0.37–1.97; P=0.711); arm 3,
tumor size <2 cm vs. tumor size >2 cm (HR, 0.850; 95% CI,
0.38–1.93; P=0.698); arm 4, lymph node metastasis-positive vs.
lymph node metastasis-negative (HR, 1.66; 95% CI, 0.83–3.33;
P=0.155); arm 5, ERα methylation vs. ERα unmethylation (HR, 19.41;
95% CI, 4.86–77.53; P<0.001).
 | Table II.Univariate and multivariate model for
cell inhibition by cisplatin in TNBC. |
Table II.
Univariate and multivariate model for
cell inhibition by cisplatin in TNBC.
|
|
| Univariate Cell
inhibition ratio | Multivariate Cell
inhibition ratio |
|---|
|
|
|
|
|
|---|
| Factor | Patients, n | Mean, % | P-value | OR | 95% CI | P-value |
|---|
| Age, years |
|
| 0.594 |
|
| 0.715 |
|
<40 | 20 | 40.40 |
| 0.715 | 0.30–1.70 |
|
|
≥40 | 15 | 53.13 |
|
|
|
|
| Menopausal
status |
|
| 0.693 |
|
| 0.711 |
|
Premenopausal | 19 | 47.12 |
| 0.85 | 0.37–1.97 |
|
|
Postmenopausal | 16 | 44.79 |
|
|
|
|
| Tumor size, cm |
|
| 0.970 |
|
| 0.698 |
|
<2 | 13 | 49.15 |
| 0.85 | 0.38–1.93 |
|
| ≥2 | 22 | 43.91 |
|
|
|
|
| LNM |
|
| 0.182 |
|
| 0.155 |
|
Positive | 18 | 51.24 |
| 1.66 | 0.83–3.33 |
|
|
Negative | 17 | 40.78 |
|
|
|
|
| ERα
methylation |
|
| 0.000 |
|
| <0.001 |
|
Positive | 8 | 20.25 |
| 19.41 | 4.86–77.53 |
|
|
Negative | 27 | 53.44 |
|
|
|
|
Analysis of ERα methylation and
cisplatin resistance in TNBC
Bisulfite-treated DNA was amplified with specific
primers for ERα. If one or more regions were positive for MSP in
the methylation labels ERα1, ERα3, ERα4 and ERα5, the tumor tissue
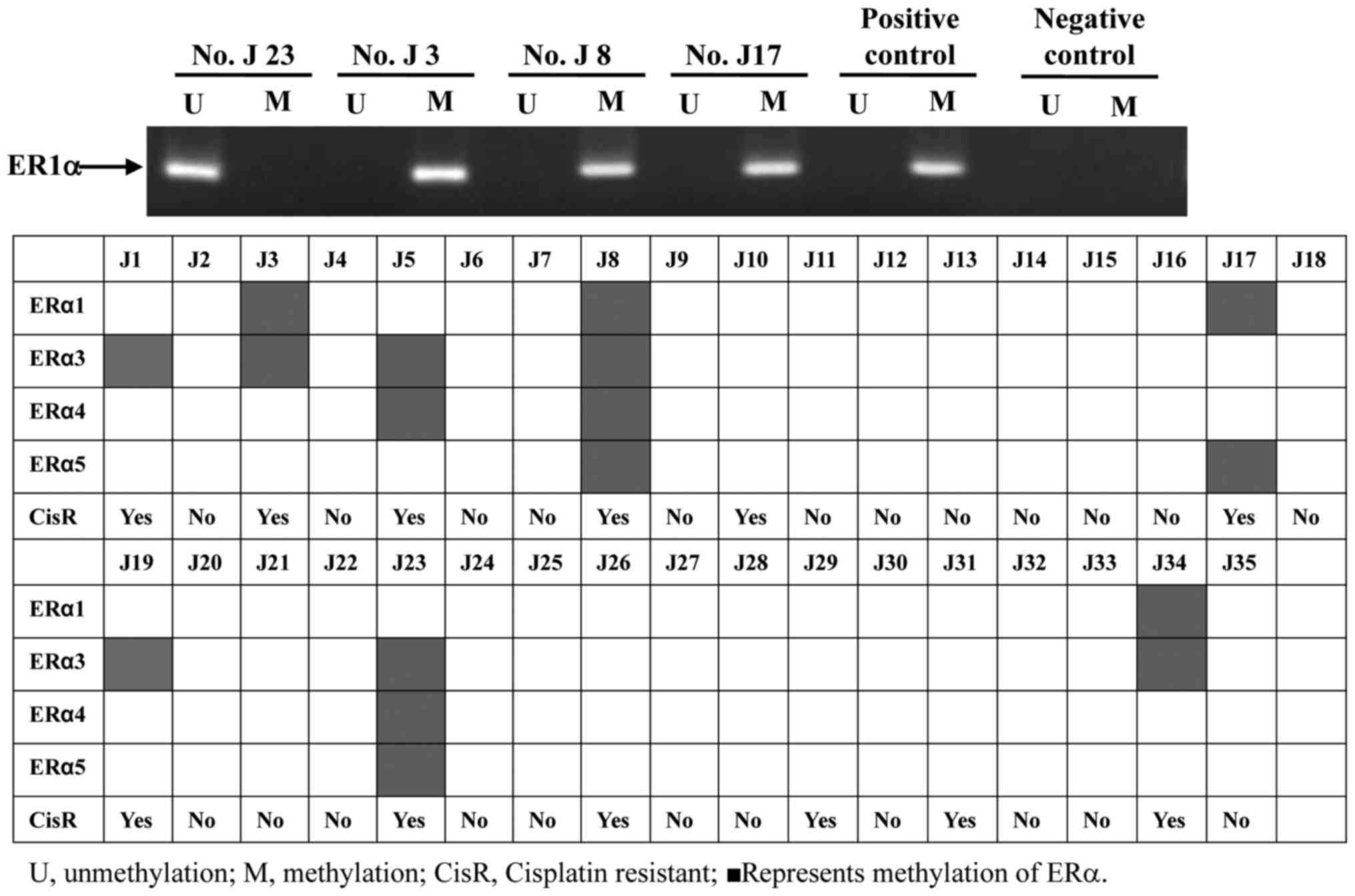
was considered to be ERα-methylated. As presented in Fig. 1, neither ERα methylation nor
unmethylation were observed in the negative control group; however,
the ERα methylation lane was present in positive group. This result
demonstrated that MSP had sensitivity and specificity for the
present study. In the 35 tumor tissue samples, 8/35 (22.9%) of
specimens were ERα-methylated (Fig.
1). The formulation of ERα methylation varied in the samples.
ERα1 methylation was positive in four samples (J3, J8, J17 and
J34), ER3 methylation was observed in 7/8 positive samples (J1, J3,
J5, J19, J23 and J34), ERα4 methylation was positive in three
samples (J5, J8 and J23) and ERα5 methylation was observed in three
samples (J8, J17 and J23). ERα3 methylation was the most widely
shared among these samples. All eight samples with ERα methylation
were resistant to cisplatin. The results of this analysis indicated
that ERα methylation was significantly associated with cisplatin
resistance in vitro. However, the formation and degree of
methylation was not associated with cell prohibition by cisplatin
in the ERα methylation group. ERα methylation may serve an
important role in primary cisplatin resistance in TNBC.
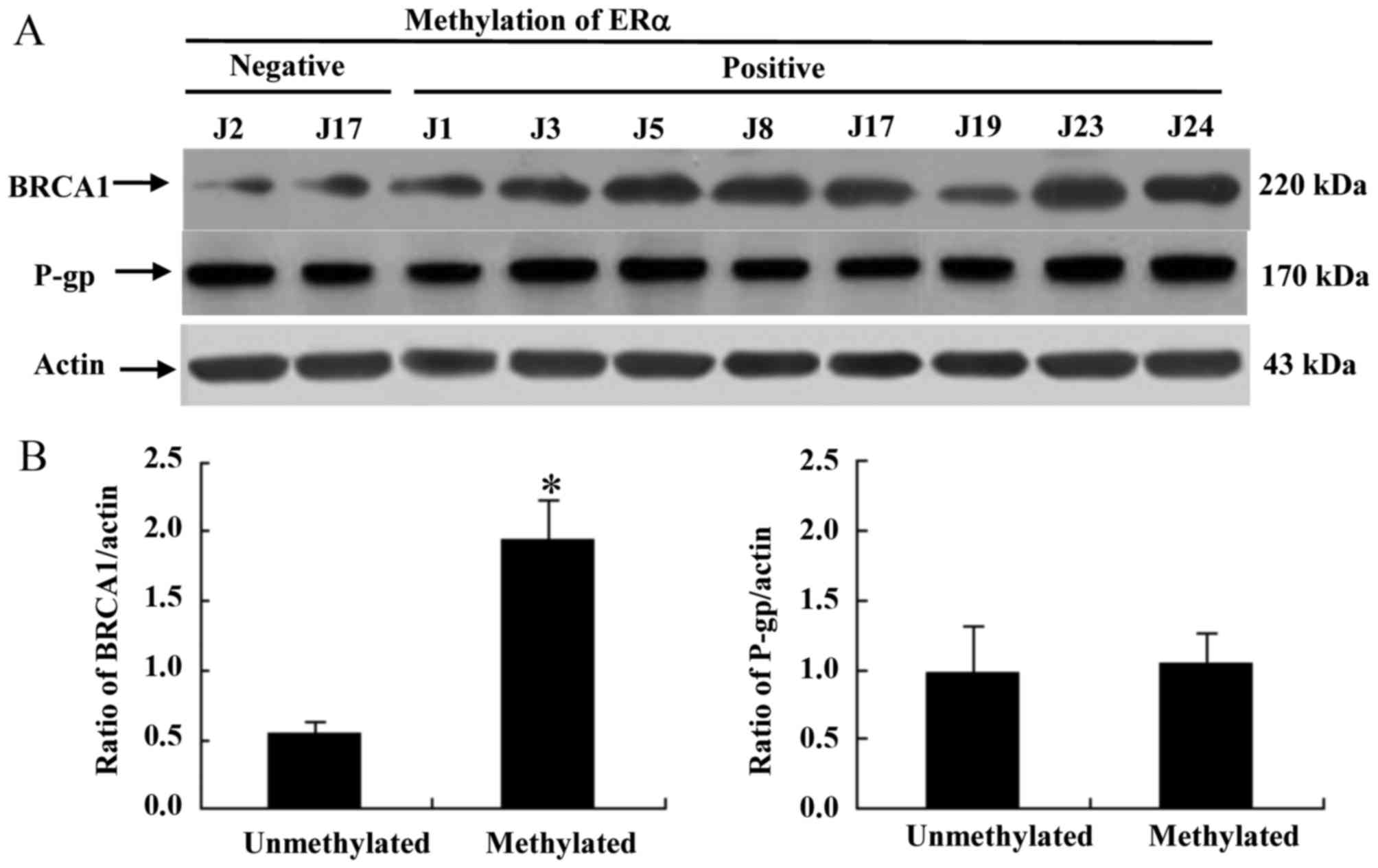
ERa methylation promotes cisplatin
resistance in TNBC by BRCA overexpression
To determine the contribution of the
multidrug-resistance protein P-gp and BRCA1 to primary cisplatin
resistance induced by ERα methylation, western blot analysis was
performed on primary cancer cells obtained from patients. As
presented in Fig. 2A, ERα methylation
was associated with an increase in the protein expression of BRCA1,
whereas the protein expression level of P-gp was not associated
with ERα methylation. These results provide evidence that ERα
methylation promotes cisplatin resistance in vitro via the
overexpression of BRCA, rather than P-gp.
Clinical prognosis of patients in
vivo
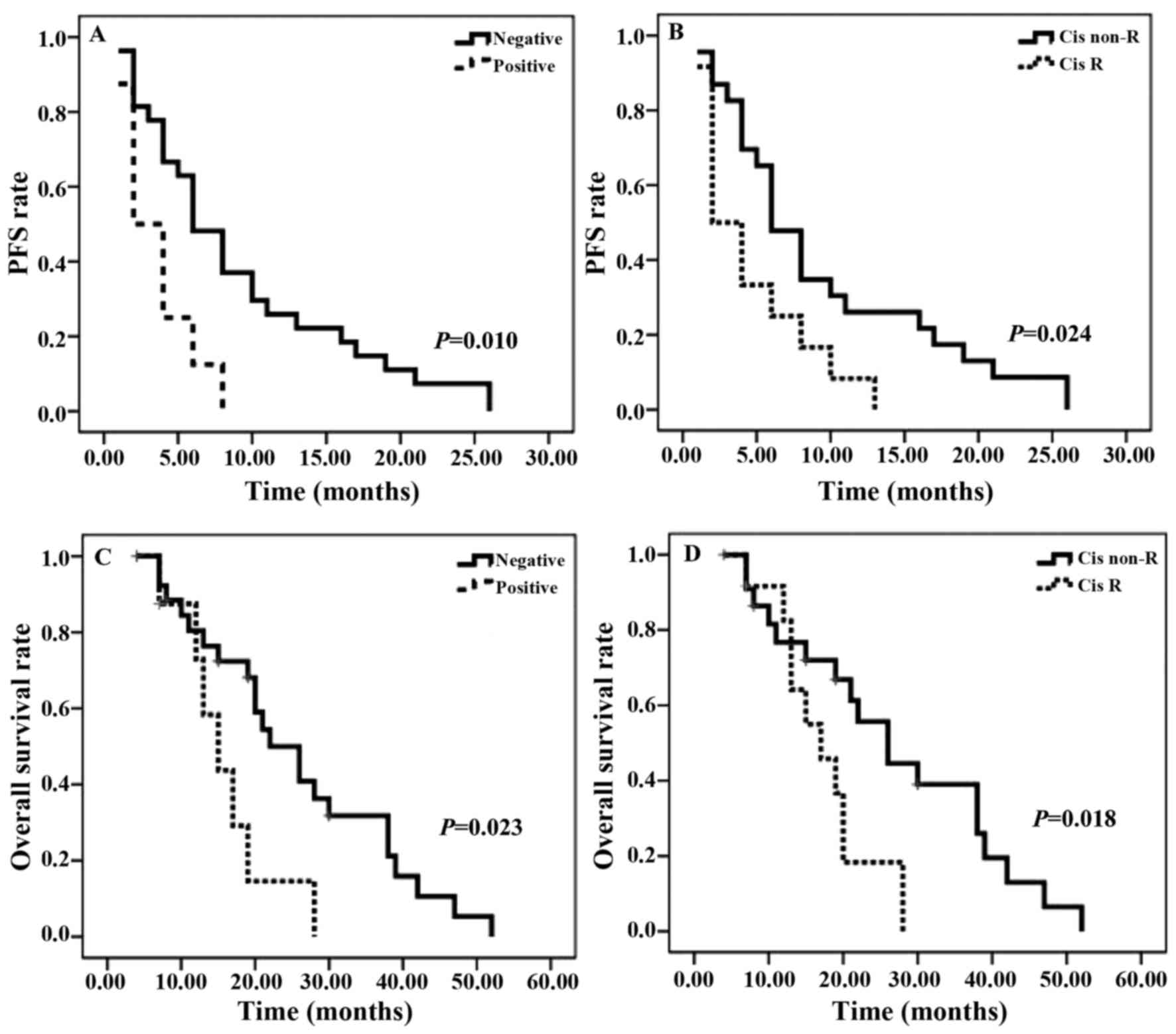
A total of 35 patients, for whom the ERα methylation
characteristics were known, were assessed (Fig. 3). As presented in Fig. 3A and C, a significant decrease in ERα
methylation was identified in PFS and OS rates. A similar pattern
was observed for cisplatin resistance: A significant decrease in
PFS rate was observed in patients with cisplatin resistance for the
follow-up duration (Fig. 3B). As
presented in Fig. 3D, OS rate with
cisplatin resistance gradually increased in the first 12 months of
follow-up, however, the curve exhibited a significantly decreased
OS rate in the subsequent months. Between the two survival curves
there is a marked crossover. The median PFS time for patients with
ERα methylation was 3 months, compared with 6 months for those in
the ERα-unmethylated arm. The median OS time was similar between
the two groups: 14 months in ERα methylation-positive arm and 20.5
months in the ERα methylation-negative arm. The median PFS and OS
times of patients with cisplatin resistance were 3 and 16 months,
respectively, compared with 7 months and 21.5 months for those in
the cisplatin-sensitive arm.
Discussion
Breast cancer cells accumulate genetic and protein
changes that allow them to evade chemotherapeutic drugs and cause
increasingly higher risks to patients, particularly in those with
TNBC (3,4). In view of effective specific target,
compared with endocrine therapy in hormone-positive breast cancer
or transtuzumab therapy in HER2-positive breast cancer, there is no
preferred standard chemotherapy to prohibit the proliferation of
cancer cells and their metastasis in patients with TNBC (8,17–19). Accurate identification of TNBC and
adequately powered prospective trials are required to identify
effective chemotherapy regimens and to assess the validity of
biomarkers. TNBC accounts for ~15% of all invasive breast cancers
with higher histological grade compared with that in other types of
molecular cancer (4).
A number of studies indicate that the weekly
addition of paclitaxel to an anthracycline-containing adjuvant
therapy may be superior to only anthacycline-containing regimens
without paclitaxel; however, the DFS time remained shorter for TNBC
compared with non-TNBC (20,21). On the basis of this inhibitory effect
on the proliferation of breast cancer cells in mouse models,
particularly in the dysfunction of BRCA1 and its pathway, the use
of cisplatin to treat TNBC is currently being assessed in clinical
trials and certain studies (7–8,22). In neoadjuvant chemotherapy, the use of
cisplatin with docetaxel exhibited a higher pathological complete
response in patients with BRCA1 mutations (23). In metastatic TNBC, compared with
docetexal, carboplatin exhibited significant improvements in the
rate of tumor response (68.0 vs. 33.3%, respectively; P=0.03),
median PFS times were 6.8 months (24,25).
Similarly, the use of olaprib, an oral poly(ADP-ribose) polymerase
(PARP) inhibitor, resulted in tumor regression in <41% in
patients carrying BRCA1 mutations (22). The tumors from patients with TNBC tend
to carry different tumor protein mutations, and tumor protein p53
(TP53) and BRCA1 mutations have been described as the most
dangerous mutant in breast cancer (26–29).
Patients with mutations in BRCA1 exhibit
deficiencies in double-stranded DNA break repair; those with
germline BRCA1 mutations have a ~20-fold increased risk of breast
cancer, with more aggressive carcinogenesis and angiogenesis
(30–32). If patients harbor these mutations, the
effective response rate to cisplatin was potentially higher
compared with that for other chemotherapy regimens (22,33).
Previous results indicated that ERα methylation was
associated with tumor stage, lymph node metastasis, nuclear
accumulation of p53 and BRCA1 expression in basal-like breast
cancer (14,34–37). The
mechanism of action of cisplatin in breast cancer cells with DNA
repair dysfunctions remains unknown. The present study indicates
that ERα methylation was significantly associated with cisplatin
resistance. Previous studies have been searching for and
identifying other biomarkers and pathways involved in TNBC
(38,39). However, non-validated biomarkers were
explicated to evaluate chemotherapy efficacy (40). The present study identified that ERα
methylation may be a meaningful biomarker for the evaluation of
cisplatin resistance. However, it is not clear whether ERα
methylation is a biomarker for PARP inhibitors, which, like
cisplatin, target DNA repair. No significant difference in the
expression level of P-gp, which serves an important role in
multidrug resistance, was identified. However, it should be noted
that there was an association between ERα methylation and
overexpression of BRCA1. Patients overexpressing BRCA1 exhibit
increased DNA repair function (41–44). There
are certain possible explanations for the puzzling effect of ERα
methylation on the overexpression of BRCA. One may be the effect on
ataxia telangiectasia mutated (ATM) protein by ERα methylation: It
has been proposed that ATM inhibition by the normal ER may provide
a strategy to sensitize tumors to DNA-damaging agents including
cisplatin (45). Low ER-expression in
patients with ERα methylation may partially counteract ATM
inhibition (46,47). In addition, elevated BRCA expression
was demonstrated to upregulate ATM-associated DNA double-strand
break repair (48,49). Other studies indicated that homogenous
repair genes were dysregulated and BRCA was overexpressed in
patients with breast cancer (50,51).
Notably, BRCA overexpression has been associated with poor outcomes
in patients with TNBC (52). Despite
this, there was not enough evidence to conclude that ERα
methylation directly affects DNA repair in the present study.
The results of the present study require
confirmation in clinical trials, as well as in other breast cancer
types and different tumors. However, these results may dictate
valid therapy. The results of the present study may lead to
alteration of the therapy regimens in patients with ERα methylation
in TNBC. If clinical trials confirm the results of the present
study, patients with TNBC with methylated ERα may benefit from
other chemotherapy in place of cisplatin to avoid cisplatin
resistance.
In conclusion, the results of the present study
indicated that ERα methylation affected not only the sensitivity of
breast cancer cells to cisplatin, but also the expression of BRCA1
protein. Further analysis of the mechanism of ERα methylation in
cisplatin resistance may aid the development of a novel therapeutic
approach to targeting the BRCA1-related signal pathway.
Acknowledgements
The present study was supported by the Liaoning
Province Doctor Startup Fund Program (grant no. 20121132 and
201501108), National Nature Science Foundation (grant no.
81502188), Central Guidance for Special Funds (2016007011) and the
Clinical Capability Construction Project for Liaoning Provincial
Hospitals (grant no. LNCCC-C05-2015).
References
|
1
|
Marmé F and Schneeweiss A: Targeted
therapies in triple-negative breast cancer. Breast Care.
10:159–166. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schmadeka R, Harmon BE and Singh M:
Triple-negative breast carcinoma: Current and emerging concepts. Am
J Clin Pathol. 141:462–477. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Elsamany S and Abdullah S: Triple-negative
breast cancer: Future prospects in diagnosis and management. Med
Oncol. 31:8342014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Merlin JL, Harlé A, Lion M, Ramacci C and
Leroux A: Expression and activation of P38 MAP kinase in invasive
ductal breast cancers: Correlation with expression of the estrogen
receptor, HER2 and downstream signaling phosphorylated proteins.
Oncol Rep. 30:1943–1948. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bachmann C, Schmidt S, Staebler A, Fehm T,
Fend F, Schittenhelm J, Wallwiener D and Grischke E: CNS metastases
in breast cancer patients: Prognostic implications of tumor
subtype. Med Oncol. 32:4002015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cho EY, Chang MH, Choi YL, Lee JE, Nam SJ,
Yang JH, Park YH, Ahn JS and Im YH: Potential candidate biomarkers
for heterogeneity in triple-negative breast cancer (TNBC). Cancer
Chemother Pharmacol. 68:753–761. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lips EH, Mulder L, Oonk A, van der Kolk
LE, Hogervorst FB, Imholz AL, Wesseling J, Rodenhuis S and Nederlof
PM: Triple-negative breast cancer: BRCAness and concordance of
clinical features with BRCA1-mutation carriers. Br J Cancer.
108:2172–2177. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hu XC, Zhang J, Xu BH, Cai L, Ragaz J,
Wang ZH, Wang BY, Teng YE, Tong ZS, Pan YY, et al: Cisplatin plus
gemcitabine versus paclitaxel plus gemcitabine as first-line
therapy for metastatic triple-negative breast cancer (CBCSG006): A
randomised, open-label, multicentre, phase 3 trial. Lancet Oncol.
16:436–446. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fan Y, Xu BH, Yuan P, Ma F, Wang JY, Ding
XY, Zhang P, Li Q and Cai RG: Doxetaxel cisplatin might be superior
to docetaxel capecitabine in the first line treatment of
metastastic triple negative breast cancer. Ann Oncol. 24:1219–1225.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gelmon K, Dent R, Mackey JR, Laing K,
McLeod D and Verma S: Target triple negative breast cancer:
Optimizing therapeutic outcomes. Ann Oncol. 23:2223–2234. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang J, Wang Z, Hu X, Wang B, Wang L,
Yang W, Liu Y, Liu G, Di G, Hu Z, et al: Cisplatin and gemcitabine
as the first line therapy in metastatic triple negative breast
cancer. Int J Cancer. 136:204–211. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Taniguchi T, Tischkowitz M, Ameziane N,
Hodgson SV, Mathew CG, Joenje H, Mok SC and D'Andrea AD: Disruption
of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian
tumors. Nat Med. 9:568–574. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wei M, Xu J, Dignam J, Nanda R, Sveen L,
Fackenthal J, Grushko TA and Olopade OI: Estrogen receptor alpha,
BRCA1, and FANCF promoter methylation occur in distinct subsets of
sporadic breast cancers. Breast Cancer Res Treat. 111:113–120.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jing MX, Mao XY, Li C, Wei J, Liu C and
Jin F: Estrogen receptor-alpha promoter methylation in sporadic
basal-like breast cancer of Chinese women. Tumor Biol. 32:713–719.
2011. View Article : Google Scholar
|
|
15
|
Taniguchi T, Kakkar AK, Tuddenham EG,
Williamson RC and Lemoine NR: Enhanced expression of urokinase
receptor induced through the tissue factor-factor via pathway in
human pancreatic cancer. Cancer Res. 58:4461–4467. 1998.PubMed/NCBI
|
|
16
|
Gradishar WJ, Anderson BO, Balassanian R,
Blair SL, Burstein HJ, Cyr A, Elias AD, Farrar WB, Forero A,
Giordano SH, et al: NCCN guidelines insights breast cancer, version
1.2016. J Natl Compr Canc Netw. 13:1475–1485. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Seitz S, Rick FG, Schally AV, Treszl A,
Hohla F, Szalontay L, Zarandi M, Ortmann O, Engel JB and Buchholz
S: Combination of GHRH antagonists and docetaxel shows experimental
effectiveness for the treatment of triple-negative breast cancers.
Oncol Rep. 30:413–418. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
van de Water W, Seynaeve C, Bastiaannet E,
Markopoulos C, Jones SE, Rea D, Hasenburg A, Putter H, Hille ET,
Paridaens R, et al: Elderly postmenopausal patients with breast
cancer are at increased risk for distant recurrence: A tamoxifen
exemestane adjuvant multinational study analysis. Oncologist.
18:8–13. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Makubate B, Donnan PT, Dewar JA, Thompson
AM and McCowan C: Cohort study of adherence to adjuvant endocrine
therapy, breast cancer recurrence and mortality. Br J Cancer.
108:1515–1524. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Adams S, Gray RJ, Demaria S, Goldstein L,
Perez EA, Shulman LN, Martino S, Wang M, Jones VE, Saphner TJ, et
al: Prognostic value of tumor-infiltrating lymphocytes in triple
negative breast cancer from two phase III randomized adjuvant
breast cancer trail: ECOG 2197 and ECOG 1199. J Clin Oncol.
32:2959–2966. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schneider BP, Zhao F, Wang M, Stearns V,
Martino S, Jones V, Perez EA, Saphner T, Wolff AC, Sledge GW Jr, et
al: Neuropathy is not associated with clinical outcomes in patients
receiving adjuvant taxane-containing therapy for operable breast
cancer. J Clin Oncol. 30:3051–3057. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Johnson N, Johnson SF, Yao W, Li YC, Choi
YE, Bernhardy AJ, Wang Y, Capelletti M, Sarosiek KA, Moreau LA, et
al: Stabilization of mutant BRCA1 protein confers PARP inhibitor
and platinum resistance. Proc Natl Acad Sci USA. 110:17041–17046.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Julka PK, Chacko RT, Nag S, Parshad R,
Nair A, Oh DS, Hu Z, Koppiker CB, Nair S, Dawar R, et al: A phase
II study of sequential neoadjuvant gemcitabine plus doxorubicin
followed by gemcitabine plus cisplatin in patients with operable
breast cancer: Prediction of response using molecular profiling. Br
J Cancer. 98:1327–1335. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Staudacher L, Cottu PH, Diéras V,
Vincent-Salomon A, Guilhaume MN, Escalup L, Dorval T, Beuzeboc P,
Mignot L and Pierga JY: Platinum-based chemotherapy in metastatic
triple-negative breast cancer: The institut curie experience. Ann
Oncol. 22:848–856. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang K, Xie S, Ren Y, Xia H, Zhang X and
He J: Establishment of a bioluminescent MDA-MB-231 cell line for
human triple-negative breast cancer research. Oncol Rep.
27:1981–1989. 2012.PubMed/NCBI
|
|
26
|
Al Dhaheri Y, Eid A, AbuQamar S, Attoub S,
Khasawneh M, Aiche G, Hisaindee S and Iratni R: Mitotic arrest and
apoptosis in breast cancer cells induced by origanum majorana
extract: Upregulation of TNF-α and downregulation of survivin and
mutant p53. PLoS One. 8:e566492013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ma CX, Cai S, Li S, Ryan CE, Guo Z,
Schaiff WT, Lin L, Lin L, Hoog J, Goiffon RJ, Prat A, et al:
Targeting CHK1 in p53-deficient triple-negative breast cancer is
therapeutically beneficial in human-in-mouse tumor models. J Clin
Invest. 122:1541–1552. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Deb S, Wong SQ, Li J, Do H, Weiss J, Byrne
D, Chakrabarti A, Bosma T; kConFab Investigators, ; Fellowes A, et
al: Mutational profiling of familial male breast cancers reveals
similarities with luminal A female breast cancer with rare TP53
mutations. Br J Cancer. 111:2351–2360. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Michils G, Hollants S, Dehaspe L, Van
Houdt J, Bidet Y, Uhrhammer N, Bignon YJ, Vermeesch JR, Cuppens H
and Matthijs G: Molecular analysis of the breast cancer genes BRCA1
and BRCA2 using amplicon-based massive parallel pyrosequencing. J
Mol Diagn. 14:623–630. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Murphy CG and Moynahan ME: BRCA gene
structure and function in tumor suppression: A repair-centric
perspective. Cancer J. 16:39–47. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Oplustilova L, Wolanin K, Mistrik M,
Korinkova G, Simkova D, Bouchal J, Lenobel R, Bartkova J, Lau A,
O'Connor MJ, et al: Evaluation of candidate biomarkers to predict
cancer cell sensitivity or resistance to PARP-1 inhibitor
treatment. Cell Cycle. 11:3837–3850. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Choi DH, Lee MH, Bale AE, Carter D and
Haffty BG: Incidence of BRCA1 and BRCA2 mutations in young korean
breast cancer patients. J Clin Oncol. 22:1638–1645. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lord CJ and Ashworth A: Mechanisms of
resistance to therapies targeting BRCA-mutant cancers. Nat Med.
19:1381–1388. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mao XY, Fan CF, Zheng HC, Wei J, Yao F and
Jin F: p53 nuclear accumulation and ERalpha expression in ductal
hyperplasia of breast in a cohort of 215 chinese women. J Exp Clin
Cancer Res. 29:1122010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mao XY, Chen H, Wang H, Wei J, Liu C,
Zheng HC, Yao F and Jin F: MTA1 expression correlates significantly
with ER-alpha methylation in breast cancer. Tumour Biol.
33:1565–1572. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao L, Wang L, Jin F, Ma W, Ren J, Wen X,
He M, Sun M, Tang H and Wei M: Silencing of estrogen receptor alpha
(ERalpha) gene by promoter hypermethylation is a frequent event in
Chinese women with sporadic breast cancer. Breast Cancer Res Treat.
117:253–259. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jin F, Liu C, Guo Y, Chen H and Wu Y:
Clinical implications of Girdin and PI3K protein expression in
breast cancer. Oncol Lett. 5:1549–1553. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sahab ZJ, Man YG, Byers SW and Sang QX:
Putative biomarkers and targets of estrogen receptor negative human
breast cancer. Int J Mol Sci. 12:4504–4521. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pachmann K, Camara O, Kroll T, Gajda M,
Gellner AK, Wotschadlo J and Runnebaum IB: Efficacy control of
therapy using circulating epithelial tumor cells (CETC) as ‘liquid
biopsy’: Trastuzumab in HER2/neu-positive breast carcinoma. J
Cancer Res Clin Oncol. 137:1317–1327. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Soria JC, Blay JY, Spano JP, Pivot X,
Coscas Y and Khayat D: Added value of molecular targeted agents in
oncology. Ann Oncol. 22:1703–1716. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
De P, Sun Y, Carlson JH, Friedman LS,
Leyland-Jones BR and Dey N: Doubling down on the PI3K-AKT-mTOR
pathway enhances the antitumor efficacy of PARP inhibitor in triple
negative breast cancer model beyond BRCA-ness. Neoplasia. 16:43–72.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Konishi H, Mohseni M, Tamaki A, Garay JP,
Croessmann S, Karnan S, Ota A, Wong HY, Konishi Y, Karakas B, et
al: Mutation of a single allele of the cancer susceptibility gene
BRCA1 leads to genomic instability in human breast epithelial
cells. Proc Natl Acad Sci USA. 108:17773–17778. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sy SM, Huen MS and Chen J: PALB2 is an
integral component of the BRCA complex required for homologous
recombination repair. Proc Natl Acad Sci USA. 106:7155–7160. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Stefansson OA, Jonasson JG, Johannsson OT,
Olafsdottir K, Steinarsdottir M, Valgeirsdottir S and Eyfjord JE:
Genomic profiling of breast tumours in relation to BRCA
abnormalities and phenotypes. Breast Cancer Res. 11:R472009.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Albarakati N, Abdel-Fatah TM, Doherty R,
Russell R, Agarwal D, Moseley P, Perry C, Arora A, Alsubhi N,
Seedhouse C, et al: Targeting BRCA1-BER deficient breast cancer by
ATM or DNA-PKcs blockade either alone or in combination with
cisplatin for personalized therapy. Mol Oncol. 9:204–217. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tommiska J, Bartkova J, Heinonen M,
Hautala L, Kilpivaara O, Eerola H, Aittomäki K, Hofstetter B, Lukas
J, von Smitten K, et al: The DNA damage signalling kinase ATM is
aberrantly reduced or lost in BRCA1/BRCA2-deficient and
ER/PR/ERBB2-triple-negative breast cancer. Oncogene. 27:2501–2506.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Murria R, Palanca S, de Juan I, Egoavil C,
Alenda C, García-Casado Z, Juan MJ, Sánchez AB, Santaballa A,
Chirivella I, et al: Methylation of tumor suppressor genes is
related with copy number aberrations in breast cancer. Am J Cancer
Res. 5:375–385. 2014.PubMed/NCBI
|
|
48
|
Bueno RC, Canevari RA, Villacis RA,
Domingues MA, Caldeira JR, Rocha RM, Drigo SA and Rogatto SR: ATM
down-regulation is associated with poor prognosis in sporadic
breast carcinomas. Ann Oncol. 25:69–75. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Jiang H, Reinhardt HC, Bartkova J,
Tommiska J, Blomgvist C, Nevanlinna H, Bartek J, Yaffe MB and
Hemann MT: The combined status of ATM and p53 link tumor
development with therapeutic response. Genes Dev. 23:1895–1909.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chan KY, Ozcelik H, Cheung AN, Ngan HY and
Khoo US: Epigenetic factors controlling the BRCA1 and BRCA2 genes
in sporadic ovarian cancer. Cancer Res. 62:4151–4156.
2002.PubMed/NCBI
|
|
51
|
Haitjema A, Brandt BW, Ameziane N, May P,
Heringa J, de Winter JP, Joenje H and Dorsman JC: A protein
prioritization approach tailored for the FA/BRCA pathway. PLoS One.
8:e620172013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Mote PA, Leary JA, Avery KA, Sandelin K,
Chenvix-Trench G, Kirk JA and Clarke CL: kConFab Investigators:
Germ-line mutations in BRCA1 or BRCA2 in the normal breast are
associated with altered expression of estrogen-responsive proteins
and the predominance of progesterone receptor A. Genes Chromosomes
Cancer. 39:236–248. 2004. View Article : Google Scholar : PubMed/NCBI
|