Introduction
A woman's health is seriously affected by cervical
cancer, which is the second most common malignancy in women.
Importantly, with more than 520,000 new cervical cancer patients
each year, 260,000 cancer-related deaths occur in women aged 15 to
44 years annually. Cervical cancer ranks as the fourth leading
cause of female cancer deaths worldwide. In developing regions, the
death toll is twice that of developed regions (1). Nevertheless, approximately 131,500 women
suffer from cervical cancer every year in China, which accounts for
approximately 28.8% of the new incident cases globally. The
incidence rate is 6 times higher than that of developed regions.
The development of cervical cancer not only exhibits a complex
multiple-factor etiology but also involves multiple phases, and
many studies suggest that the paramount factor in the development
of cervical cancer is a persistent human papillomavirus (HPV)
infection (2). However, according to
estimates, 30 to 60% of women infected with genital HPV do not
develop cancer (3). In most women who
have been infected with HPV, the infection is a significant rather
than sufficient risk factor for the development of cervical cancer.
In recent years, several studies have revealed that changes in
individual genetic factors, such as the immune regulatory genes,
oncogenes and tumor suppressor genes, play a very important role in
the pathogenesis of cervical cancer (4). Increasing evidence has indicated that
genetic polymorphisms are associated with the occurrence of
cervical cancer, and single nucleotide polymorphisms (SNPs) of the
cell surface molecules of the CD83 gene, including rs750749,
rs9296925 and rs9370729, are associated with cervical cancer
susceptibility (5,6). In this context, it should be mentioned
that the polymorphisms of other genes may also be associated with
cervical cancer susceptibility, including p53 codon 72 and
the IL-8 and MMP genes (7,8).
Macroautophagy (hereafter referred to as autophagy)
is an extreme degradative process that is necessary to conserve
homeostasis in eukaryotic cells (9).
Relevant studies show autophagy could modulate the expression level
of PA proteins in cervical cancer. Currently, it is widely accepted
that the autophagy system might have ambiguous effects in the
development of cancer, such as promotion and antagonization
(10). Furthermore, autophagy is
mainly regulated by autophagy-related genes (ATGs). To date,
more than 30 ATGs have been reported. Previous studies have
suggested that initiation of autophagosomes requires the
participation of the two ubiquitin-like conjugated systems,
ATG12-ATG5 and ATG8-PE (11). This process is indispensable for
ATG4 splitting, permitting conjugation of ATG8 to PE
on phagophore membranes, deconjugation of the compound
ATG8-PE, and acceleration of autophagosome maturation
(12). Therefore, ATG4 is a
significant factor for the autophagy process and may be a target
for cancer prevention and intervention. To date, ATG4A has
been reported to be related to breast tumorigenesis (13) and the risk of lung cancer (14). The same vital functions of
ATG4A have been well-documented in ovarian cancer (15). While numerous steps of autophagy have
been well described at the molecular level, the role of
ATG4A in cervical cancer is less well understood. Our group
conducted a case-control study to evaluate the association of
ATG4A variants with the risk of cervical cancer.
Materials and methods
Study samples
For this study, a total of 542 peripheral blood
samples from 285 incident cervical cancer patients and 257 healthy
control subjects were collected and analyzed. We used the
guidelines of the International Federation of Gynecology and
Obstetrics to assess the tumor type, stage and histological
features of the cervical cancer cases. The control subjects in this
study were ThinPrep cytology test (TCT)-negative in a physical
examination. All subjects participated in specialized tests such as
the TCT and HPV subtype tests. A definitive diagnosis was based on
histopathology results.
Study population
Between January 2012 and December 2016, subjects
from The First and Second Affiliated Hospitals of Chongqing Medical
University and Chongqing Cancer Institute (Chongqing, China) were
enrolled in this study; all subjects were members of the Han
Chinese population. The types of cancers in the cervical cancer
group were as follows: squamous cell carcinoma (SCC) (n=236),
adenocarcinoma (n=39) and other histologic types of tumors (n=10)
(Table I). All patients were females,
aged 27.0 to 67.0 years. Information on demographic
characteristics, family history of cancer, occupational exposure,
tobacco smoking, pregnancy, oral contraceptive use, induced
abortion, embryo number, menarche and menopause, and reproductive
histories were obtained for all participants using a
quasi-questionnaire survey. Informed consent was obtained from all
participants. Control group frequency matching of cases was
conducted using 257 age- and region-matched married females, aged
27.0 to 67.0 years, consisting of healthy individuals with no
history of cancer who were randomly selected during the same time
period as the case study.
 | Table I.Clinical and demographic
characteristics of patients and controls. |
Table I.
Clinical and demographic
characteristics of patients and controls.
| Characteristic | Patients, n=285
(%) | Controls, n=257
(%) |
|---|
| Age, years (mean ±
SDa) | 47.3±9.5 | 47.1±8.7 |
| Tumor stage |
|
|
| I | 70
(24.8) |
|
| II | 141 (49.6) |
|
|
III | 63
(21.8) |
|
| IV | 11 (3.8) |
|
| Labor
presentation |
|
|
|
Eutocia | 237 (83.2) | 182 (70.8) |
|
Caesarean section | 48
(16.8) | 75
(29.2) |
| Histological
type |
|
|
|
Squamous cell carcinoma | 236 (82.9) |
|
|
Adenocarcinoma | 39
(13.7) |
|
|
Other | 10
(3.4) |
|
| Parity |
|
|
|
Never | 10
(3.5) | 15
(5.8) |
| 1 | 132 (46.6) | 159 (61.9) |
| 2 | 94
(32.9) | 56
(21.8) |
| 3 | 36
(12.6) | 19
(7.4) |
| ≥4 | 13
(4.4) |
8 (3.1) |
| Abortion
frequency |
|
|
|
Never | 98
(34.4) | 112 (43.6) |
| 1 | 49
(17.2) | 51
(19.8) |
| 2 | 53
(18.6) | 57
(22.2) |
| ≥3 | 85
(29.8) | 37
(14.4) |
| Oral contraceptive
pill use |
|
|
|
Never | 239 (83.8) | 235 (91.4) |
|
Ever | 46
(16.2) | 22
(8.6) |
| Liquor and
tobacco |
|
|
|
Never | 177 (62.1) | 168 (65.4) |
|
Ever | 108 (37.9) | 89
(34.6) |
| Menopausal
status |
|
|
|
Premenopausal | 205 (71.9) | 187 (72.7) |
|
Postmenopausal | 80
(28.1) | 70
(27.3) |
| Blood type |
|
|
| O | 54
(18.9) | 49
(19.1) |
| A | 95
(33.3) | 78
(30.3) |
| B | 116 (40.7) | 85
(33.1) |
| AB | 20
(7.1) | 45
(17.5) |
| HPV
genotypesb |
|
|
|
Negative | 79
(27.7) | 198 (77.0) |
| 16 or
18 | 158 (55.5) | 31
(12.1) |
|
Othersc | 48
(16.8) | 28
(10.9) |
| BMI |
|
|
|
<18.5 |
6 (2.1) |
5 (1.9) |
|
18.5–23.9 | 203 (71.2) | 211 (82.1) |
|
≥24 | 76
(26.7) | 41
(17.0) |
| Family history of
cancer |
|
|
| No | 220 (77.1) | 223 (86.8) |
|
Yes | 65
(22.9) | 34
(13.2) |
This study was approved by the ethics committees of
the hospitals. Data obtained using structured questionnaires were
saved into databases. All procedures performed in studies were in
accordance with the ethical standards of the institutional or
national research committee and with the 1964 Helsinki declaration
and its later amendments or comparable ethical standards.
DNA extraction and genotyping
The genomic DNA of all patients was isolated from
the clinical peripheral blood samples using the phenol-chloroform
extraction method. The DNA was sent to Bio Miao Biological Science
and Technology Co., Ltd. (Beijing, China) to detect the SNP
genotype. A large number of literature and site related researches
were analyzed and used to assist with the process of genotyping. We
selected candidate gene expression studies via bioinformatics and
examined ATG4, LC3, ATG9, and ATG16; we found a total of 39 donor
splice sites, including rs4036579: A>G, rs5973822: A>G,
rs807181: G>C, rs807182: A>C, rs807183: G>A, and
others.
Analysis of SNP-SNP interactions
Multiple dimension reduction (MDR) is a method used
to reduce the dimensionality of multilocus information (16); this method was used to improve the
identification of polymorphism combinations associated with the
risk of cervical cancer. MDR analysis was also used to help
determine the correlation between SNPs and cervical cancer.
Similarly, the Bonferroni correction was used, and P<0.05 was
considered to indicate a statistically significant difference for
the permutation test. All the SNP combinations are evaluated in the
training dataset based on 10-fold CV. Two selection criteria are
used to choose the best SNP combination, the cross validation
consistency (CVC) and the average of test balanced accuracy (TA),
consequently, the chosen SNP combination has the highest CVC and/or
test TA. This gene predictor summarizes the effects of individual
genes on each gene. Meta-analyses were performed to determine
significant SNP-SNP interactions associated with HPV infection, and
multiple testing corrections were performed using the Bonferroni
method.
Statistical analysis
All statistical analyses were performed using PLINK
version 1.07 (17). Allele and
genotype frequencies of the SNPs were compared using the chi-square
test and Fisher's exact test. Odds Ratio (OR) and 95% Confidence
Interval (CI) were calculated using unconditional logistic
regression and by adjusting the relevant confounding factors, such
as age, pregnancy, abortion, BMI, menopausal status, and family
history of cancer (15). A P-value
less than 0.05 was considered statistically significant. Because
all participants in the experiment were Han Chinese, the population
was not stratified.
Results
Clinical features
The basic information of these subjects is listed in
Table I. Cases were, on average, a
greater frequency of eutocia (83.2/70.8%) and fertility births
(49.9/32.3%) were observed than control. And the proportion of
blood type B (40.7/33.1%) was also higher than that of the control.
HPV16 and 18 were associated with the highest rate of infection
(55.5%), with HPV16 accounting for 32% of infections and HPV18
accounting for 23.5% of infections. We found that HPV types were
related to pathology, and HPV16 was the predominant strain
associated with SCC (72%). However, the major strain involved in
adenocarcinomas was HPV18 (53%). A descriptive analysis of selected
polymorphisms was carried out (Table
II), establishing the polymorphisms of the ATG4A gene and their
association with cervical cancer. In addition, polymorphic loci
were mainly located in intron, and most of them were mutated from A
to G.
 | Table II.Polymorphisms of the ATG4A gene and
their relationship to cervical cancer. |
Table II.
Polymorphisms of the ATG4A gene and
their relationship to cervical cancer.
| Gene | Chromosomal
region | Region | Function | Chromosomal
Location | Nucleotide
position | changea | dbSNP
IDb |
|---|
| ATG4A | Xq22.1–22.3 | 5′near
genee | TFBSc | intron | 108090623 | A/G | rs4036579 |
|
|
|
| miRNA | 3′UTRd | 108153728 | A/G | rs5973822 |
|
|
|
| TFBS | intron | 108090354 | C/G | rs807181 |
|
|
|
| TFBS | intron | 108092031 | A/C | rs807182 |
|
|
|
| TFBS | intron | 108094263 | A/G | rs807183 |
Gene sequencing
Among the 39 loci tested, the statistically
significant loci are shown in Fig. 1,
and the remaining sites were not statistically significant. These
statistically significant sites after screening will be followed by
subsequent tests to determine their effect.
Characterization of SNPs in the ATG4A
gene
As shown in Table
III, for rs4036579, the frequency of the dominant model allele
A in the case group was higher than that in the control group
(94.39 vs. 90.66%, respectively, P=0.0057). The OR was 0.461 (95%
CI 0.154 to 0.697). In the recessive model study, the frequency of
the variant allele G was higher in control subjects than in the
case group (38.91 vs. 26.67%, respectively, P=0.0098). The OR was
0.682 (95% CI 0.386 to 0.991). Compared with the homozygous
wildtype genotype (AA), the homozygous variant genotype (GG) was
associated with a significant decrease in the incidence of cervical
cancer (P=0.0013, OR=0.427, 95% CI 0.184 to 0.848), whereas no
significant correlation was found among carriers of the
heterozygous variant genotype (AG). Unconditional logical analysis
showed that the wildtype A allele of rs4036579 was associated with
a reduced risk of cervical cancer (P<0.001, OR=0.504, 95% CI
0.401 to 0.707).
 | Table III.Association of the SNPs of ATG4A with
the risk of cervical cancer in the case and control groups. |
Table III.
Association of the SNPs of ATG4A with
the risk of cervical cancer in the case and control groups.
| SNP | Genotype | Cases [n (%)] | Controls [n
(%)] | P-value | Crude OR | (95% CI) | Adjusted P | Adjusted
ORa | (95% CI) |
|---|
| ATG4A
rs4036579 |
| 285 | 257 |
|
|
|
|
|
|
|
| GG | 16 (5.61) | 34
(13.23) |
|
|
|
|
|
|
|
| AA | 209 (73.33) | 175 (68.09) | 0.0028 | 0.394 | 0.2104 to
0.7379 | 0.0013 | 0.427 | 0.184 to 0.848 |
|
| AG | 60
(21.05) | 48
(18.68) | 0.8351 | 0.9554 | 0.6218 to
1.468 | 0.796 | 0.997 | 0.603 to 1.521 |
|
Dominant model | Any A vs. GG | 269 (94.39) | 233 (90.66) | 0.0002 | 0.33 | 0.1807 to
0.6025 | 0.0057 | 0.461 | 0.154 to 0.697 |
|
Recessive model | Any G vs. AA | 76
(26.67) | 100 (38.91) | 0.0135 | 0.6364 | 0.4441 to
0.9118 | 0.0098 | 0.682 | 0.386 to 0.991 |
|
Additive model | AA vs. AG vs.
GG | 209/60/16 | 58/42/175 | <0.0001 | 0.64 | 0.526 to 0.807 | 0.001 | 0.603 | 0.463 to 0.801 |
| Allele
OR | A vs. G | 478/92 | 408/142 | <0.0001 | 0.553 | 0.4122 to
0.7418 | <0.0001 | 0.504 | 0.401 to 0.707 |
| ATG4A
rs5973822 |
|
|
|
|
|
|
|
|
|
|
| GG | 11 (3.86) | 29
(11.28) |
|
|
|
|
|
|
|
| AA | 234 (82.11) | 202 (78.60) | 0.0015 | 0.3274 | 0.1595 to
0.6722 | 0.0046 | 0.384 | 0.148 to 0.733 |
|
| AG | 40
(14.04) | 26
(10.12) | 0.2915 | 0.753 | 0.4438 to
1.277 | 0.254 | 0.802 | 0.407 to 1.288 |
|
Dominant model | Any A vs. GG | 274 (96.14) | 238 (92.61) | <0.0001 | 3.872 | 1.932 to 7.762 | <0.0001 | 3.927 | 1.896 to 7.652 |
|
Recessive model | Any G vs. AA | 51
(17.89) | 73 (28.4) | 0.0137 | 0.6031 | 0.4025 to
0.9036 | 0.096 | 0.684 | 0.364 to 0.949 |
|
Additive model | AA vs. AG vs.
GG | 234/40/11 | 202/26/29 | 0.003 | 1.803 | 1.204 to 2.622 | 0.006 | 1.987 | 1.224 to 3.132 |
| Allele
OR | A vs. G | 478/92 | 398/116 | <0.0001 | 0.4882 | 0.3487 to
0.6835 | <0.0001 | 0.407 | 0.253 to 0.648 |
| ATG4A rs807181 |
|
|
|
|
|
|
|
|
|
|
| CC | 11 (3.86) | 42
(16.34) |
|
|
|
|
|
|
|
| GG | 204 (71.58) | 170 (66.15) | <0.0001 | 4.582 | 2.288 to 9.176 | <0.0001 | 4.245 | 2.023 to 9.003 |
|
| CG | 70
(24.56) | 45
(17.51) | 0.2321 | 1.296 | 0.8463 to
1.985 | 0.216 | 1.313 | 0.805 to 1.998 |
|
Dominant model | Any C vs. GG | 81
(28.42) | 87
(33.85) | 0.1722 | 0.7759 | 0.5387 to
1.117 | 0.138 | 0.814 | 0.485 to 1.377 |
|
Recessive model | Any G vs. CC | 274 (96.14) | 215 (83.66) | <0.0001 | 4.866 | 2.446 to 9.678 | <0.0001 | 4.657 | 2.326 to 9.453 |
|
Additive model | CC vs. CG vs.
GG | 11/70/204 | 42/45/170 | 0.297 | 1.203 | 1.056 to 2.799 | 0.225 | 1.323 | 1.003 to 2.826 |
| Allele
OR | C vs. G | 92/478 | 129/385 | 0.0003 | 0.5744 | 0.4258 to
0.7749 | 0.0051 | 0.596 | 0.403 to 0.821 |
| ATG4A rs807182 |
|
|
|
|
|
|
|
|
|
|
| AA | 202 (70.88) | 175 (68.09) |
|
|
|
|
|
|
|
| CC | 11 (3.86) | 36
(14.01) | <0.0001 | 0.2647 | 0.1308 to
0.5358 | <0.0001 | 0.225 | 0.104 to 0.497 |
|
| AC | 72
(25.26) | 46 (17.9) | <0.0001 | 5.123 | 2.372 to 11.06 | <0.0001 | 5.021 | 2.113 to
10.866 |
|
Dominant model | Any A vs. CC | 274 (96.14) | 221 (85.99) | <0.0001 | 4.058 | 2.018 to 8.157 | <0.0001 | 4.002 | 1.996 to 8.022 |
|
Recessive model | Any C vs. AA | 83
(29.12) | 82
(31.91) | 0.4819 | 1.14 | 0.7906 to
1.645 | 0.448 | 1.185 | 0.732 to 1.694 |
|
Additive model | AA vs. AC vs.
CC | 202/72/11 | 175/46/36 | 0.007 | 2.335 | 1.857 to 4.536 | 0.024 | 2.383 | 1.824 to 4.776 |
| Allele
OR | A vs. C | 476/94 | 396/118 | 0.0074 | 0.6627 | 0.4900 to
0.8964 | 0.012 | 0.715 | 0.446 to 0.913 |
| ATG4A rs807183 |
|
|
|
|
|
|
|
|
|
|
| GG | 206 (72.28) | 166 (64.59) |
|
|
|
|
|
|
|
| AA | 13 (4.56) | 42
(16.34) | <0.0001 | 0.2494 | 0.1296 to
0.4801 | <0.0001 | 0.215 | 0.069 to 0.427 |
|
| AG | 66
(23.16) | 49
(19.07) | <0.0001 | 4.352 | 2.110 to 8.974 | <0.0001 | 4.068 | 1.995 to 8.484 |
|
Dominant model | Any A vs. GG | 79
(27.72) | 91
(35.41) | 0.054 | 0.6996 | 0.4860 to
1.007 | 0.035 | 0.722 | 0.443 to 1.356 |
|
Recessive model | Any G vs. AA | 272 (95.44) | 215 (83.66) | <0.0001 | 4.087 | 2.139 to 7.809 | <0.0001 | 4.025 | 2.039 to 7.402 |
|
Additive model | AA vs. AG vs.
GG | 13/66/206 | 42/49/166 | 0.066 | 0.746 | 0.577 to 0.896 | 0.101 | 0.769 | 0.514 to 0.931 |
| Allele
OR | A vs. G | 92/478 | 133/381 | <0.0001 | 1.814 | 1.346 to 2.443 | <0.0001 | 1.573 | 1.305 to 2.085 |
Similarly, for the donor splice site of rs5973822,
the frequency of the wildtype A allele in the case group was higher
than that of the control group (96.14 vs. 92.61%, respectively,
P<0.0001, OR=3.927, 95% CI 1.896 to 7.652). In the recessive
model, the frequency of the variant G allele in control subjects
was higher than that in the case group (28.40 vs.17.89%,
respectively, P=0.096, OR 0.684, 95% CI 0.364 to 0.949). Compared
with the homozygous variant genotype (GG), the homozygous wildtype
genotype (AA) was associated with a significantly decreased
incidence of cervical cancer (P=0.0046, OR=0.384, 95% CI 0.148 to
0.733). Analogously, the unconditional logical analysis
demonstrated that the A allele of rs4036579 was associated with a
decreased risk of cervical cancer (P<0.001, OR=0.407, 95% CI
0.253 to 0.648). The results for rs807182 were identical to the
above-mentioned results in that the wildtype A allele was
associated with a decreased risk of cervical cancer (P=0.0074,
OR=2.6627, 95% CI 2.4900 to 2.8964).
An analysis of rs807181 indicated that the variant C
allele was associated with a lower risk of cervical cancer
(P=0.0051, OR=0.596, 95% CI 0.403 to 0.821). A greater frequency
was observed for the recessive model of polymorphism among case
(96.14%) than among control (83.66%). No significant deviations
were observed for dominant model. Furthermore, to exclude the
influence of other factors, we used a stratified analysis to
separately evaluate the influence of the homozygous variant
genotype (CC) of rs807181 on the risk of cervical cancer in
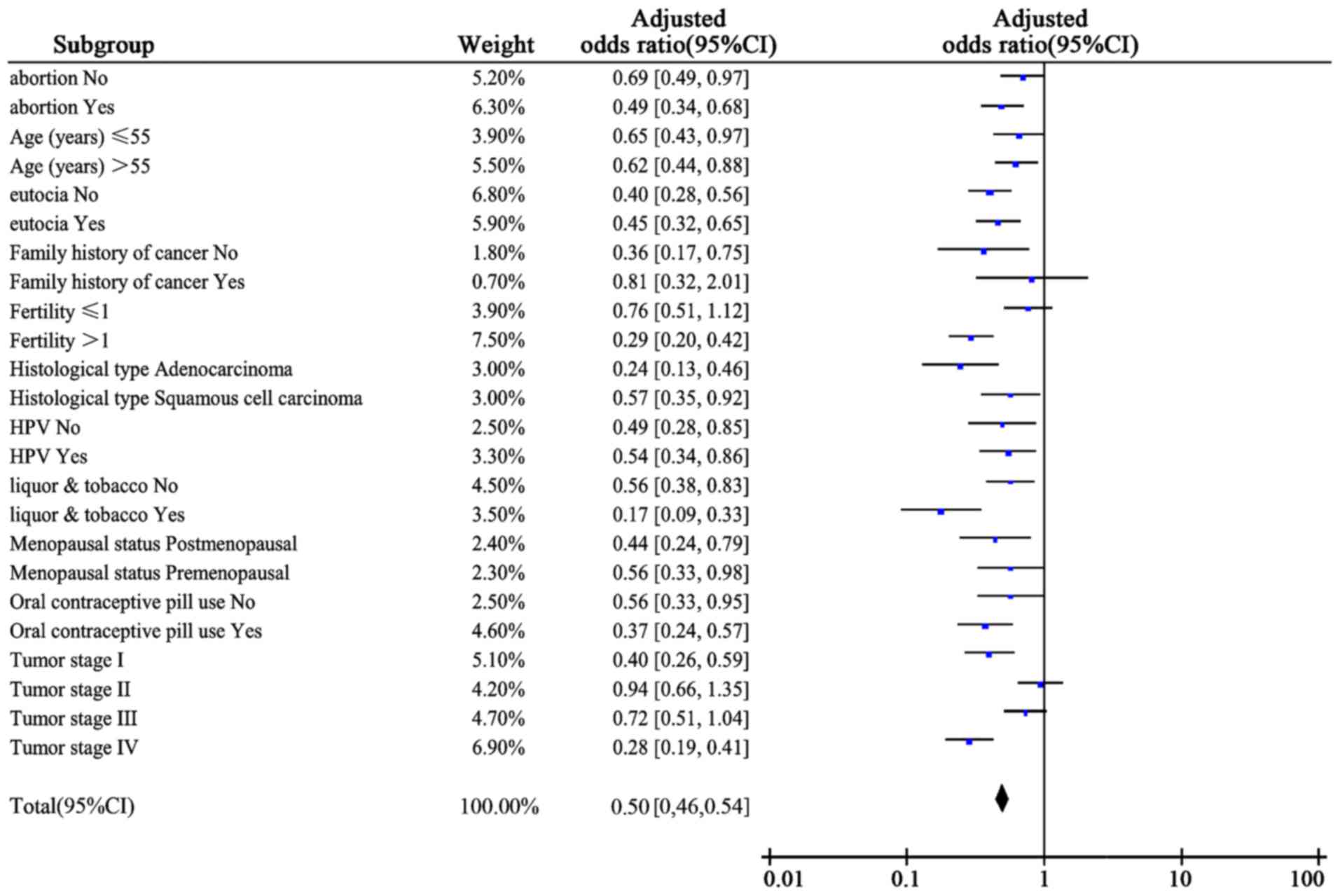
different subgroups in a dominant model (Fig. 2).
For rs807183, the variant A allele was associated
with an increased risk of cervical cancer (P<0.0001, OR=1.573,
95% CI 1.305 to 2.085). In assessing the correlation in the
recessive model shown that the frequency of the wildtype G allele
was higher in the case group than in the control group (95.44 vs.
83.66%, P<0.0001, OR=4.025, 95% CI 2.039 to 7.402). Others did
not show significant differences. When we performed the association
analysis of SNPs with the risk of cervical cancer, the stratified
analysis did not lead to considerable changes in any of the
results, no statistically significant association was found for any
of the models analyzed (data no show).
The association between ATG4A SNPs and
HPV infection
In order to explore whether the SNPs could act
together to increase HPV infection risk, a total of 29 SNPs were
identified with an average density of 1 SNP per 0.55 kb. An
r2 threshold of 0.8 was used to tag pairs with Haploview
(18), with the ultimate goal of
selecting 3 tagged SNPs. The results of haplotype analysis are
shown in Fig. 1. When we evaluated
the interaction between each of the SNPs expression at the systemic
level, as well as the SNP-SNP interaction, we conducted 9 single
SNP analyses, and 27 (3×9) SNP-SNP analyses shown different
results. No significant associations of SNP-SNP interactions were
observed (P>0.05) after multiple testing corrections. As shown
in Table IV, the MDR program
(v3.0.2) was used to analyze the interactions between ATG4A SNPs
and HPV infection. Four SNPs (rs807181, rs807182, rs807183, and
rs807185) showed significant interactions with HPV infection
(permutation P=0.002). In addition, the results of the
meta-analyses that revealed a correlation between SNP rs807181 and
rs807183 were verified by multiple testing corrections (combined
P=2.00 ×10−4, Table V).
Both SNPs are located in the intron of ATG4A. The results showed
that the interaction of rs807181 and rs807183 was significantly
associated with HPV infection (P=0.0120).
 | Table IV.The interactions of ATG4A SNPs and
their relationship to HPV infection. |
Table IV.
The interactions of ATG4A SNPs and
their relationship to HPV infection.
| Factor model | TA | CVC | Permutation
P-value |
|---|
| CCC vs. HC |
|
|
|
|
rs1130905, rs1130906 | 0.4613 | 9/10 | 0.992 |
|
rs26532, rs26534, | 0.4832 | 10/10 | 0.166 |
|
rs26537 |
|
|
|
|
rs807181, rs807182, | 0.4771 | 10/10 | 0.002 |
|
rs807183,
rs807185a |
|
|
|
 | Table V.Significant SNP-SNP interactions in
ATG4A associated with HPV infection. |
Table V.
Significant SNP-SNP interactions in
ATG4A associated with HPV infection.
| SNPa-SNPb | Subjects | Combined P | Allele A | Allele B | MAF A | MAF B | Beta | SE | P-value |
|---|
|
rs807181-rs807183 | CCC |
2.00×10−3 | C/G | G/A | 0.242 | 0.275 | 0.6245 | 0.0286 | 0.012 |
|
| HC |
3.22×10−2 | C/G | G/A | 0.136 | 0.154 | 2.153 | 0.0058 | 0.037 |
Discussion
Autophagy is a well-conserved and functionally
complex pathway that is essential for cellular homeostasis. Many
studies have suggested that autophagy has a tumor-suppressive
function in cancer. In contrast, other studies have argued that
autophagy might have beneficial effects for the survival of cancer
cells (19). The process of autophagy
in the maintenance of coordination of cell functions depends on
complex reaction systems, including ATG4, which is a
cysteine protease that plays diverse functional roles. The
ATG4 family includes four members, ATG4A, ATG4B,
ATG4C and ATG4D, which are orthologs of yeast
ATG4 (20). Recent reports
have shown that ATG4B efficiently binds to and cleaves
LC3 proteins, and the C-terminal LC3-interacting region of
ATG4B plays an important role in this process (21). Furthermore, dysregulation proteins of
ATG4 have been observed in other diseases. One study found
that ATG4B is a biomarker, has a latent function for
predicting the response of therapeutic treatment using CML
stem/progenitor cells and may serve as a drug target for these
cells (22). SNPs have become the
third generation of genetic markers. The human body exhibits many
phenotypic differences, different susceptibilities to a drug or a
disease and other differences. All of these differences may be
mainly associated with SNPs.
Dysregulation of cysteine protease activity has
great relevance as a therapeutic target in some diseases, such as
cancer and inflammatory bowel disease (23). Some researchers have examined the
dysfunctional expression of these proteases in cervical cancer and
recommended potential therapies based on their modulation (24). The rs5973822 SNP of ATG4A has
been associated with granulomas in Crohn's disease (25). Furthermore, little is known about the
relationship between the locus mutations of ATG4 and
cervical cancer. In this study, we highlighted the key role of
human ATG4 in autophagy and its relationship with cervical
cancer. In addition, ATG4A has the potential to become a new
target for cancer treatment.
Our observations offer meaningful insight into five
SNPs of ATG4A, including rs4036579, rs5973822, rs807181,
rs807182 and rs807183, and the role that they play in the
susceptibility to cervical cancer. Carriers with a homozygous
wildtype genotype of AA or an A allele have a lower the risk of
morbidity from cervical cancer. Similar phenomena have been
observed with the rs4036579, rs5973822 and rs807182 SNPs. However,
the homozygous variant genotype CC and the C allele are associated
with a decreased risk of cervical cancer in rs807181 carriers. In
contrast, for rs807183, the homozygous variant genotype AA and the
A allele are associated with an increased risk of cervical cancer.
According to the results of recent studies, intron and
transcription factor binding site polymorphisms are associated with
the risk of cancer or other diseases due to a coupling imbalance
with functional or genetic transcription. Currently, the sequences
in TP53 intron 1 encode transcripts that may play a
significant role in modulating the R249S mutation rate in
HCC, and the TCF21 rs12190287 polymorphism may increase the
genetic susceptibility to breast cancer by regulating the
expression of TCF21 (26,27).
Moreover, some studies report a focus on gene targeting therapy for
tumors; for example, a target therapy drug, bevacizumab, targets
the VEGF signaling pathway and is currently used to treat advanced
cervical cancer. Scholars agree that ROS (reactive oxygen
species)-modulated core autophagic pathways involved in
ATG4-ATG8/LC3, Beclin-1, p53, and MAPK signaling in cancer. It is
worth noting that Beclin1 could enhance the expression levels of
ATG4, overexpression of Beclin1 inhibited proliferation, migration
and invasion. Furthermore, VEGF was involved in Beclin1-mediated
inhibition of migration and invasion. The current studies have
shown that, the MAPK and P53 pathways should be targeted by drugs
(28,29). We believe that ATG4, which has an
intricate relevance, may become a target gene for cancer drug
therapy. Additional treatments for cervical cancer related to the
gene targets have not been researched and discussed; gene target
therapies cannot be replaced by other treatments.
Notably, our study is the first exploration that
reveals the correlation among the intron SNPs rs4036579, rs807182,
rs807181 and rs807183 and the untranslated region SNP rs5973822 in
ATG4A and the risk of cervical cancer in the Han Chinese
population. The variants of intron SNPs rs4036579 and rs807182 and
the untranslated region SNP rs5973822 might be non-conservation
factors, and the variant of the intron SNP rs807181 might be a
protective factor against cervical cancer. Notably, the variant of
the intron SNP rs807183 might be a risk factor for cervical cancer.
Intriguingly, the SNPs rs4036579, rs807182, rs807181 and rs807183
of ATG4A are located in the intron, and all are
transcription factor binding sites. An miRNA binding site is
located in the untranslated region SNP rs5973822. Some studies
revealed that protein expression can be controlled via AUG
sequences located upstream to the initiation codon in the 5′ UTR of
autophagy related genes. Thus, the ATG4A protein level may be
correlated with the SNPs. Possibly, these SNPs exert their effects
by regulating gene transcription or function. These variants SNPs
might regulate ATG4A and indirectly increase or reduce the
expression of proteins that directly influence autophagy.
Although ATG4A is involved in autophagy, we
aimed to determine whether SNPs in ATG4A contribute to HPV
infection in the Han Chinese population. Based on the results, we
speculated that the interaction of rs807181 and 807183 affected the
ATG4A gene and down-regulated or inhibited its role in
autophagy, which led to an increased susceptibility and ability of
HPV to bind to the host. Some studies have shown that SNP-SNP
interactions were notably associated with BMI (30,31), as
well as colon cancer. The association between ATG4A genetic
variations and HPV infection is still unknown. Analyses of SNP-SNP
interactions and the association with HPV infection were performed.
According to our study, rs807181 and rs807183 interaction, which is
a haplotype of ATG4A SNPs, is significantly associated with
HPV infection. Many types for HPV, the experiment mainly involves
16/18 type. At present, no research reports correlation between
ATG4A and HPV, based on our results, genetic variants of
ATG4A is likely to be tightly related to 16 or 18, and this
view needs further exploration. HPV infection is the most important
prerequisite for the occurrence and development of most cervical
cancers in clinical practice, therefore, ATG4A SNPs are also
a great correlation with cervical cancer susceptibility. Our
results reveal the association between genes and HPV in terms of
disease mechanisms, therefore, we suggest that could aid in
disease-related genetic diagnosis and providing clinical control,
prevention and treatment strategies for HPV-related cervical
cancer. In the future, we will examine the genetic variants of the
ATGs and their relationship to cervical cancer. Furthermore,
these studies will confirm the fundamental mechanism by which these
SNPs regulate gene expression. The sample size of this study was
limited, and further analysis to address the prognosis of cervical
cancer in relation to different mutations was not performed.
In summary, our study determined the main role of
several SNPs, which may serve as direct inducers and intermediates
in different modalities of autophagy associated with ATG4A.
The results of our study may serve to provide a new approach for
cervical cancer prevention and treatment strategies.
Acknowledgements
This study was jointly supported by the grant from
National Natural Science Foundation of China (no. 81672080) and
Municipal Health and Family Planning Commission Medical Research
Fund of Chongqing (no. 20141005).
References
|
1
|
ICO Information Centre on HPV and Cancer
(HPV Information Centre), . Human Papillomavirus and Related
Diseases Report. Summary Report, 2017. http://www.hpvcentre.net/statistics/reports/XWX.pdfJuly
27–2017
|
|
2
|
Zur Hausen H: Papillomavirus
infections-major cause of human cancers. Biochim Biophys Acta.
1288:F55–F78. 1996.PubMed/NCBI
|
|
3
|
Saslow D, Castle PE, Cox JT, Davey DD,
Einstein MH, Ferris DG, Goldie SJ, Harper DM, Kinney W, Moscicki
AB, et al: American cancer society guideline for human
papillomavirus (HPV) vaccine use to prevent cervical cancer and its
precursors. CA Cancer J Clin. 57:7–28. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Horn LC, Raptis G and Fischer U: Familial
cancer history in patients with carcinoma of the cervix uteri. Eur
J Obstet Gynecol Reprod Biol. 101:54–57. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang Z, Borecki I, Nguyen L, Ma D, Smith
K, Huettner PC, Mutch DG, Herzog TJ, Gibb RK, Powell MA, et al:
CD83 gene polymorphisms increase susceptibility to human invasive
cervical cancer. Cancer Res. 67:11202–11208. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yu KJ, Rader JS, Borecki I, Zhang Z and
Hildesheim A: CD83 Polymorphisms and cervical cancer risk. Gynecol
Oncol. 114:319–322. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shintani T and Klionsky DJ: Autophagy in
health and disease: A double-edged sword. Science. 306:990–995.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ojeda JM, Ampuero S, Rojas P, Prado R,
Allende JE, Barton SA, Chakraborty R and Rothhammer F: p53 codon 72
polymorphism and risk of cervical cancer. Biol Res. 36:279–283.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu S, Lu S, Tao H, Zhang L, Lin W, Shang H
and Xie J: Correlation of polymorphism of IL-8 and MMP-7 with
occurrence and lymph node metastasis of early stage cervical
cancer. J Huazhong Univ Sci Technolog Med Sci. 31:114–119. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Levine B and Kroemer G: Autophagy in the
pathogenesis of disease. Cell. 132:27–42. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ichimura Y, Imamura Y, Emoto K, Umeda M,
Noda T and Ohsumi Y: In vivo and in vitro reconstitution of ATG8
conjugation essential for autophagy. J Biol Chem. 279:40584–40592.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Betin VM, Singleton BK, Parsons SF, Anstee
DJ and Lane JD: Autophagy facilitates organelle clearance during
differentiation of human erythroblasts: Evidence for a role for
ATG4 paralogs during autophagosome maturation. Autophagy.
9:881–893. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wolf J, Dewi DL, Fredebohm J,
Müller-Decker K, Flechtenmacher C, Hoheisel JD and Boettcher M: A
mammosphere formation RNAi screen reveals that ATG4A promotes a
breast cancer stem-like phenotype. Breast Cancer Res. 15:R1092013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
He Q, Lu Y, Hu S, Huang Q, Li S, Huang Y,
Hu Q, Wu L and Chen W: An intron SNP rs807185 in ATG4A decreases
the risk of lung cancer in a southwest Chinese population. Eur J
Cancer Prev. 25:255–258. 2015. View Article : Google Scholar
|
|
15
|
Liao YP, Chen LY, Huang RL, Su PH, Chan
MW, Chang CC, Yu MH, Wang PH, Yen MS, Nephew KP and Lai HC:
Hypomethylation signature of tumor-initiating cells predicts poor
prognosis of ovarian cancer patients. Hum Mol Genet. 23:1894–1906.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ritchie MD, Hahn LW, Roodi N, Bailey LR,
Dupont WD, Parl FF and Moore JH: Multifactor-dimensionality
reduction reveals high-order interactions among estrogen-metabolism
genes in sporadic breast cancer. Am J Hum Genet. 69:138–147. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Purcell S, Neale B, Todd-Brown K, Thomas
L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ
and Sham PC: PLINK: A tool set for whole-genome association and
population-based linkage analyses. Am J Hum Genet. 81:559–575.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Barrett JC, Fry B, Maller J and Daly MJ:
Haploview: Analysis and visualization of LD and haplotype maps.
Bioinformatics. 21:263–265. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Degenhardt K, Mathew R, Beaudoin B, Bray
K, Anderson D, Chen G, Mukherjee C, Shi Y, Gélinas C, Fan Y, et al:
Autophagy promotes tumor cell survival and restricts necrosis,
inflammation, and tumorigenesis. Cancer Cell. 10:51–64. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Quesada V, Ordóñez GR, Sánchez LM, Puente
XS and López-Otín C: The Degradome database: Mammalian proteases
and diseases of proteolysis. Nucleic Acids Res. 37(Database Issue):
D239–D243. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Skytte Rasmussen M, Mouilleron S, Kumar
Shrestha B, Wirth M, Lee R, Bowitz Larsen K, Abudu Princely Y,
O'Reilly N, Sjøttem E, Tooze SA, et al: ATG4B contains a C-terminal
LIR motif important for binding and efficient cleavage of mammalian
orthologs of yeast ATG8. Autophagy. 13:834–853. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rothe K, Lin H, Lin KB, Leung A, Wang HM,
Malekesmaeili M, Brinkman RR, Forrest DL, Gorski SM and Jiang X:
The core autophagy protein ATG4B is a potential biomarker and
therapeutic target in CML stem/progenitor cells. Blood.
123:3622–3634. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fernández ÁF and López-Otín C: The
functional and pathologic relevance of autophagy proteases. J Clin
Invest. 125:33–41. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen Y, Liu XR, Yin YQ, Lee CJ, Wang FT,
Liu HQ, Wu XT and Liu J: Unravelling the multifaceted roles of Atg
proteins to improve cancer therapy. Cell Prolif. 47:105–112. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Brinar M, Vermeire S, Cleynen I, Lemmens
B, Sagaert X, Henckaerts L, Van Assche G, Geboes K, Rutgeerts P and
De Hertogh G: Genetic variants in autophagy-related genes and
granuloma formation in a cohort of surgically treated Crohn's
disease patients. J Crohns Colitis. 6:43–50. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ortiz-Cuaran S, Cox D, Villar S, Friesen
MD, Durand G, Chabrier A, Khuhaprema T, Sangrajrang S, Ognjanovic
S, Groopman JD, et al: Association between TP53 R249S mutation and
polymorphisms in TP53 intron 1 in hepatocellular carcinoma. Genes
Chromosomes Cancer. 52:912–919. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gao X, Yang J, Wang M and Zhang J: TCF21
genetic polymorphisms and breast cancer risk in Chinese women.
Oncotarget. 7:55757–55764. 2016.PubMed/NCBI
|
|
28
|
Zagouri F, Sergentanis TN, Chrysikos D,
Filipits M and Bartsch R: Molecularly targeted therapies in
cervical cancer. A systematic review. Gynecol Oncol. 126:291–303.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu B, Ding JF, Luo J, Lu L, Yang F and
Tan XD: Seven protective miRNA signatures for prognosis of cervical
cancer. Oncotarget. 7:56690–56698. 2016.PubMed/NCBI
|
|
30
|
Dong SS, Hu WX, Yang TL, Chen XF, Yan H,
Chen XD, Tan LJ, Tian Q, Deng HW and Guo Y: SNP-SNP interactions
between WNT4 and WNT5A were associated with obesity related traits
in Han Chinese Population. Sci Rep. 7:439392017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Molano M, Moreno-Acosta P, Morales N,
Burgos M, Buitrago L, Gamboa O, Alvarez R, Garland SM, Tabrizi SN,
Steenbergen RD and Mejía JC: Association between type-specific HPV
Infections and hTERT DNA methylation in patients with invasive
cervical cancer. Cancer Genomics Proteomics. 13:483–491. 2016.
View Article : Google Scholar : PubMed/NCBI
|